Abstract
Menin functions as a critical oncogenic cofactor of mixed lineage leukemia (MLL) fusion proteins in the development of acute leukemias, and inhibition of the menin interaction with MLL fusion proteins represents a very promising strategy to reverse their oncogenic activity. MLL interacts with menin in a bivalent mode involving 2 N-terminal fragments of MLL. In the present study, we reveal the first high-resolution crystal structure of human menin in complex with a small-molecule inhibitor of the menin-MLL interaction, MI-2. The structure shows that the compound binds to the MLL pocket in menin and mimics the key interactions of MLL with menin. Based on the menin–MI-2 structure, we developed MI-2-2, a compound that binds to menin with low nanomolar affinity (Kd = 22nM) and very effectively disrupts the bivalent protein-protein interaction between menin and MLL. MI-2-2 demonstrated specific and very pronounced activity in MLL leukemia cells, including inhibition of cell proliferation, down-regulation of Hoxa9 expression, and differentiation. Our results provide the rational and essential structural basis to design next generation of inhibitors for effective targeting of the menin-MLL interaction in leukemia and demonstrate a proof of concept that inhibition of complex multivalent protein-protein interactions can be achieved by a small-molecule inhibitor.
Introduction
Translocations of the MLL (mixed lineage leukemia) gene frequently occur in aggressive human acute myeloid and lymphoid leukemias in both children and adults.1 Fusion of MLL with 1 of more than 60 different genes results in chimeric MLL fusion proteins that enhance proliferation and block hematopoietic differentiation, ultimately leading to acute leukemia.2 Patients with leukemias harboring MLL translocations have very unfavorable prognoses and respond poorly to currently available treatments.2,3 The relapse risk is very high using conventional chemotherapy and stem cell transplantation,2 leading to an overall 5-year survival rate of only approximately 35%.4
All MLL fusion proteins preserve an N-terminal MLL fragment approximately 1400 amino acids in length fused in-frame with the C-terminus of the fusion partner.3,5–7 Two regions in this fragment of MLL have been shown to be indispensable for leukemogenic transformation: the N-terminal region, which binds to menin8 and to lens epithelium–derived growth factor (LEDGF),9 and the conserved region encompassing the CXXC domain, which mediates binding to nonmethylated CpG DNA10–12 and interacts with the polymerase associated factor complex (PAFc).13,14 Targeting these interactions provides new opportunities for the development of new therapeutic agents for the MLL leukemias.15
Menin is a tumor-suppressor protein encoded by the MEN1 (multiple endocrine neoplasia 1) gene.16 Mutations of MEN1 are associated with tumors of the parathyroid glands, pancreatic islet cells, and anterior pituitary gland.17 Menin is also a highly specific binding partner for MLL and MLL fusion proteins and is required to regulate the expression of MLL target genes, including HOXA9 and MEIS1.8 Loss of the ability to bind menin abolishes the oncogenic potential of MLL fusion proteins both in vitro and in vivo.8 Disruption of the interaction between menin and MLL fusion proteins using genetic methods blocks the development of acute leukemia in mice,8 indicating that menin functions as a critical oncogenic cofactor of MLL fusion proteins and is required for their leukemogenic activity. The menin-MLL interaction represents an attractive therapeutic target for the development of novel drugs for acute leukemias with MLL rearrangements.
The development of small-molecule inhibitors of protein-protein interactions (PPIs) is a challenging task.18 Menin interacts with 2 fragments of MLL, the high-affinity motif MBM1 (menin-binding motif 1, Kd = 53nM) and low-affinity MBM2 (Kd = 1.4μM), located within the intrinsically unstructured 43-amino acid fragment at the N-terminus of MLL.19 MBM1 and MBM2 are separated by a 7-glycine linker and most likely bind to adjacent sites on menin. The development of small molecules effectively targeting the menin-MLL interaction would require the disruption of this bivalent interaction. A point mutation within MBM1 is sufficient to abolish the oncogenic properties of MLL fusion proteins in vivo.8 We have recently developed small molecules that bind directly to menin and inhibit the menin-MLL interaction in vitro and in human cells.20
To understand the molecular mechanism of the menin-MLL interaction and to facilitate the development of novel drugs targeting this interaction, we have determined the high-resolution (1.46 Å) crystal structure of human menin, as well as menin in complex with the high-affinity binding motif of MLL, MBM1. We have also obtained the first crystal structure of menin in complex with MI-2, our recently developed small-molecule inhibitor of the menin-MLL interaction,20 and found that this compound binds to the MLL site on menin and mimics the key interactions of MBM1. Based on the structure of the menin–MI-2 complex, we developed a very potent second-generation inhibitor, MI-2-2, which binds to menin with low nanomolar affinity and is capable of potently inhibiting the MLL-fusion protein–mediated leukemogenic transformation. Our work provides an important structural insight into menin's function as a critical cofactor of oncogenic MLL fusion proteins in leukemogenesis. The structures of menin complexes with the small-molecule inhibitors MI-2 and MI-2-2 shed light on inhibitor binding to the MLL binding site on menin and provide the rational and molecular basis for the development of the next generation of menin-MLL inhibitors as novel therapies in acute leukemias with MLL translocation.
Methods
Cloning, protein engineering, expression, and purification of human menin
The gene encoding human menin was subcloned into pET32a vector (Novagen) and the generation of deletion constructs was performed by mutagenesis according to the QuikChange Site-Directed Mutagenesis kit protocol to introduce different internal deletions and a stop codon. Proteins were expressed and purified using a protocol described previously.21 For crystallization experiments, proteins were purified using the size-exclusion column HiLoad 16/60 Superdex 75 pg resin (GE Healthcare) and 50mM Tris-HCl, pH 8.0, 50mM NaCl, and 1mM tris(2-carboxyethyl)phosphine as the mobile phase.
Crystallization of menin and menin complexes
For crystallization experiments, 2.5 mg/mL of menin was incubated with MLL MBM1 peptide (MLL residues 4-15; GenScript) in a 1:1 molar ratio. Crystals were obtained using a sitting-drop technique at 10°C in 0.2M ammonium acetate, 0.1M HEPES, pH 7.5, and 25% wt/vol PEG 3350. Before data collection, crystals were transferred into a cryosolution containing 20% PEG550 MME and flash-cooled in liquid nitrogen. A similar procedure was used for crystallization of the complexes with MI-2 and MI-2-2.
Crystallographic data collection and structure determination
Diffraction data for menin and menin complexes were collected at the 21-ID-D and 21-ID-F beam lines at the Life Sciences Collaborative Access Team at the Advanced Photon Source. Data were processed with HKL-2000.22 Structure of the free protein was determined using HKL-300023 and MOLREP24 using menin from Nematostella vectensis (PDB code: 3RE2) as a search model in molecular replacement. The model was rebuilt with BUCCANNER25 and RESOLVE26 and the refinement was carried out using HKL-3000, REFMAC,27 COOT,28 and the CCP4 package.29 In the final stages, refinement was performed with addition of the TLS groups defined by the TLMSD server30 or using anisotropic B-factors in the case of the high-resolution structures. Validation of the structures was performed using MOLPROBITY31 and ADIT.32 Details of data processing and refinement are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4GPQ, 4GQ6, 4GQ3, and 4GQ4.
MLL-binding experiments
Dissociation constants for binding of MBM1 (MLL residues 4-15), MBM2 (residues 23-40), and MLL4-43 (MLL residues 4-43) to full-length human menin were determined by the fluorescence polarization method using a protocol published previously.19
Biochemical characterization of menin-MLL inhibitors
The ability of small molecules to inhibit menin-MBM1 and menin-MLL4-43 interaction was assessed using the fluorescence polarization assay in a protocol described previously.19,20 Details of isothermal titration calorimetry experiments for the measurement of dissociation constants for menin inhibitors were also described previously.20
Coimmunoprecipitation experiments
HEK293 cells were transfected with β-actin Flag-MLL-AF9 plasmid using Fugene 6 (Roche). Forty-eight hours after transfection, cells were treated with compounds (0.25% final DMSO concentration) or DMSO for 12 hours. Whole-cell lysates were immunoprecipitated with anti-FLAG M-2 Magnetic Beads (Sigma-Aldrich) and analyzed by SDS-PAGE and Western blotting. For more details, see Grembecka et al.20
Viability assays
MLL-AF9–transduced mouse bone marrow cells (BMCs) were prepared as described previously.13 MV4;11, KOPN-8, ML-2, and MOLM-13 cells were cultured in RPMI 1640 medium with 10% FBS, 1% penicillin/streptomycin, and nonessential amino acids. The MTT viability assay was carried out using a recently published protocol.20 For growth curves, 3 × 105/mL cells were plated (1 mL/well) and treated with compounds or 0.25% DMSO. Media were changed every 48 hours with viable cell concentration restored to 3 × 105 cells/mL and compound resupplied. At a designated time point, cell culture samples were mixed with Trypan blue (GIBCO/Invitrogen) and counted.
Colony formation assay
The MLL-AF9–transduced murine BMCs were plated in 12-well plates at a concentration of 5 × 103 cells/mL with 1 mL of methylcellulose medium M3234 (StemCell Technologies) containing 20% IMDM, 1% penicillin/streptomycin, IL-3 and 0.25% DMSO, or compounds. Six days later, colonies were stained with 100 μL of iodonitrotetrazolium chloride (Sigma-Aldrich) at a final concentration of 1 mg/mL, incubated at 37°C for 30 minutes, and counted. Pictures of colonies were taken at room temperature using an Olympus IX50 microscope with LCPIanFI 10×/0.3 Ph1 objective lenses and an Olympus DP70 camera with Olympus DP Controller software. Pictures were resized using Adobe Photoshop CS2.
Real-time PCR
Total RNA was extracted from cells using the RNeasy mini kit (QIAGEN), and then 100-1000 ng of total RNA was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Real-time PCR was performed using the ABI Prism 7700 sequence detection system. TaqMan Gene Expression Master Mix and TaqMan Gene Expression Assays for mouse Hoxa9 (Mm00439364_m1), Meis1 (Mm00487664_m1), β-Actin (4352933), human HOXA9 (Hs00365956_m1), MEIS1 (Hs00180020_m1), and 18S RNA (Hs99999901_s1) were purchased from Applied Biosystems. Relative quantification of each gene transcript was carried out using the comparative Ct method as described in the Applied Biosystems User Bulletin no. 2.
Annexin V/propidium iodide assay of inhibitor effects on apoptosis
A total of 5 × 105 cells/mL were plated in 12-well plates (1 mL/well) and treated with compounds (0.25% final concentration of DMSO for each condition) or 0.25% DMSO control and incubated for 48 hours at 37°C in a 5% CO2 incubator. After incubation, 1.5 × 105 cells were harvested and resuspended in 100 μL of 1× annexin V–binding buffer from the Annexin V–FITC Apoptosis Kit (BD Biosciences), incubated with 4 μL of annexin V–FITC and 6 μL of propidium iodide (Sigma-Aldrich) at room temperature in the dark for 10 minutes, and analyzed by flow cytometry on an LSR II instrument. Data analysis was performed using WinList Version 3.0 software (Verity Software House Inc).
Expression of CD11b
MV4;11 cells or MLL-AF9 transduced BMCs were plated in 12-well plates at an initial concentration of 5 × 105 cells/mL and treated with compounds or 0.25% DMSO. Media were changed every 48 hours with viable cell concentration restored to 5 × 105 cells/mL and compounds resupplied. Seven days after the experiment was set up, 1.5 × 105 cells were harvested and washed with FACS buffer (PBS, 1% FBS, and 0.1% NaN3). Cells were resuspended in 100 μL of FACS buffer and incubated with 2 μL of Pacific blue mouse anti–human CD11b Ab (BD Biosciences) or 1 μL of Pacific Blue rat anti–mouse CD11b Ab (BioLegend) at 4°C for 30 minutes. Cells were then washed, resuspended in 100 μL of annexin V binding buffer, and incubated with 4 μL of annexin V–FITC (BD Biosciences) and 6 μL of propidium iodide (1 mg/mL; Sigma-Aldrich) at room temperature for 10 minutes before being analyzed by flow cytometry.
Cytospin/Wright-Giemsa staining
MV4;11 and mouse BMCs transduced with MLL-AF9 were plated in 12-well plates (1 mL/well) at an initial concentration of 5 × 105/mL cells, treated with compounds (0.25% final DMSO concentration) or 0.25% DMSO control, and incubated at 37°C in a 5% CO2 incubator. At a designated time point, 1 × 105 cells were harvested and placed in Shandon EZ Single Cytofunnel (Thermo Electron). Samples were centrifuged at 550g for 5 minutes. The slides were air-dried before staining with the Hema-3 kit (Fisher Scientific). Cytospin pictures were taken at room temperature using an Olympus BX41 microscope with UPIanFLN 100×/1.30 Oil objective lenses, immersion oil Immersol 518N (Carl Zeiss Microscopy) and an Olympus DP71 camera with Olympus DP Controller software. Pictures were resized using Adobe Photoshop CS2.
Inhibitor effects on cell cycle of MV4;11 leukemia cells
MV4;11 cells (5 × 105/mL) were plated in 12-well plates (1 mL/well) and treated with MI-2-2 compound (0.25% final concentration of DMSO for each condition) or 0.25% DMSO control and incubated for 48 hours at 37°C in a 5% CO2 incubator. After incubation, 5 × 105 cells were harvested, washed in PBS buffer, resuspended in 1 mL of PBS buffer, and mixed with 9 mL of 70% ethanol. Cells were kept at −20°C for at least 24 hours, washed and resuspended in FACS buffer, and then incubated with 100 μg/mL RNase (QIAGEN) and 10 μg/mL of propidium iodide at 37°C for 30 minutes before flow cytometry.
Chemistry
Synthesis and characterization of the second generation of menin-MLL inhibitors (MI-2-2 and analogs) is provided in supplemental Methods.
Results
Crystal structure of human menin and menin-MLL complex
Our initial attempts to crystallize full-length human menin failed, most likely because of the presence of several internal fragments predicted to be unstructured. Based on the structure of menin homolog from N vectensis,21 we deleted 3 internal fragments and the C-terminus in human menin, which we predicted to correspond to loops and unstructured regions (supplemental Figure 1A). This resulted in a construct that yielded protein crystals diffracting to a 1.46 Å resolution (supplemental Table 1). The engineered protein retains the ability to bind MLL with a similar affinity as the wild-type menin, indicating that deletion of these fragments does not alter the MLL-binding site (supplemental Figure 1B). Human menin is predominantly an α-helical protein (supplemental Figure 1C) and closely resembles the structure of N vectensis menin that we described previously,21 with only minor structural differences, localized to the peripheral fragments and loop regions. Very recently, the structure of a longer human menin construct has been reported.33 Interestingly, deletion of the loops resulted in a very significant improvement in the resolution of the structure.
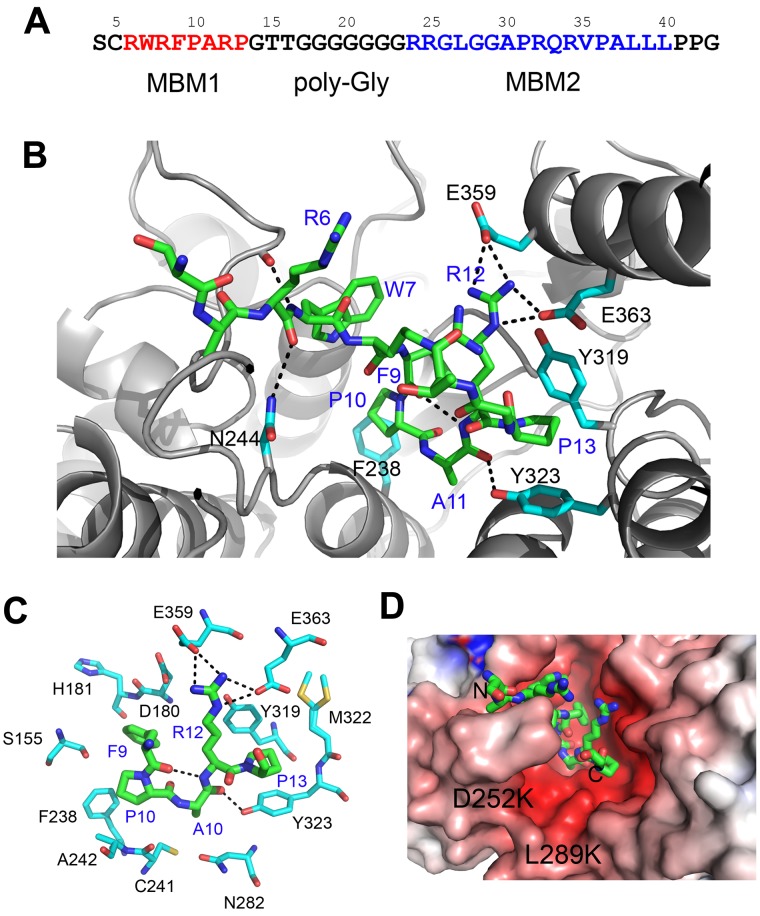
MLL associates with menin in a bivalent mode using 2 short motifs, MBM1 and MBM2, separated by a polyglycine linker (Figure 1A).19 We previously reported that MBM1 and MBM2 peptides bind to menin with, respectively, 50nM and 1.4μM affinities. The longer fragment containing both motifs interacts with menin with 10nM affinity.19 To provide an insight into the recognition of MLL by menin, we have determined the structure of menin in complex with the high-affinity motif, MBM1 (supplemental Table 1). The structure revealed that MBM1 binds to the large central cavity on menin. The peptide is well ordered and its backbone adopts a U-shaped conformation, with a single β-turn comprising residues 9-12 (Figure 1B and supplemental Figure 1C). The β-turn is stabilized by an intramolecular hydrogen bond between the carbonyl of Phe9MLL and the backbone amide of Arg12MLL (Figure 1C). The binding of MBM1 to menin is maintained by several hydrophobic contacts involving the side chains of Phe9MLL, Pro10MLL, and Pro13MLL (Figure 1B-C), which we demonstrated to play the most important role for MBM1 binding to menin.19 Phe9MLL is entirely buried in the complex and fits between the protein backbone (residues 179-181) and the side chain of Phe238 (Figure 1C). Pro10MLL binds to the site adjacent to Phe9MLL and interacts with Phe238 and Ala242, whereas Pro13MLL fits into a hydrophobic pocket formed by Tyr319, Tyr323, and Met322. Mutations of Phe9MLL, Pro10MLL, and Pro13MLL to alanine residues result in a dramatic decrease of MBM1 binding to menin (2000-, 30-, and 50-fold decrease, respectively),19 validating the importance of these hydrophobic interactions. Additional stabilization of the MBM1-menin complex results from a salt bridge between Arg12MLL and Glu359 and Glu363, as well as 3 intermolecular hydrogen bonds involving the MLL backbone (residues Arg6MLL, Trp7MLL, and Ala11MLL) and menin side chains (Asn244, Asp136, and Tyr323; Figures 1B-C). Mutation of Arg12MLL to alanine has a relatively modest effect on MBM1 binding (ie, a 4-fold decrease).19 Remarkably, binding of the 12–amino acid fragment of MLL does not cause any significant changes within the menin structure. Very recently, the lower-resolution crystal structure of the menin-MLL complex has been shown to have a similar interaction mode.33
Figure 1.
Structure of the menin-MLL complex. (A) Sequence of the N-terminal fragment of MLL with MBM1 and MBM2 motifs. (B) Details of the menin-MBM1 interaction. Structure of the MBM1 is shown in stick representation (green carbon atoms) and MLL residues are labeled in blue. Menin is presented as a gray ribbon and selected side chains involved in contacts with MBM1 are shown as sticks (cyan carbon atoms); hydrogen bonds are shown as dashed lines. (C) The most significant contacts between MBM1 (green carbons) and selected menin side chains (cyan carbons). (D) Probing the MBM2-binding site on menin. MBM1 (shown in sticks) occupies a negatively charged central cavity on menin. The positions of D252 and L289 that were mutated to lysines are labeled. The electrostatic potential was calculated using APBS and mapped onto menin structure.41
We were not able to determine the crystal structure of menin with the MBM2 fragment of MLL, presumably because of the crystal contacts interfering with the binding. MBM1 and MBM2 are separated by a linker comprising 7 glycine residues (Figure 1A), and therefore it is possible that MBM2 occupies the space in a close proximity to MBM1. The MBM2 is positively charged and, using site-directed mutagenesis in menin, we probed whether it binds to the negatively charged site in a close proximity to the MBM1-binding site (Figure 1D). Introduction of 2 point mutations in menin, D252K and L289K, which we assumed would cause electrostatic repulsion with the MBM2 (Figure 1D), interfered with binding of MBM2 without affecting protein stability or the interaction with MBM1 (supplemental Figure 2). This indicates that the low-affinity motif, MBM2, most likely binds in the proximity of the MBM1-binding site and provides an additional anchor enhancing the interaction of MLL with menin.
Thienopyrimidine inhibitors mimic the binding mode of MBM1
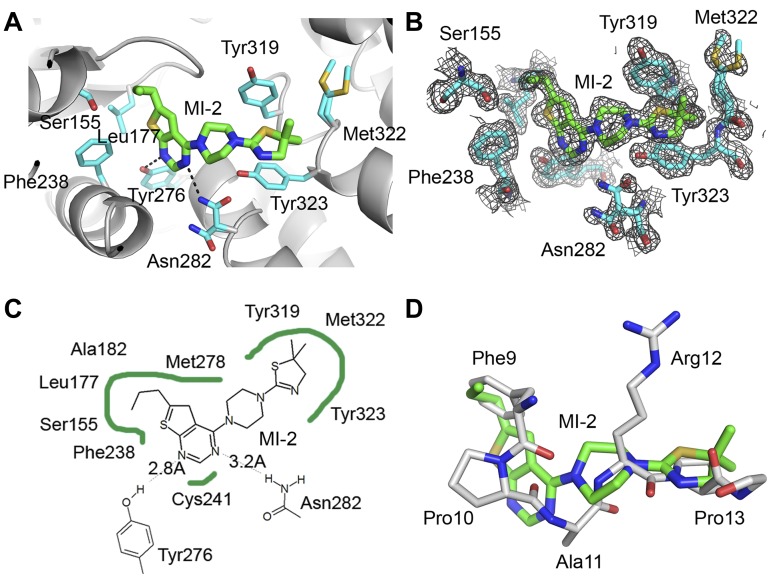
We have recently developed small molecules with a thienopyrimidine scaffold that bind to menin with submicromolar affinities and disrupt the menin-MLL interaction.20 One of these compounds, MI-2 (IC50 = 446 nM), showed pronounced effects in MLL leukemia cells, representing a very promising lead compound for the development of new antileukemic agents.20 However, lack of structural data for the MI-2 interaction with menin impeded the development of more potent inhibitors of the menin-MLL interaction. To overcome this limitation, we determined the 1.56 Å resolution structure of MI-2 bound to menin (Figure 2A-B, supplemental Figure 11, and supplemental Table 1), which represents the first determination of the crystal structure of menin in complex with a small-molecule inhibitor. The electron density for MI-2 inhibitor is very well defined (Figure 2B), enabling detailed analysis of its interactions with menin.
Figure 2.
Crystal structure of the menin–MI-2 complex. (A) Details of MI-2 interaction with menin. Selected menin side chains are shown in sticks (cyan carbons) and hydrogen bonds are shown as dashed lines. (B) Menin-MI-2 complex determined at 1.56 Å resolution with corresponding 2Fo-Fc electron density map contoured at the 1σ level. (C) Diagram depicting VdW contacts and hydrogen bonds (dashed lines) between MI-2 and menin. (D) Superposition of MI-2 (green carbons) with a fragment of the MBM1 motif (MLL residues 9-13, gray carbons) in a menin-bound conformation.
We found that MI-2 binds to the same central cavity on menin that is occupied by the MBM1 fragment of MLL. The structure of MI-2 contains an n-propyl–substituted thienopyrimidine ring connected by a piperazine linker to a dimethyl-thiazoline ring (Figure 2A-C). The interaction of MI-2 with menin is predominantly stabilized via hydrophobic interactions and by 2 hydrogen bonds between N1 and N3 nitrogen atoms of the thienopyrimidine ring and the side chains of Tyr276 and Asn282, respectively (Figure 2A and C). The n-propyl-thienopyrimidine fragment fits into a pocket formed by the main chain residues Ser178 to His181 and the side chains of mostly hydrophobic residues (Phe238, Leu177, Ala182, Tyr276, Met278, Cys241, and Ser155), whereas the piperazine ring serves mostly as a linker and approaches the side chains of Met278 and Tyr319. The dimethyl-thiazoline moiety of MI-2 fits into a site formed by 2 orthogonally oriented tyrosine side chains (Tyr319 and Tyr323) and the side chain of Met322 (Figure 2A).
Strikingly, MI-2 mimics the binding mode and the key interactions formed by the MBM1 with menin (Figure 2D). The n-propyl group of MI-2 fits into the pocket occupied by Phe9MLL, whereas the dimethyl thiazoline ring binds to the same pocket as Pro13MLL. The piperazine ring in MI-2 also mimics a part of the MBM1 backbone by overlapping with the main chain atoms of Arg12MLL (Figure 2D). Overall, MI-2 is a relatively rigid molecule, and its high binding affinity to menin is likely because of the strong shape complementarity of this compound to the binding site on menin. Despite its relatively small size (375 Da), MI-2 binds to menin with only a 3-fold lower affinity than the 12–amino acid MBM1 fragment of MLL and mimics the key interactions of MBM1 with menin. Therefore, MI-2 is a very promising candidate for further development into a more potent inhibitor of the menin-MLL interaction.
Structure-based design of a nanomolar inhibitor of the menin-MLL interaction
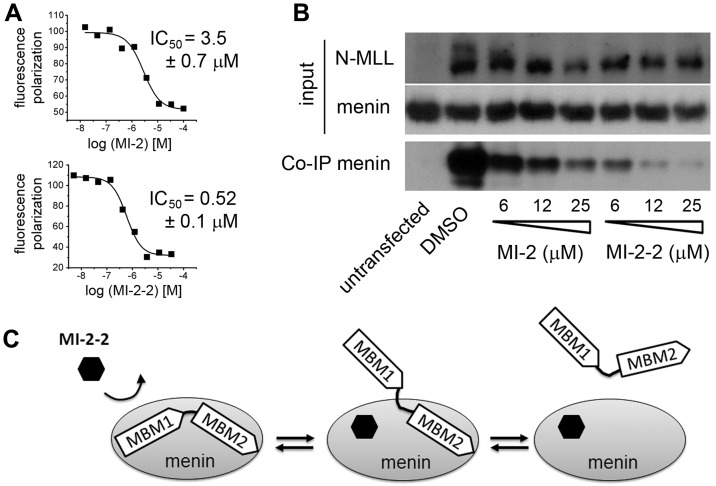
We have exploited the structure of the menin–MI-2 complex to design new analogs with improved binding affinities. Inspection of the structure revealed that the n-propyl group of MI-2 does not represent an optimal substituent for interacting with the binding pocket on menin. The structure also rationalizes why substitution of the n-propyl by bulky hydrophobic groups did not improve binding affinity.20 To develop more potent compounds, we used structural information and designed several modifications of the R1 substituent on the thienopyrimidine ring (Figure 3A). First, we introduced oxygen, which could form a hydrogen bond with the side chain hydroxyl of Ser155. However, addition of the methoxy-propyl group at R1 (MI-2-1) resulted in a 3-fold decreased activity (Figure 3A). As an alternative strategy, we synthesized several analogs by addition of fluorine atoms to R1 to achieve optimal shape complementarity with the binding site on menin (Figure 3A). Indeed, the replacement of n-propyl by trifluoroethyl group (MI-2-2) substantially improved the activity (Figure 3A-B). MI-2-2, which has been designed based on the structure of the menin–MI-2 complex, represents a second generation of menin inhibitors and binds to menin with Kd = 22nM as assessed by isothermal titration calorimetry (Figure 3C). The MI-2-2 inhibits the menin-MBM1 interaction with IC50 = 46nM (Figure 3B), representing an approximately 7- to 9-fold improvement compared with MI-2 (IC50 = 446nM, Kd = 158nM). Remarkably, MI-2-2 binds to menin with a more than 2-fold higher affinity than the MBM1 fragment of MLL (Figure 3B).
Figure 3.
Development of second-generation menin-MLL inhibitors. (A) Structures and activities of new compounds designed based on the structure of the menin–MI-2 complex. IC50 values for the inhibition of the menin-MBM1 complex are provided in parentheses. (B) FP experiments comparing activities of MI-2, MI-2-2 and MBM1 for disruption of the menin-MBM1 interaction demonstrating that MI-2-2 is a more potent inhibitor than an MLL-derived peptide. (C) Isothermal titration calorimetry showing the binding of MI-2-2 to menin. N represents a stoichiometry of binding. (D) Crystal structure of the menin–MI-2-2 complex determined at 1.27 Å resolution with corresponding 2Fo-Fc electron density map contoured at 1σ level. Water molecules were omitted for clarity. (E) Orthogonal dipolar interactions between MI-2-2 fluorine and backbone atoms of His181. The distances are shown in Å.
To understand the molecular basis of a significant effect caused by introduction of the trifluoroethyl group, we have determined the high-resolution structure (1.27 Å; supplemental Table 1) of the MI-2-2-menin complex (Figure 3D and supplemental Figure 11). We found that binding mode of MI-2-2 to menin is the same as that observed for MI-2. Interestingly, one of the fluorine atoms resides in a very close proximity (3.0-3.1 Å) to the backbone atoms of His181 (Figure 3E). Such an orthogonal orientation of the fluorine relative to the protein backbone results in a favorable C-F⋯C=O dipolar interaction that has previously been identified to significantly enhance protein-ligand interactions.34 Introducing fluorine atoms has become common in medicinal chemistry to enhance the binding affinity of protein ligands and to improve their drug-like properties.35 Our high-resolution crystal structure of MI-2-2 bound to menin allows for detailed analysis of the protein interactions with fluorine, and may represent a valuable model system to better understand such interactions and to improve the design of potent small-molecule inhibitors of other protein-protein interactions.
Small molecules are capable of inhibiting the bivalent menin-MLL interaction
The intrinsically unstructured N-terminus of MLL interacts with menin via a complex mechanism, with 2 short MLL fragments (MBM1 and MBM2) involved in binding.19 Deletion of a high-affinity motif MBM1 is sufficient to abolish transformation by MLL-ENL,8 which shows that this interaction constitutes a hot spot for small-molecule development. Because MBM2 also contributes to the binding to menin, it was necessary to establish whether small molecules targeting the MBM1 site on menin are sufficient to efficiently inhibit the bivalent interaction of menin with MLL. We found that both MI-2 and MI-2-2 can inhibit the interaction of menin with MLL4-43, which comprises the intact menin-binding fragment.19,36 As expected, the second-generation inhibitor MI-2-2 was approximately 7-fold more potent in disrupting the menin-MLL4-43 interaction, with an IC50 = 520nM (Figure 4A). To assess whether these inhibitors can dissociate menin interaction with the full-length MLL-AF9 fusion protein in cells, we performed coimmunoprecipitation experiments. We found that both MI-2 and MI-2-2 inhibit the interaction of menin with MLL-AF9 in HEK293 cells in a dose-dependent manner at low micromolar concentrations (Figure 4B), with MI-2-2 representing an approximately 4-fold improvement over MI-2 (Figure 4B). These experiments provide important evidence that small molecules that bind to menin with high affinity at the MBM1 site are capable of fully dissociating the entire menin-MLL fusion protein complex (Figure 4C).
Figure 4.
Small molecules targeting the MBM1 site efficiently disrupt bivalent menin-MLL interaction. (A) Fluorescence polarization experiments demonstrating displacement of MLL4-43 from menin by MI-2 and MI-2-2. Data represent mean values from 2 experiments ± SD. (B) Coimmunoprecipitation experiment in HEK293 cells transfected with Flag-MLL-AF9 showing that MI-2 and MI-2-2 disrupt the interaction of menin with MLL-AF9 in human cells. Input shows the levels of menin and MLL. The amount of menin bound to Flag-MLL-AF9 was detected by coimmunoprecipitation using anti-Flag Ab followed by immunoblotting using menin Ab. (C) Model of the disruption of bivalent MLL-menin interaction by MI-2-2 via binding to MBM1 site in menin.
MI-2-2 has significantly improved cellular activity compared with MI-2
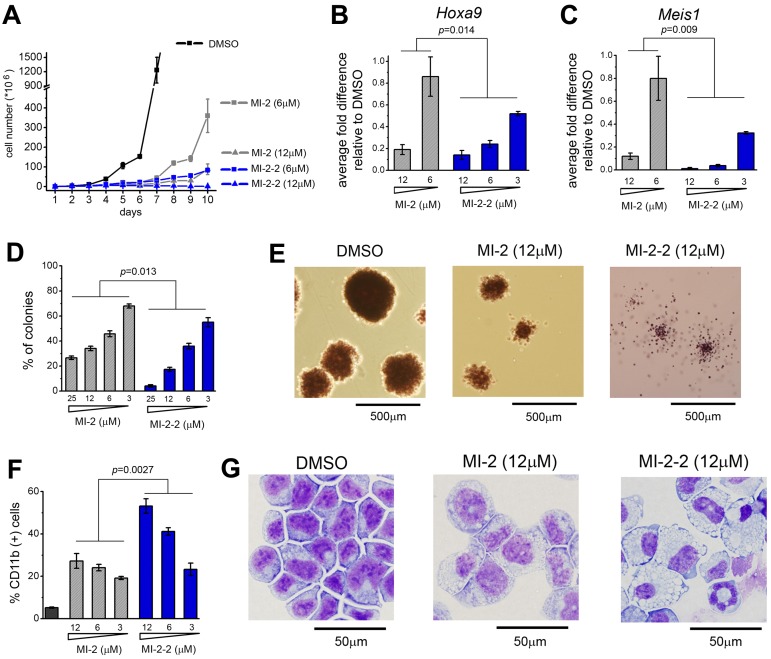
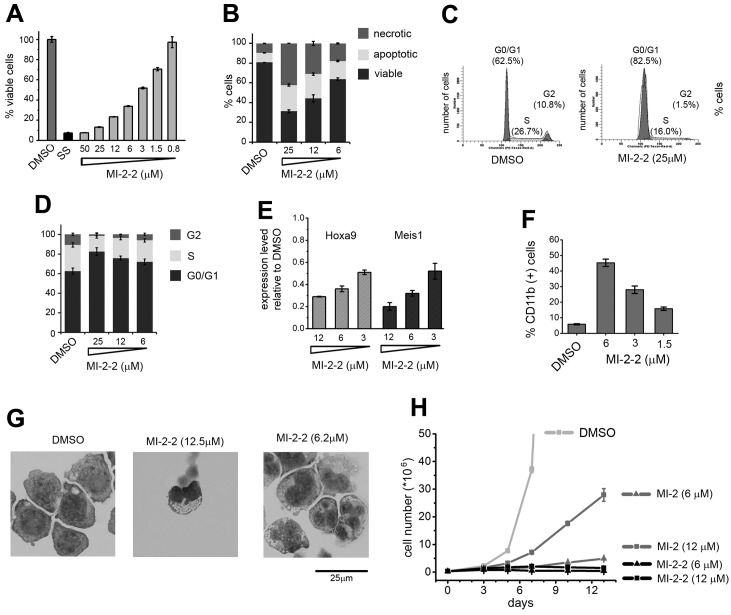
Our second-generation compound, MI-2-2, demonstrates substantially improved inhibition of the menin-MLL interaction over MI-2, and therefore we compared the effects of these 2 compounds in mouse BMCs transformed with MLL-AF9. Both compounds caused very significant growth inhibition at low micromolar doses (Figure 5A). Interestingly, treatment for longer than 6 days revealed that only the more potent compound, MI-2-2, stably suppressed growth of MLL-AF9–transformed BMCs (Figure 5A). We also assessed the capability of MI-2 and MI-2-2 to down-regulate the expression of MLL fusion protein target genes. Although treatment with 6μM MI-2 had a small effect, the same dose of MI-2-2 caused greater than 80% down-regulation of Hoxa9 and Meis1 expression (Figure 5B-C). In the colony formation assay, treatment with MI-2-2 resulted in a more pronounced effect on colony number compared with MI-2, with essentially no colonies formed at 25μM MI-2-2 (Figure 5D-E). The colonies were also smaller and much more diffused upon treatment with MI-2-2, reflecting a loss of transforming properties by MLL-AF9 (Figure 5E and supplemental Figure 3). Furthermore, treatment with MI-2-2 resulted in a more pronounced hematopoietic differentiation than what was observed for MI-2, as reflected by a substantial increase in the level of CD11b, a differentiation marker of myeloid cells (Figure 5F), and very pronounced change in cell morphology. Treatment for 7 days with 12μM MI-2-2 was sufficient to cause terminal monocytic differentiation of MLL-AF9–transformed BMCs, whereas MI-2 did not cause such a pronounced effect (Figure 5G and supplemental Figure 4). In summary, the structure-based design inhibitor MI-2-2 represents a significant improvement over MI-2, as reflected by its substantially more potent cellular activity in MLL-AF9–transduced BMCs.
Figure 5.
Second-generation inhibitor MI-2-2 exhibits strongly enhanced cellular activities compared with MI-2. (A) Growth curves for MLL-AF9 transduced BMC grown in liquid culture treated with DMSO, MI-2, and MI-2-2. The experiment was performed 2 times. (B-C) Quantitative real-time PCR showing the expression of Hoxa9 (B) and Meis1 (C) in MLL-AF9–transduced BMCs over the 6 days of treatment with MI-2 and MI-2-2. Expression of Hoxa9 and Meis1 has been normalized to β-actin and is referenced to the DMSO-treated cells. Data represent the mean values for duplicates ± SD. The experiment was performed 3 times. (D) Colony counts for methylcellulose colony assay performed with MLL-AF9–transduced BMCs treated for 7 days with MI-2-2 and MI-2-2. Error bars indicate SD from duplicate experiments; experiments were performed 2 times. (E) Representative colonies shown for DMSO-, MI-2-, and MI-2-2–treated MLL-AF9–transduced BMCs plated on methylcellulose. Black lines represent the scale bars (500 μm). (F) Quantification of CD11b expression in MLL-AF9 transduced BMC treated for 6 days with the menin-MLL inhibitors as detected by flow cytometry. Data represent the mean values for duplicates ± SD. The experiment was performed 2 times. (G) Wright-Giemsa–stained cytospins for MLL-AF9–transformed BMCs after 7 days of treatment with DMSO, MI-2 (12μM), and MI-2-2 (12μM). Black lines represent the scale bars (50 μm). Statistical analysis and calculation of P values was performed using 2-way ANOVA.
MI-2-2 exhibits potent activities in human MLL leukemia cells
We also tested the activity of MI-2-2 in the MV4;11 human MLL leukemia cell line, which harbors a MLL-AF4 translocation. Similar to the effects observed in MLL-AF9–transduced BMCs, treatment of MV4;11 cells with MI-2-2 caused growth inhibition of these cells (GI50 = 3μM; Figure 6A), whereas weak or no activity was shown in non-MLL leukemia cells (supplemental Figure 5). The MI-2-2 compound also resulted in a substantial and dose-dependent increase in the number of cells undergoing apoptosis (Figure 6B) and in G0/G1 cell-cycle arrest (Figure 6C-D). The MI-2-2 also exhibited strong down-regulation of HOXA9 and MEIS1 expression in MV4;11 cells (Figure 6E) and induced differentiation in these cells, as manifested by an increase in the expression of CD11b and the formation of multilobed nuclei and highly vacuolated cytoplasm (Figure 6F-G and supplemental Figure 6). Remarkably, the effect of MI-2-2 on growth inhibition (Figure 6H) and differentiation of MV4;11 cells was significantly more pronounced compared with the first-generation compound MI-2.20 We also observed very similar activities of MI-2-2 on growth inhibition and differentiation in 3 other cell lines harboring various MLL translocations: ML-2, MOLM-13, and KOPN8 (supplemental Figures 8-10).Overall, these results demonstrate that the second-generation inhibitor MI-2-2 exhibits very pronounced effects in human leukemia cells carrying the MLL translocation, which is consistent with the improved inhibition of the menin-MLL interaction.
Figure 6.
MI-2-2 exhibits pronounced activity in MV4;11 human leukemia cells with MLL-AF4 translocation. (A) Inhibition of cell proliferation in MV4;11 cells induced by MI-2-2 after 72 hours of treatment, as detected by the MTT cell viability assay. Data represent mean values for 4 samples ± SD. The experiment was performed 3 times. (B) Apoptosis and cell death induced by MI-2-2 in MV4;11 cells as detected by flow cytometry using Annexin V/propidium iodide (PI) staining. Data represent mean values for duplicates ± SD. (C) Histograms from cell-cycle analysis performed by FACS after PI staining in MV4;11 cells treated with DMSO and MI-2-2. (D) Dose-dependent effect of MI-2-2 on cell-cycle progression measured by FACS in MV4;11 cells after PI staining. Data represent the mean values for 4 experiments ± SD. (E) Expression of the HOXA9 and MEIS1 genes normalized to 18S rRNA determined by quantitative RT-PCR in MV4;11 cells treated for 4 days with MI-2-2 referenced to DMSO-treated cells. Data represent the mean values for duplicates ± SD. (F) Quantification of CD11b expression in MV4;11cells treated for 7 days with the MI-2-2 as detected by flow cytometry. Data represent the mean values for duplicates ± SD. The experiment was performed 2 times. (G) Wright-Giemsa–stained cytospins demonstrating differentiation of MV4; 11 cells after 10 days of treatment with MI-2-2 and compared with DMSO. (H). Comparison of growth curves for MV4;11 cells treated with DMSO, MI-2, and MI-2-2.
Discussion
The development of low-molecular-weight compounds targeting PPIs is generally considered a challenging task.18 Recently, we have provided a proof of principle that targeting the PPI between menin and MLL by small molecules is feasible.20 In the present study, we carried out structural studies to characterize human menin and its interaction with MLL, which provides an essential molecular basis to understand the function of menin as an oncogenic cofactor of MLL fusion proteins in acute leukemias. We have also determined the crystal structure of menin in complex with the small-molecule inhibitor MI-2, and found that this compound binds to menin in a mode that mimics the key interactions of MBM1 with menin. The structure of the menin–MI-2 complex was crucial to the development of a second-generation inhibitor, MI-2-2, with 8-fold improved binding affinity toward menin. The MI-2-2 compound binds to menin with Kd = 22nM and is capable of inhibiting both the interaction of menin with MBM1 (IC50 = 46nM) and with the bivalent fragment of MLL that comprises both MBM1 and MBM2 (IC50 = 520 nM). The MI-2-2 exhibits very pronounced activities at low micromolar concentrations in BMCs transformed with MLL-AF9 and in MV4;11, a human leukemia cell line harboring the MLL-AF4 translocation. This also indicates that this compound has high cellular permeability and might be used as a valuable chemical probe with which to study the function of the menin-MLL and menin-MLL fusion protein interactions.
The difficulty in developing potent small-molecule inhibitors of menin-MLL arises from the relatively high binding affinity (Kd = 10 nM) due to the bivalent interaction mode, where 2 MLL fragments (MBM1 and MBM2) are involved in binding to menin.19 Our previous studies demonstrated that the entire N-terminal fragment of MLL binds to menin with an approximately 5-fold stronger affinity than MBM1 alone due to the presence of a second, low-affinity binding motif.19 Mutational analysis indicates that deletion of MBM1 is sufficient to abolish development of leukemia in vivo,8 and therefore the menin-MBM1 interaction constitutes a primary target for inhibitor development. The small-molecule compound we developed, MI-2-2, binds to menin with an affinity comparable to the 40–amino acid–long MLL fragment encompassing both MBM1 and MBM2.19 More importantly, MI-2-2 can efficiently disrupt not only the binding of MBM1, but also the entire MLL fusion protein to menin (Figure 4).
Multisite binding modes have also been described for the interactions of other intrinsically unstructured proteins.37 Therefore, the development of small molecules that would efficiently target such interactions represents an additional difficulty for PPI inhibitors. Our work provides an important proof of concept that the inhibition of a complex with multivalent interactions can be achieved by a low-molecular-weight compound if that the ligand binds with high affinity to the PPI hot spot.
Structural analysis of protein-ligand complexes revealed that inhibitors of PPIs tend to be large molecules forming extensive hydrophobic contacts with relatively few hydrogen bonds.38 The MI-2-2 has relatively low molecular weight (415 Da) and lipophilicity (cLogP = 3.9), and is fully compliant with the Lipinski rule of 5 for orally bioavailable drugs.39 Furthermore, MI-2-2 has a very favorable ligand efficiency index (LE = 0.39),40 which is much better than the average value of 0.24 reported for PPI inhibitors.38 In summary, MI-2-2 has several advantageous qualities as a protein-protein inhibitor, including nanomolar affinity and favorable drug-like properties. The availability of the high-resolution (1.27 Å) crystal structure of the MI-2-2 in complex with menin provides an excellent foundation to develop novel drugs for effective targeting of menin-MLL interaction in acute leukemias with MLL rearrangements.
Supplementary Material
Acknowledgments
The authors thank Jay L. Hess and Andrew G. Muntean for cell lines.
This work was funded by an American Cancer Society Research Scholar Grant (RSG-11-082-01-DMC to T.C.), a Leukemia & Lymphoma Society TRP grant (6116-12 to J.G.), and a National Institutes of Health grant (1R01CA160467 to J.G.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (grant 085P1000817).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.S. synthesized the compounds; M.J.M., T.H., and G.R. crystallized the menin and performed the biochemical experiments; S.H. and T.P. performed the cellular assays; G.L. and M.C. determined the crystal structures; and J.G. and T.C. planned the experiments and wrote the manuscript with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.C. is Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC.
Correspondence: Tomasz Cierpicki or Jolanta Grembecka, University of Michigan, Department of Pathology, 1150 W Medical Center Dr, MSRB1, Rm 4510, Ann Arbor, MI 48109; e-mail: tomaszc@umich.edu or jolantag@umich.edu.
References
- 1.Cox MC, Panetta P, Lo-Coco F, et al. Chromosomal aberration of the 11q23 locus in acute leukemia and frequency of MLL gene translocation: results in 378 adult patients. Am J Clin Pathol. 2004;122(2):298–306. doi: 10.1309/RX27-R8GJ-QM33-0C22. [DOI] [PubMed] [Google Scholar]
- 2.Slany RK. When epigenetics kills: MLL fusion proteins in leukemia. Hematol Oncol. 2005;23(1):1–9. doi: 10.1002/hon.739. [DOI] [PubMed] [Google Scholar]
- 3.Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95(2):234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 4.Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106(3):614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 5.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10(10):500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 7.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94(7):984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cierpicki T, Risner LE, Grembecka J, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17(1):62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30(4):958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen MD, Grummitt CG, Hilcenko C, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25(19):4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muntean AG, Tan J, Sitwala K, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17(6):609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milne TA, Kim J, Wang GG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrasekharappa SC, Guru SC, Manickam P, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276(5311):404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 17.Marx SJ. Molecular genetics of multiple endocrine neoplasia types 1 and 2. Nat Rev Cancer. 2005;5(5):367–375. doi: 10.1038/nrc1610. [DOI] [PubMed] [Google Scholar]
- 18.White AW, Westwell AD, Brahemi G. Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med. 2008;10:e8. doi: 10.1017/S1462399408000641. [DOI] [PubMed] [Google Scholar]
- 19.Grembecka J, Belcher AM, Hartley T, Cierpicki T. Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias. J Biol Chem. 2010;285(52):40690–40698. doi: 10.1074/jbc.M110.172783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grembecka J, He S, Shi A, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J Biol Chem. 2011;286(36):31742–31748. doi: 10.1074/jbc.M111.258186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 24.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 25.Cowtan KD, Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr D Biol Crystallogr. 1993;49(1):148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11(1):49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- 31.Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. Web server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60(10):1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Gurung B, Wan B, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482(7386):542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen JA, Banner DW, Seiler P, et al. Fluorine interactions at the thrombin active site: protein backbone fragments H-C(alpha)-C=O comprise a favorable C-F environment and interactions of C-F with electrophiles. Chembiochem. 2004;5(5):666–675. doi: 10.1002/cbic.200300907. [DOI] [PubMed] [Google Scholar]
- 35.Müller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: looking beyond intuition. Science. 2007;317(5846):1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 36.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arai M, Ferreon JC, Wright PE. Quantitative analysis of multisite protein-ligand interactions by NMR: binding of intrinsically disordered p53 transactivation subdomains with the TAZ2 domain of CBP. J Am Chem Soc. 2012;134(8):3792–3803. doi: 10.1021/ja209936u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higueruelo AP, Schreyer A, Bickerton GR, Pitt WR, Groom CR, Blundell TL. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem Biol Drug Des. 2009;74(5):457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 39.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 40.Hopkins AL, Groom CR, Alex A. Ligand efficiency:a useful metric for lead selection. Drug Discov Today. 2004;9(10):430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.