Abstract
Tumour necrosis factor alpha (TNFα) is a potent cytokine that signals through nuclear factor kappa B (NFκB) to activate a subset of human genes. It is usually assumed that this involves RNA polymerases transcribing responsive genes wherever they might be in the nucleus. Using primary human endothelial cells, variants of chromosome conformation capture (including 4C and chromatin interaction analysis with paired-end tag sequencing), and fluorescence in situ hybridization to detect single nascent transcripts, we show that TNFα induces responsive genes to congregate in discrete ‘NFκB factories’. Some factories further specialize in transcribing responsive genes encoding micro-RNAs that target downregulated mRNAs. We expect all signalling pathways to contain this extra leg, where responding genes are transcribed in analogous specialized factories.
Keywords: chromosome conformation capture, cytokine signalling, nuclear factor kappa B (NFκB), RNA polymerase, tumour necrosis factor alpha (TNFα), transcription factory
Introduction
It is widely assumed that RNA polymerases transcribe by initiating on genes wherever they might be in a nucleus. However, accumulating evidence is consistent with an alternative: genes diffuse to dedicated sites—‘transcription factories’—to be transcribed (Chakalova and Fraser, 2010; Cook, 2010). Transcription and associated RNA processing are enhanced by the high local concentration of relevant machinery in such a factory, which we define as a site containing at least two polymerases engaged on different templates. Strong support for this alternative is provided by chromosome conformation capture (3C) and fluorescence in situ hybridization (FISH): sequences distant on the genetic map often lie together in 3D nuclear space, and they are usually transcribed and/or associated with transcription factors (Osborne et al, 2004; Simonis et al, 2006; Fullwood et al, 2009; Göndör and Ohlsson, 2009; Yaffe and Tanay, 2011; Li et al, 2012). Moreover, each of the three nuclear RNA polymerases is concentrated in its own dedicated factories (Pombo et al, 1999), which can be purified as complexes of >8 MDa (Melnik et al, 2011). Polymerase II factories further specialize to transcribe different genes; two mini-chromosomes carrying identical units are transcribed in the same factories, but inserting into one a different promoter (or an intron) targets it to a different factory (Xu and Cook, 2008). In addition, factories transcribing genes encoding interleukins (Cai et al, 2006), cytochrome c subunits (Dhar et al, 2010), Hox genes (Noordermeer et al, 2011a), steroid receptor-binding genes (Fullwood et al, 2009; Grøntved and Hager, 2012), and factors involved in globin production (Brown et al, 2008; Schoenfelder et al, 2010; Soler et al, 2010; Noordermeer et al, 2011b) have been uncovered, as have associations of non-coding elements (Robyr et al, 2011). Here, we examine whether genes activated by a canonical signalling pathway congregate in factories specializing in transcribing responsive genes.
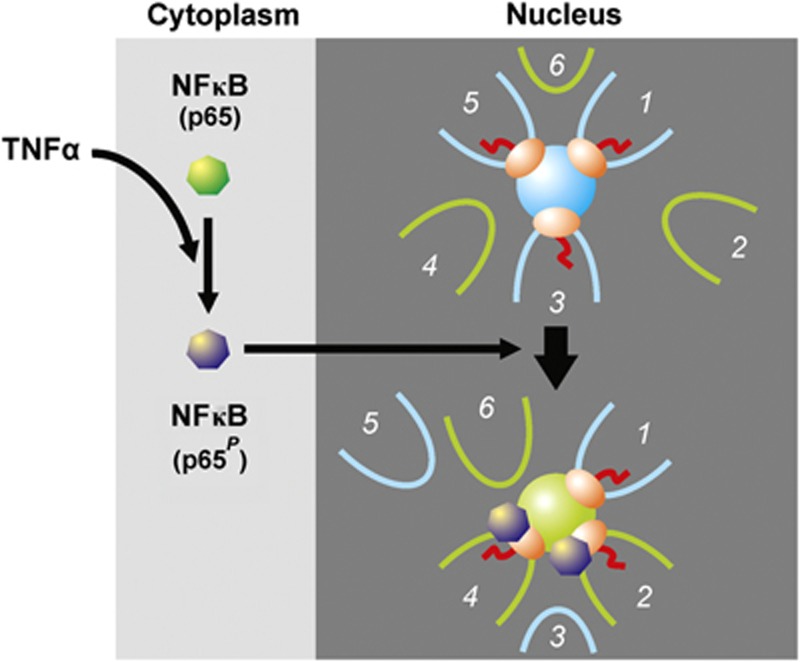
Tumour necrosis factor alpha (TNFα) is a potent cytokine that signals through nuclear factor kappa B (NFκB) to orchestrate the inflammatory response (Smale, 2010). NFκB is normally sequestered in the cytoplasm, but TNFα induces (via IKK-mediated phosphorylation and degradation of IκBs) phosphorylation of its p65 subunit, nuclear import, binding to cognate cis elements, and activation of responding genes (Ashall et al, 2009; Smale, 2010). Several hundred genes are activated within minutes, including SAMD4A and EXT1 (Wada et al, 2009; Papantonis et al, 2010). If the traditional model for transcription applies, then there is no reason to expect responsive genes carried on different chromosomes to lie near these two genes in 3D space, either before or after TNFα induction. But if responsive genes are transcribed in specialized ‘NFκB’ factories, we would expect them to associate preferentially on stimulation (Figure 1). Using derivatives of 3C (de Wit and de Laat, 2012; Ethier et al, 2012)—a focussed one called variously ‘circular 3C’, ‘4C’, ‘3C-inverse PCR’, or ‘cACT’ (Simonis et al, 2006; Zhao et al, 2006; Würtele and Chartrand, 2006; Papantonis et al, 2010) and one detecting a wider interactome called ‘chromatin interaction analysis with paired-end tag sequencing’ (ChIA-PET; Li et al, 2010)—we find most genes contacted by these two genes after stimulation to be TNFα responsive. Results are consistent with TNFα signalling through specialized ‘NFκB’ factories. As another cytokine—transforming growth factor β (TGFβ; Meulmeeste and Ten Dijke, 2011)—induces its responsive genes to associate, we suggest all cytokines will signal through specialized factories.
Figure 1.
Hypothesis. NFκB (green) is usually cytoplasmic, and genes 1, 3, and 5 are transcribed in a factory (blue sphere) while TNFα-responsive genes 2, 4 and 6 are unattached and inactive. Only 3 of the ∼16 sequences attached to a factory are shown (Cook, 2010). TNFα induces phosphorylation of the p65 subunit of NFκB (now purple), import into the nucleus, binding to responsive promoters and/or the factory, and—once relevant promoters diffuse through the nucleoplasm and collide with the factory—transcription of responsive genes in what has become a ‘specialized’ factory (green sphere). As a result, gene 2 now lies near other responsive NFκB-binding genes. Gene 1 is still attached and transcribed, but may later be replaced by responsive gene 6. If this model applies, then TNFα stimulation should bring gene 2 close to other responsive genes.
TNFα stimulation also downregulates many genes. As miRNAs are powerful downregulators, and as the nuclease (Drosha) involved in the initial step of miRNA processing acts co-transcriptionally (Morlando et al, 2008; Pawlicki and Steitz, 2008), we speculated that relevant pre-miRNAs are produced in ‘miRNA’ factories. We used the same strategy to see if genes hosting responsive miRNAs (Suárez et al, 2010) not only co-associated with other responsive genes, but also with genes hosting miRNAs; they did. This suggests that some ‘NFκB’ factories further specialize in producing non-coding transcripts regulating the inflammatory response.
Results
SAMD4A and EXT1 develop new contacts on stimulation
We apply 3C (Dekker et al, 2002) to detect proximity of two DNA sequences in 3D nuclear space, plus two variants producing more complete interactomes—a 4C variant using nested PCR (Papantonis et al, 2010) and ChIA-PET (Li et al, 2010). Our purpose is not to compile a complete interactome of responsive genes, but to focus on the principles governing co-association. Each approach has its own bias (introduced during amplification, cloning, immunoprecipitation, and/or DNA size selection prior to sequencing), but none should enrich for or against TNFα-responsive genes.
We first applied 4C to screen contacts made by two genes that respond promptly and synchronously to TNFα—SAMD4A (on HSA 14) and EXT1 (on 8). Sixteen different 4C libraries were prepared at four different times after adding TNFα to HUVECs (i.e., 0, 10, 30, and 60 min), using SacI or HindIII, and one of two reference points (the transcription start site, TSS, of SAMD4A or EXT1). Four more libraries were prepared after pretreatment with BAY 11-7085 (BAY), an indirect inhibitor of NFκB phosphorylation and so the signalling cascade (Pierce et al, 1997). 4C libraries were generated, cloned, and ∼80 inserts per library sequenced (∼48 for BAY libraries). This allowed analysis of inserts varying in length from 40 to >1000, bp, and—as conventional sequencing reads across ligation junctions—bona fide 3C products were verified. Less than 1% sequences in each library lacked appropriate restriction sites and were discarded. We also re-analysed EXT1 and SAMD4A libraries (prepared with HindIII 0–60 and 30 min after stimulation, respectively) using ‘next-generation’ sequencing; amplified 4C products were re-cut, linkers attached, DNA fragments of 300±100 bp selected, and ∼107 (36-bp single-end) reads per library uniquely mapped to the genome.
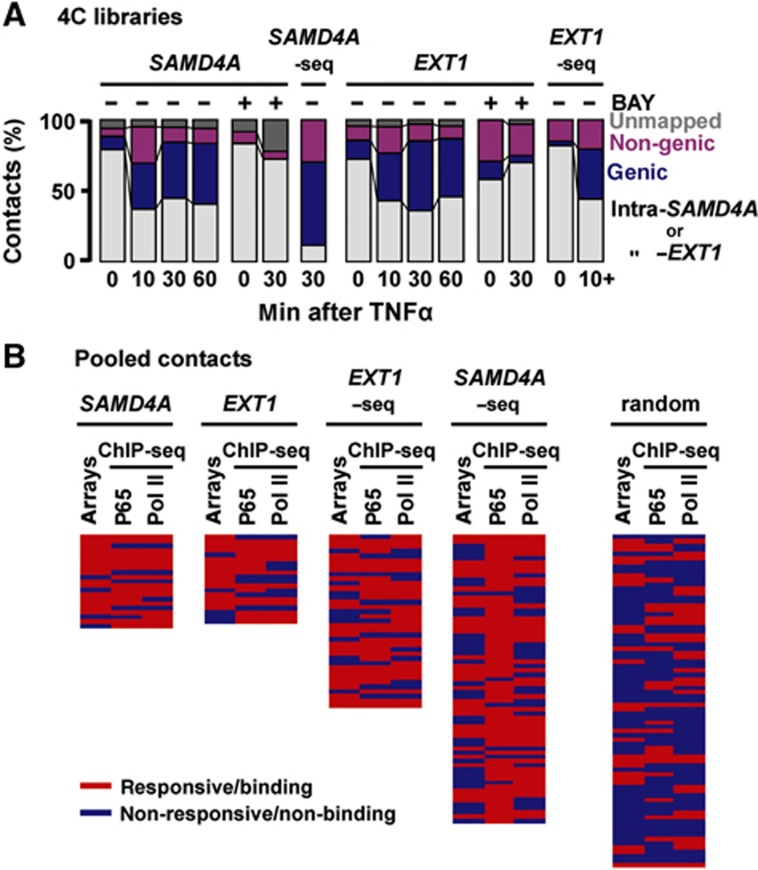
The profile of SAMD4A and EXT1 contacts changes on stimulation (Figure 2A). A minority of sequences in all libraries were ‘unmapped’ (mainly inserts <40 bp). At 0 min, most 4C products arise by self-ligation (ligation restores the original genomic sequence) or by ligation to nearby restriction sites within reference genes; we call all these ‘intra-SAMD4A/-EXT1’ contacts. Their presence is consistent with cut ends of each reference TSS lying far from other genes but close to other ends produced in these long genes of 221 and 312 kbp (as in Figure 1, top). This applies to most regions of the genome that make many local (cis) contacts on the same, but few (trans) contacts with other chromosomes (Lieberman-Aiden et al, 2009). But after 10 min, each (now-active) TSS often becomes ligated to DNA sequences lying within (or between) other RefSeq genes, many on different chromosomes (Supplementary Table S1). We call such contacts ‘genic’ (or ‘non-genic’), and attribute the increase to the reference TSS binding to a factory surrounded by other genes (as in Figure 1, bottom). By 30 min, most contacts are genic ones (e.g., 21 and 26 different genic contacts were made by SAMD4A and EXT1, respectively, compared to 11 and 10 non-genic ones). BAY prevents development of these contacts (Figure 2A). We did not analyse non-genic contacts in detail, but they typically bind NFκB and possess histone marks indicative of active enhancers (Discussion).
Figure 2.
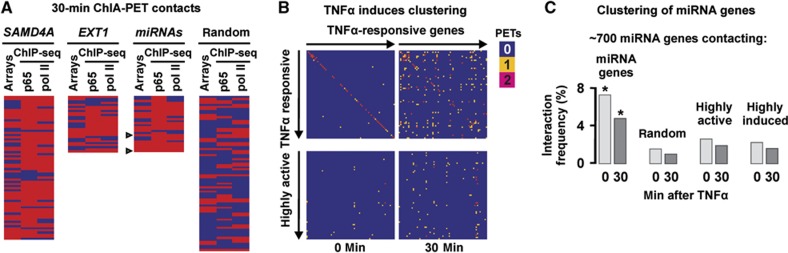
4C shows that TNFα induces SAMD4A and EXT1 to associate with other responsive genes. HUVECs were grown in TNFα for 0–60 min (±BAY 11-7085, ‘BAY’—an inhibitor of p65 phosphorylation), and 4C applied. (A) Evolving contacts in 20 4C libraries. Reference gene, BAY pretreatment, and time after stimulation are indicated. Results from libraries prepared using SacI or HindIII are pooled. Some HindIII libraries were also analysed by high-throughput sequencing (‘EXT1-/SAMD4A-seq’, with results of 10–60 min EXT1 libraries pooled and labelled as ‘10+’). At 0 min, most contacts are ‘intra-SAMD4A/-EXT1’. After 10 min, contacts develop with non-genic and genic regions; by 30 and 60 min, most are with other genic regions. Pretreatment with BAY prevents this evolution. (B) Most contacts (detected on ⩾2 independent occasions between 10 and 60 min) are with responding, p65-binding, genes. Genes were scored as TNFα responsive (⩾1.5-fold change compared to 0-min levels, determined using publicly available microarray data) and able to bind the p65 subunit of NFκB or RNA polymerase II (red), or non-responsive/non-binding (blue; determined using ChIP-seq here). Each row gives results for one gene; a set of 75 randomly selected human genes is presented for comparison (genes listed in rank order of decreasing number of contacts, as in Supplementary Tables S1 and S2). In all, 57% contacts (n=21) made by SAMD4A (39% by EXT1-seq; n=57) are with genes that are responsive and bind both p65 and the polymerase; the value for the random set (7%; n=75) is significantly lower (P<0.0001 in both cases; two-tailed Fisher’s exact test).
On stimulation, contacts are with other TNFα-responsive genes
If factories specialize in transcribing responsive genes, then many new contacts should be with upregulated genes that bind both NFκB and active RNA polymerase II. To focus on frequent contacts, and mindful that 4C involves amplification that generates multiple identical clones of one 3C product, we require that all genic contacts analysed are seen on more than one occasion. Thus, after conventional sequencing, contacts must be seen in ⩾2 libraries or contact ⩾2 different parts of one gene; as expected, the two approaches detect partially overlapping contacts (Supplementary Tables S1 and S2). We determined whether each contacted gene was TNFα responsive using microarray data (Materials and methods), and chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq), was used to assess whether contacted genes also bind the p65 subunit of NFκB and phosphorylated isoforms of polymerase II; a randomly generated set of 75 human genes (which represent the complete range of activity) serves as a control (Figure 2B; Supplementary Tables S1 and S2). As significantly more contacts made by SAMD4A and EXT1 are with genes that are both responsive and/or have p65 and/or the polymerase bound to their promoters, the two reference genes mostly contact other responsive/p65-binding genes.
Responsiveness assessed using microarray data reflects changes in steady-state mRNA levels occurring over hours; however, changes in nascent RNAs occurring within minutes are of interest here. Therefore, we monitored such changes using intronic qRT–PCR; we also assessed p65 and RNA polymerase binding by ChIP. Three sets of 12 genic contacts seen after stimulation were randomly selected for detailed analysis; a random set provides a control (Supplementary Table S1). Almost all genes in the experimental sets were upregulated and had p65 and the polymerase bound to their promoters, and BAY abolished this (Supplementary Figures S1A, B and S2A, C, D). Note also that binding of p65 to ∼1/3 of these genes depends on ongoing transcription (it is inhibited by elongation-inhibitor DRB; Supplementary Figure S2B), and that most of the genes are expressed at levels comparable to GAPDH (Supplementary Figure S3A). Moreover, 4C contacts strongly correlate with the number of p65 binding sites on different chromosomes (R=0.75; Supplementary Figure S3B). Taken together, results are consistent with responsive genes associating on activation, and with their contacts evolving thereafter (for an overview, see Supplementary Figure S4).
TNFα-responsive genes encoding miRNAs co-associate
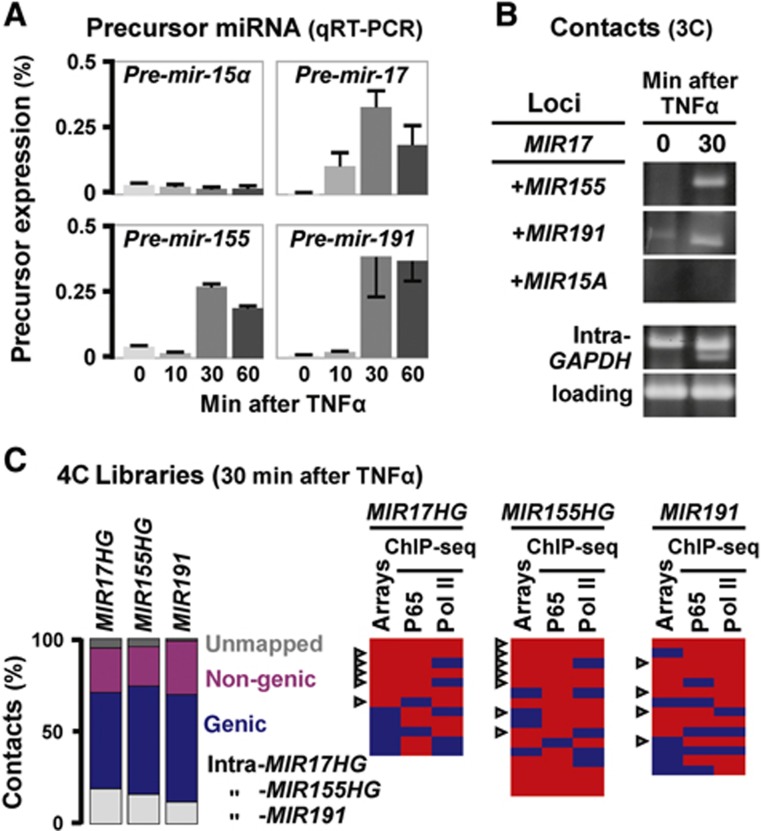
The above analysis concentrates on coding genes; what of non-coding ones? Speculating that TNFα-responsive pre-miRNAs are produced in discrete ‘miRNA’ factories, we applied the same strategy to see if three genes hosting responsive miRNAs (encoding miR-17, -155, and -191; Suárez et al, 2010) not only came together with other responsive genes, but also with ones hosting miRNAs. As only ∼1500 of the ∼22 000 human genes are currently known to host miRNAs (Dweep et al, 2011), random co-association of miRNA genes is unlikely. First, qRT–PCR confirmed that TNFα induces transcription of miR-17, -155, and -191 precursors in HUVECs, but not of non-responsive miR-15α (Figure 3A). 3C then revealed that MIR17HG (on HSA 13) contacts none of the others before adding TNFα. However, 30 min after stimulation it contacts MIR155HG (on 21) and MIR191 (it lies within DALRD3 on 3), but not non-responsive MIR15A (on 13; Figure 3B).
Figure 3.
TNFα-responsive genes hosting miRNAs co-associate. HUVECs were grown in TNFα (0–60 min), total nucleic acids purified and 3C/4C applied. (A) Levels of precursor miRNAs assessed by qRT–PCR (normalized relative to RNU6 RNA; ±s.d.; n=3). Levels of (non-responsive) miR-15α remain unchanged; those of the three other miRNAs peak after 30 min. (B) Responsive miRNA host genes co-associate. 3C reveals that MIR17 (on HSA 13) contacts MIR155 (on 21) and MIR191 (on 3) after 30 min (but not at 0 min); it does not contact MIR15A (on 13) at either time. Bands reflect contacts, and controls for intra-GAPDH contacts and loading are shown. (C) MIR17HG, MIR155HG, and MIR191 often contact other responsive genes that bind p65. 4C libraries were generated 30 min after stimulation and ∼96 inserts in each sequenced. Left: Bar graphs illustrate types of contact seen. Right: Genic contacts seen at least twice are listed in rank order of those seen most frequently (see also Supplementary Table S3). These were then scored (as in Figure 2B) as responsive and/or able to bind the p65 subunit of NFκB and/or RNA polymerase II (red), or unresponsive/non-binding (blue). Arrowhead: miRNA host gene. The observed high frequency of contacts with genes hosting miRNAs (43%; n=42) is significant, as only 6 such contacts are seen in the 228 contacts of the SAMD4A and EXT1 protein-coding genes (Supplementary Tables S1 and S2), and as only 1424 such genes are known (out of ∼22 000 RefSeq genes; P<0.0001 in both cases; two-tailed Fisher’s exact test). In all, 33% contacts made by MIR17HG (n=12), 50% by MIR155HG (n=16), and 36% by MIR191 (n=14) are with genes that are TNFα responsive and bind both p65 and the polymerase—significantly more than the 7% (n=75) seen with the random set in Figure 2B (P=0.019, 0.0001, and 0.0076, respectively; two-tailed Fisher’s exact test). Figure source data can be found with the Supplementary data.
We next used 4C to screen for other contacting sequences. Three new libraries were generated 30 min after stimulation using MIR17HG, MIR155HG, or MIR191 as reference points, and 96 inserts in each sequenced. Most contacts (seen at least twice) made by each of the three references were with genic sequences that were TNFα responsive and/or able to bind p65 and/or the polymerase (Figure 3C); this confirms the principle that responding genes co-associate on stimulation. Moreover, ∼1/3 contacts encoded miRNAs (Figure 3C, arrowheads), and 70% of these were upregulated by TNFα in a BAY-sensitive manner (Supplementary Figure S1C). Results indicate that a subset of responding genes encoding miRNAs also co-associate on activation (for an overview, see Supplementary Figure S4 and Supplementary Table S3).
Finally, if TNFα signalling led to mRNA downregulation by activating production of miRNAs encoded by our three references and their contacts, downregulated mRNAs should be enriched with binding sites for these miRNAs (Guo et al, 2010). They appear to be, as the 100 mRNAs most downregulated 1 and 4 h after stimulation contain more potential binding sites than a random set (Supplementary Table S4). This supports the idea that miRNAs encoded by co-associating genes are functionally relevant.
Nascent RNAs and pre-miRNAs encoded by responsive genes colocalize
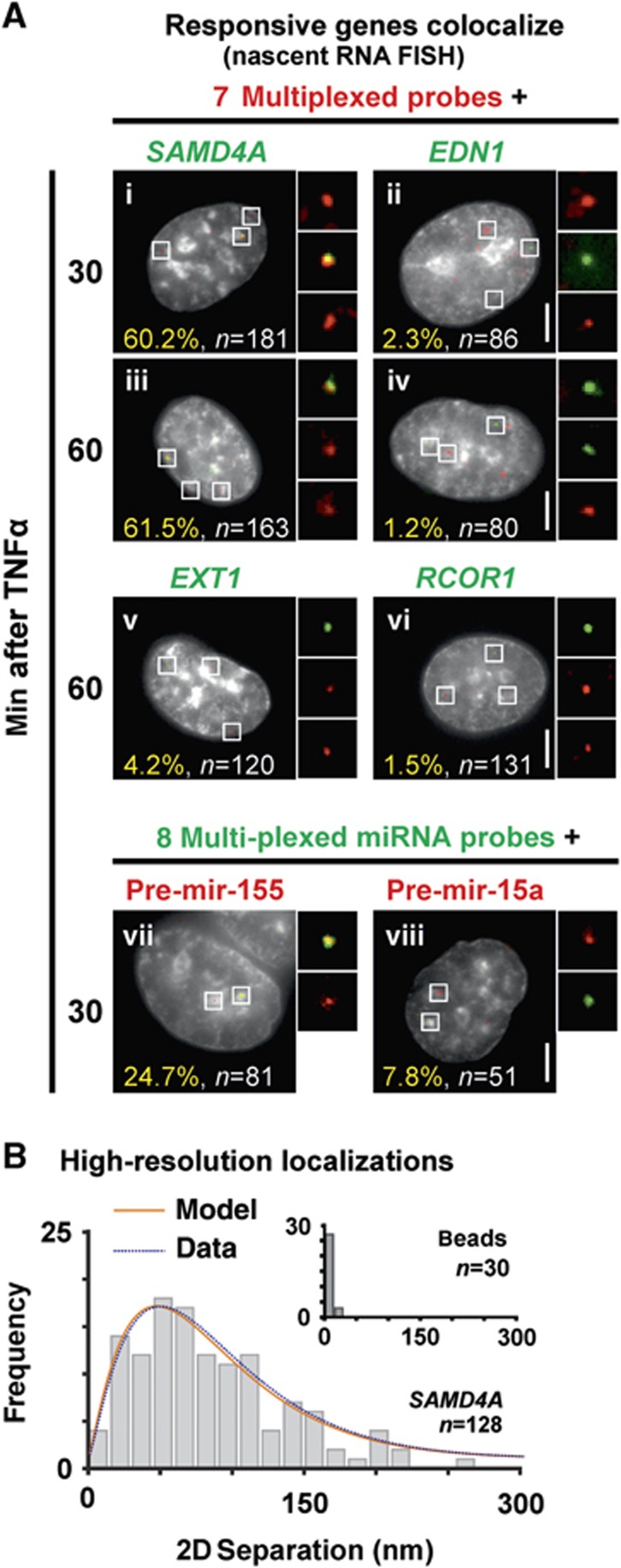
We used RNA FISH with intronic probes to confirm that nascent RNAs encoded by frequently contacting genes lie together. As stochasticity in transcription ensures both alleles of most human genes are rarely transcribed simultaneously (Chubb and Liverpool, 2010)—including SAMD4A and EXT1 (Wada et al, 2009; Papantonis et al, 2010)—we used a multiplexed set of probes targeting intronic RNA encoded by seven different genes (three cis and four trans, to SAMD4A; Materials and methods) paired with: (i) a probe targeting SAMD4A intron 1 or (ii) a (control) probe targeting EDN1 intron 2 (a constitutive, non-responsive, gene with comparable activity to SAMD4A; Wada et al, 2009). Note that (diploid) HUVECs are starved prior to stimulation so essentially all cells are in the G0 phase of the cell cycle (Larkin et al, 2012), and ∼35% SAMD4A alleles plus ∼10–30% alleles of each of the seven contacted genes in a population are active after stimulation. RNA FISH yields three types of foci: red ones mark nascent transcripts copied from alleles of one or more of the seven multiplexed targets, green ones mark SAMD4A nascent RNA, and yellow ones indicate colocalization of two (rarely two pairs of two) nascent transcripts (see Materials and methods for colocalization criteria). In all, 60% green foci colocalized with at least one red focus to give such yellow foci, significantly higher than the ∼2% colocalization seen with EDN1 (Figure 4A, i and ii). Similar results are seen with the same probes 60 min after stimulation (Figure 4A, iii–vi), with a responsive gene with a different interactome—EXT1 (Supplementary Tables S1 and S2), and another constitutively expressed control gene—RCOR1—on the same chromosome as SAMD4A (with activity comparable to GAPDH; Papantonis et al, 2010). Analogous results were obtained when SAMD4A was paired with four multiplexed probes targeting genes on four other chromosomes (29% colocalization), and by DNA FISH on two responsive loci (5% colocalization; Supplementary Figure S5A). Similarly, nascent RNA encoded by responsive MIR155HG colocalizes with nascent RNAs from eight MIR155HG contacts (Figure 4A, vii and viii). These confirm 4C results; if genes co-associate, so do their nascent transcripts. As many red and green foci do not colocalize (e.g., of 181 SAMD4A green foci analysed 30 min post induction, 27% did not overlap any red focus, 57% overlapped a single red focus, and only 16% colocalized with ⩾2 red foci), it also follows there must be many factories specializing in transcribing responsive genes in one cell (see Discussion).
Figure 4.
RNA FISH shows colocalization of nascent RNAs encoded by responsive genes in structures the size of factories. HUVECs were stimulated with TNFα, fixed, nascent transcripts detected using RNA FISH, DAPI-stained cells imaged, and distances between overlapping red and green signals measured using high-resolution localization. (A) Typical images (insets show magnifications of selected foci). Bars: 2 μm. (i–iv) A green probe targets nascent RNA encoded by SAMD4A (or EDN1, a non-contacted, non-responsive, control on HSA 6 with a comparable activity to SAMD4A), while a multiplexed set of red probes targets intronic RNA encoded by seven different genes (targeting regions >0.5 Mbp away from SAMD4A, or on different chromosomes). Approximately 60% SAMD4A foci (green) colocalize with a red focus (which might contain 1–7 targets) to give a yellow one; significantly fewer EDN1 foci (green) colocalize with red foci (P<0.0001; two-tailed Fisher’s exact test). (v, vi) Similar results were obtained when EXT1 (a responsive gene on HSA 8 that has a different interactome from SAMD4A; see Supplementary Tables S1 and S2) or RCOR1 (a non-contacted, non-responsive control on the same chromosome as SAMD4A that has an activity comparable to GAPDH) was used together with the seven multiplexed SAMD4A-contacting targets (P<0.0001 in both cases; two-tailed Fisher’s exact test). (vii, viii) A red probe targets nascent pre-miR-155 or pre-miR-15a RNA (a non-responsive control), while a multiplexed set of green probes targets eight different nascent pre-miRNAs (all 4C contacts of MIR155HG). Approximately 25% pre-miR-155 foci (red) colocalize with green foci; significantly fewer pre-miR-15a (red) foci colocalize with green foci (P=0.019; two-tailed Fisher’s exact test). (B) High-resolution localization (22-nm precision). Yellow foci like those in (A) were selected (n gives number of foci analysed), and separations between peaks in red and green channels measured. The histogram gives frequencies of separations, the blue curve is a gamma-fit to this histogram, and the orange one the distribution expected if pairs of red and green points are randomly distributed in a 35-nm shell around a 90-nm sphere (Papantonis et al, 2010). Inset: separations between peaks given by red/green fluorescent 110-nm beads used as colocalizing controls.
Nascent SAMD4A and EXT1 transcripts are found in ‘NFκB’ factories
Electron and high-resolution light microscopy reveal that nascent transcripts lie on the surface of ∼90 nm factories (Eskiw et al, 2008; Papantonis et al, 2010; Larkin et al, 2012). We measured separations between colocalizing red and green signals (like those in Figure 4A) given by SAMD4A and the multiplexed genes using high-resolution microscopy (with 22-nm precision, and so well below the resolution limit). We assume a yellow focus marks subdiffraction red/green spots, fit Gaussian profiles to intensities, and measure the separation between peaks. A ‘perfectly’ colocalizing control—red/green fluorescent beads—yields separations of <25 nm (Figure 4B, inset), as expected of randomly oriented fluors in a small sphere localized with the measured precision. Experimentally measured separations were broadly distributed up to 160 nm (Figure 4B). This distribution is the one expected of a model (Papantonis et al, 2010) where pairs of red and green spots are randomly and repeatedly distributed in a 35-nm shell around a 90-nm core—the known dimensions of a factory (Figure 4B, compare orange and blue curves).
As TNFα induces phosphorylation of the p65 subunit of NFκB and nuclear import (Ashall et al, 2009), we might expect to find p65P in factories transcribing responsive genes (Figure 1). Four results support this: p65P appears within 10 min in discrete nuclear foci in a BAY-sensitive manner (Supplementary Figure S5B), it co-purifies with large fragments of factories released from nuclei by caspases (Melnik et al, 2011; Supplementary Figure S5C), and it is associated with both nascent RNAs encoded by SAMD4A and EXT1 (Supplementary Figure S5D) and foci containing nascent BrRNA (Supplementary Figure S5E). Thus, nascent RNAs are produced in transcriptionally active sites rich in ‘active’ NFκB.
ChIA-PET confirms co-association of responsive genes
4C reveals interactomes of selected genes; ChIA-PET permits a wider analysis. We prepared two more libraries (0 and 30 min after adding TNFα) by immunoselecting chromatin bound to phosphorylated isoforms of RNA polymerase II, and generated genome-wide interactomes using next-generation sequencing (∼3.5 × 107 paired-end reads per library, from which ∼107 were uniquely mapped to the genome). To minimize amplification effects, two or more reads that were identical (or mapped ±2 bp of one another) were classified as one PET; thus, most PETs/contacts are represented by numerous reads. We analysed frequent contacts made by SAMD4A, EXT1, plus the three miRNA reference genes. (Results for the miRNA genes were pooled as each made few contacts—which we attribute to poor immunoselection due to low transcription rates (which are 1:0.45:0.2 for SAMD4A, EXT1, and MIR155HG as assessed by nascent RNA FISH)). On stimulation, the fraction of genic contacts increases, with the number of contacts seen being in the order SAMD4A>EXT1>miRNAs (again reflecting transcription rates; Supplementary Figure S6A). Most contacts after stimulation are again with responsive genes that bind p65 and/or the polymerase (Figure 5A), and a number of these encode miRNAs and/or ncRNAs (Figure 5A, arrowheads). These results confirm that stimulation induces responding genes to co-associate (for an overview, see Supplementary Figure S6B and Supplementary Tables S5 and S6).
Figure 5.
ChIA-PET confirms that TNFα-responsive genes, and those encoding miRNAs, co-associate. HUVECs were grown in TNFα for 0 or 30 min, active RNA polymerase II immuno-selected, ChIA-PET performed, and the interactions between selected genes were analysed. The statistical significance of differences was assessed using two-tailed Fisher’s exact test in (A, B) or χ2 test with Yates’ correction in (C). (A) Most genic contacts made by SAMD4A, EXT1, and three miRNA genes at 30 min are TNFα responsive and p65 binding. Contacted genes (with ⩾3 or ⩾2 PETs/contacts each, for SAMD4A/EXT1 and miRNA genes, respectively) are shown in rank order (of those more frequently seen) and scored as responsive/binding (red) or non-responsive/non-binding (blue) as in Figure 2B. Arrowhead: gene hosting two miRNAs. In all, 49% contacts made by SAMD4A (n=68), 36% by EXT1 (n=28), and 46% by miRNA genes (n=28) are TNFα responsive and bind both p65 and the polymerase—a significant enrichment (P<0.0001, 0.0006, and <0.0001, respectively) compared to the random set (7%; n=75). The observed frequency of contacts hosting miRNAs (14%; n=28) is significantly higher than for SAMD4A and EXT1 (3%; n=228; P=0.015). (B) Contacts made by the 69 genes most upregulated by TNFα. Coloured boxes indicate 0 (blue), 1 (yellow), or ⩾2 (red) contacts/PETs between two genes. Top: Genes are ranked in order of upregulation by TNFα (left to right and top to bottom; all ⩾1.9-fold, determined using microarrays 0 and 60 min after stimulation). Significantly, more contacts develop after 30 min (P<0.0001). Bottom: The same 69 most upregulated genes versus the 69 most highly expressed, but non-responsive genes (all ±1.5-fold, determined as above); after 30 min, there are significantly fewer contacts (P<0.0001) than in the 30-min matrix above (additional controls in Supplementary Figure S6). (C) Genes encoding miRNAs tend to contact each other before and after stimulation (assessed using PETs/contacts made by >700 genes encoding miRNAs). The interaction frequency (%) is the number of PETs divided by the number of possible pairwise combinations. *Significantly more contacts are seen between the ∼700 miRNA genes (left; P<0.0001), compared to the ∼700 miRNA genes with the same number of randomly selected, highly expressed, or induced genes.
To extend analysis to more genes, we selected the 69 most upregulated by TNFα after 60 min (using microarray data), and determined whether stimulation increased contacts between them; it did (Figure 5B). These contacts do not simply result from transfer of active genes to an open (active) chromatin compartment, as the 69 made significantly fewer contacts with the 69 most highly active, but non-responsive, genes (Figure 5B), or with sets of 69 constitutive or TGFβ-responsive genes (Supplementary Figure S6C). Importantly, the number of interactions between the 69 genes most upregulated by TNFα was significantly higher than that seen between the 69 highly active, non-responsive, genes (Supplementary Figure S6C), suggesting some sort of specialized spatial coordination of associations.
Unfortunately, there remains no genome-wide data on miRNAs upregulated by TNFα, so we could only use the ChIA-PET data to examine whether genes hosting miRNAs co-associate before and after stimulation (and not how stimulation influences co-association). We compiled a list of 20 upregulated miRNA-encoding genes from microarray (Suárez et al, 2010) and qRT–PCR data (Supplementary Figure S1C) and examined their interactomes. They yielded many more contacts amongst themselves, compared to those they made with 20 constitutively expressed miRNA genes, or with 20 miRNA-hosting genes upregulated by another cytokine—vascular endothelial growth factor (VEGF; Supplementary Figure S6E; Suárez et al, 2008). Finally, we examined all ∼700 miRNA-hosting genes that yielded ⩾1 PET/contact. We compared contacts they made at 0 or 30 min either with each other, or with a set of randomly selected genes. At both times, the interaction frequency between the ∼700 was higher than between the ∼700 and control groups of highly active or highly induced coding genes (Figure 5C). These results confirm that genes hosting miRNAs tend to congregate (see additional controls in Supplementary Figure S6F).
Does TGFβ signal through specialized factories?
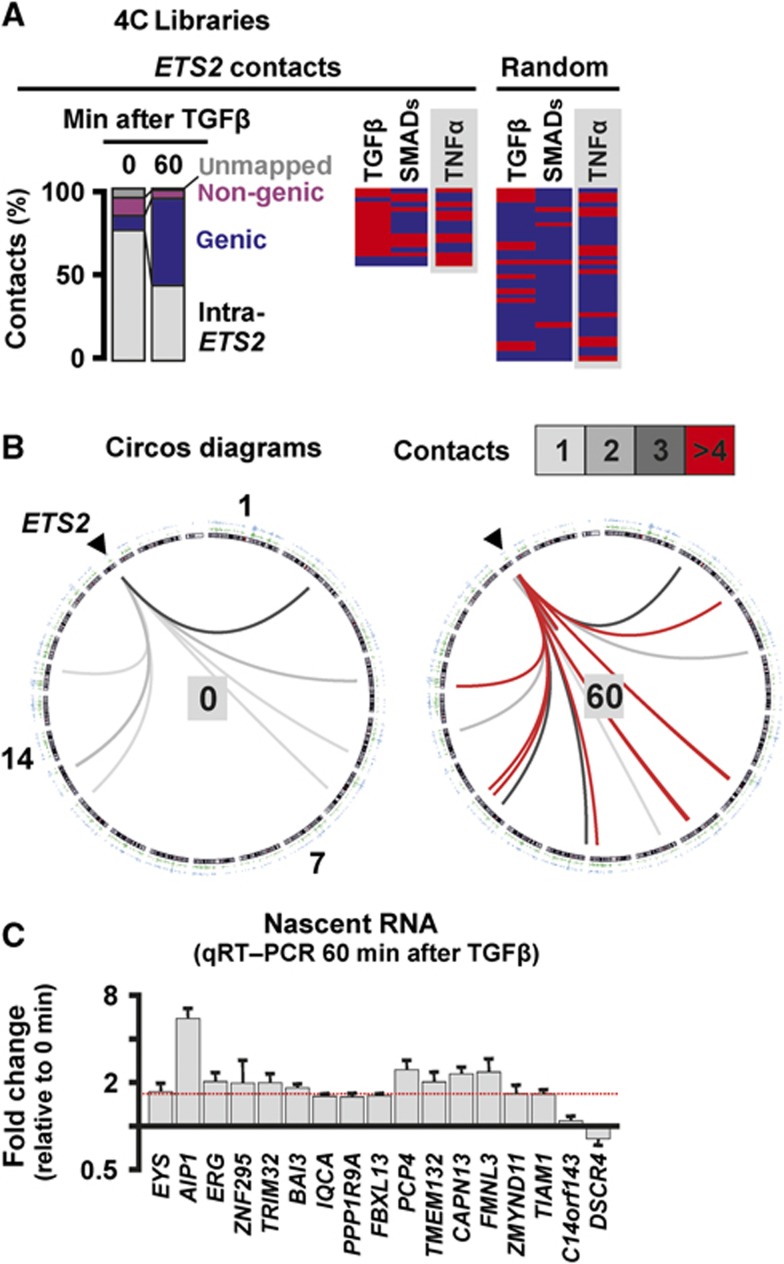
Finally, we examined whether a different cytokine—TGFβ1—uses the same strategy as TNFα. TGFβ plays a critical role in tumour development (Meulmeeste and Ten Dijke, 2011) and signals through the SMAD family of transcription factors to activate transcription of many genes, including ETS2 on HSA 21 (Koinuma et al, 2009a, 2009b). Two new 4C libraries were prepared from HUVECs harvested 0 and 60 min after TGFβ1 stimulation, using the TSS of ETS2 as a reference point. Once again, genic contacts develop (Figure 6A and B), many contacts bound SMADs (Figure 6A; binding assessed using published data; Koinuma et al, 2009a, 2009b), and most contacts seen at least twice were TGFβ responsive (Figure 6C). Moreover, one-third of these contacted genes also respond to TNFα, consistent with some overlap between the two pathways (Sullivan et al, 2009; Supplementary Table S7). In contrast, a random (control) set contains significantly fewer genes able to respond to either cytokine or bind SMADs (Figure 6A). These results support the idea that TGFβ signals through specialized ‘SMAD’ factories.
Figure 6.
TGFβ induces responsive ETS2 to associate with other TGFβ-responsive genes. 4C libraries were prepared from HUVECs harvested 0 or 60 min after stimulation with TGFβ, using HindIII and the TSS of ETS2 as a reference point; 95 and 196 inserts from the 0- and 60-min libraries were sequenced, respectively. Total RNA was also isolated, and levels of nascent RNA encoded by some contacts assessed (using qRT–PCR with intronic probes). (A) 4C libraries. Left: Contacts classified as in Figure 2A; initially most are ‘intra-ETS2’, but then more genic contacts develop. Right: Genic contacts seen at least twice (listed in rank order of number of contacts) were scored as responsive/binding (red), or unresponsive/non-binding (blue) as in Figure 2B. In all, 35% contacts (n=17) both respond to TGFβ and associate with SMADs (assessed using published ChIP data; Koinuma et al, 2009a, 2009b), significantly more than the 3% (n=36; Supplementary Table S7) in the control set (P=0.003; two-tailed Fisher’s exact test), consistent with TGFβ-responsive genes being co-transcribed in specialized ‘SMAD’ factories. Grey rectangles: some TGFβ-responsive contacts also respond to TNFα. (B) Circos software was used to depict ETS2 (on HSA 21, arrowhead) contacts evolving over time. Inner circle: chromosome ideograms drawn clockwise from 1 to Y. Outer circles: two sets of SMAD ChIP data (Koinuma et al, 2009a, 2009b). Interior: contacts made by ETS2 (box indicates numbers) increase after 60 min, and are mainly with sites binding SMADs. (C) Responsiveness of all genes contacted by ETS2 after stimulation (and seen at least twice) assessed using qRT–PCR. The fold change in nascent RNA expression levels (relative to 0 min) is presented. In all, 71% contacted genes are upregulated ⩾1.5-fold (indicated by red dotted line).
Discussion
TNFα orchestrates the inflammatory response by signalling through NFκB to activate and repress many genes (Smale, 2010). Two protein-coding (SAMD4A and EXT1) and three miRNA-hosting genes (MIR17HG, MIR155HG, and MIR191) are among the first to respond; we use these as reference points and examine whether TNFα-responsive genes are co-transcribed in ‘NFκB’ factories (Figure 1). Before stimulation, 4C and ChIA-PET reveal reference genes contact few others, but after 10–60 min they mainly contact genes that are responsive and/or bind NFκB and/or active isoforms of the polymerase (Figures 2, 3). Contacts do not simply result from transfer of active genes to an active chromatin compartment, as there were significantly fewer contacts with other highly active, non-responsive, genes (Figure 5B; Supplementary Figure S6C). RNA FISH (applied with intronic probes and coupled to high-resolution microscopy) confirms that nascent transcripts encoded by these responsive genes often lie together on the surface of 90-nm factories (Figure 4). Could these genes/transcripts associate with some structure other than a factory—for example, a nuclear ‘speckle’ that is known to lie near nascent RNA (Brown et al, 2008; Spector and Lamond, 2011)? It seems unlikely, as speckles are themselves transcriptionally inactive (Pombo and Cook, 1996) and have larger diameters (i.e., 0.5–3 μm; Hall et al, 2006) inconsistent with the profile seen in Figure 4B. While incompletely spliced transcripts can associate with speckles (Hall et al, 2006; Spector and Lamond, 2011), SAMD4A transcripts are spliced co-transcriptionally (Wada et al, 2009) so FISH signals in Figure 4A should mark transcription sites.
Some factories further specialize in producing miRNAs that target downregulated mRNAs (Figure 3; Supplementary Table S4). And even before stimulation, about half the genes encoding miRNAs tend to be co-transcribed in factories specializing in miRNA production (Figure 5C) which—presumably—contains high concentrations of Drosha, the nuclease that co-transcriptionally cleaves the miRNA precursor (Morlando et al, 2008).
These results beg many questions, such as: (i) are responsive genes poised prior to activation at/near factories (Nechaev and Adelman, 2008; Ferrai et al, 2010) so as to respond rapidly? Although SAMD4A and EXT1 make few genic contacts before stimulation, almost half are also detected after stimulation (Supplementary Table S1). Most are preloaded with RNA polymerase II (Supplementary Figure S2D) and have the potential to respond to TNFα and bind p65 (Supplementary Figures S1A, B and S2A, C). Before stimulation, we imagine that potentially responding genes lie near preexisting ‘naive’ factories, which they visit every few minutes as they diffuse through the nucleoplasm. Occasionally, promoters might transiently bind to polymerases in a factory, but few initiate as the concentration of relevant transcription factors is low. When stimulation induces nuclear influx of phospho-NFκB (Supplementary Figure S5B), the factor binds to responsive promoters and stabilizes attachment to a factory. Once productive transcription begins, responsive genes become tethered to the factory, and so our reference genes have a high probability of contacting them. As more responsive genes bind and the local NFκB concentration increases, the factory evolves into one that predominantly—but not exclusively—transcribes TNFα-responding genes. We currently favour this model over one involving de novo formation of specialized factories as some responding genes are associated with existing factories before stimulation (see also Mitchell and Fraser, 2008), and as transcription seems to be required for p65 binding to some responsive promoters (Supplementary Figure S2B). (ii) Do non-genic contacts differ from genic ones? Due to better annotation we concentrated on genic contacts, but preliminary analysis indicates that non-genic contacts are transcriptionally active and bind p65, and so might be enhancers. In all, 8 of the 10 non-genic contacts seen most frequently in all 20 (4C) SAMD4A/EXT1 libraries both possess histone marks characteristic of active enhancers (i.e., H3K27ac and H3K4me1; Zentner et al, 2011) and p65 binding increases on stimulation (Supplementary Figure S3C). We suggest such contacts reflect promoter–enhancer interactions (Lomvardas et al, 2006; Apostolou and Thanos, 2008) tethering TNFα-responsive promoters close to relevant ‘NFκB’ factories (Kolovos et al, 2012). Note also that ∼9% non-genic contacts made by the three reference genes hosting miRNAs encode non-coding RNAs (Supplementary Tables S3 and S6). (iii) How many ‘NFκB’ factories might SAMD4A access, and what is the total number of ‘NFκB’ factories per nucleus? We can only speculate, but estimate SAMD4A can access ∼8 of the 150–250 ‘NFκB’ factories—a subset of the ∼2200 polymerase II factories (for details, see Supplementary Methods and Supplementary Figure S6D). (iv) How many different types of specialized factories might there be? Again, we can only speculate, but suggest a factory specializing in transcribing genes responding to one cytokine may also transcribe some genes responding to a related cytokine, as TNFα- and TGFβ-responding reference genes contact some responding to both (Figure 6; Supplementary Table S7). Such overlapping specialization clearly allows outputs of inter-linked pathways to be integrated through transcription of genes encoding co-regulators. And as the clusters of genes associated with factories evolve after stimulation (Supplementary Figures S4 and S6B) there are rich possibilities for multi-tiered temporal regulation.
Materials and methods
Cell culture
HUVECs from pooled donors (Lonza) were grown to ∼90% confluence in Endothelial Basal Medium 2-MV with supplements (EBM; Lonza), starved (16 h) in EBM+0.5% FBS, and treated with TNFα or TGFβ1 (10 or 50 ng/ml, respectively; Peprotech) for 0–60 min. In some cases, 10 μM BAY 11-7085 (Sigma-Aldrich) or 50 μΜ 5,6-dichloro-1-β-D-ribo-furanosyl-benzimidazole (DRB; Sigma-Aldrich) was added 1 h before and retained after stimulation.
Chromosome conformation capture (3C)
3C was performed as described, until DNA purification (step 23; Göndör et al, 2008). In brief, 107 cells were fixed (1% paraformaldehyde; 10 min; 20°C; Electron Microscopy Sciences), aliquots of 106 cells in 0.125 M glycine/PBS spun, cells resuspended in the appropriate restriction enzyme buffer and lysed (16 h; 37°C) in 0.3% SDS. After sequestering SDS using 1.8% Triton X-100 (1.5 h; 37°C), cells were treated overnight with HindIII or SacI (800 units added in 4 sequential steps; New England Biolabs), the enzyme heat inactivated (25 min; 65°C), and digestion efficiency determined by quantitative PCR (qPCR); samples with digestion efficiencies >75% were ligated using T4 ligase (6000 units; New England Biolabs; DNA concentration <0.6 ng/μl; 72 h, 4°C to minimize unwanted ligations) and crosslinks reversed (16 h; 65°C) in proteinase K (10 mg/ml; New England Biolabs) before DNA was purified using an EZNA MicroElute DNA clean-up kit (Omega BioTek) and a PCR Purification kit (Qiagen). Non-digested/ligated and digested/non-ligated templates were also prepared. For 3C-PCRs, amplification efficiency controls using bacterial artificial chromosomes were as before (Papantonis et al, 2010). Identity of 3C bands was verified by sequencing (SourceBioscience, Oxford).
4C and 4C-seq
4C was based on 3C-inverse PCR/circular 3C (Simonis et al, 2006; Zhao et al, 2006; Würtele and Chartrand, 2006). Approximately, 1 μg 3C template (i.e., isolated DNA with crosslinks reversed, prepared as above) was cut with Csp6I (80 units; Invitrogen), diluted and self-ligated in 1 ml (16 h; 4°C) using T4 DNA ligase (400 units), and purified (EZNA MicroElute DNA clean-up kit); 1/50th eluate was then used in nested inverse PCR (for external primers: 95°C/2 min, plus 13 cycles at 95°C/50 s, 56°C/45 s, and 72°C/2 min, followed by a cycle at 72°C/5 min; for internal primers: 95°C/2 min, 18 cycles at 95°C/50 s, 60°C/35 s, and 72°C/1.5 min, followed by a cycle at 72°C/4 min) using GoTaq polymerase (Promega) in 2.5% dimethylsulphoxide. For conventional sequencing, amplimers were resolved on 1.5% agarose gels stained with SYBR Green stain I (Invitrogen), gel slices spanning sizes 0–100, 100–250, 250–500, 500–750, and >750 bp excised, DNA purified from each (EZNA MicroElute kit) and cloned into pGEM-T (Promega). TOP10 cells (Invitrogen) were then transformed with a 1:1:1:1:1 (1 μl each) mixture of ligation reactions, and plasmid inserts sequenced (SourceBioscience, Oxford). Inserts flanked by SAMD4A/EXT1 sequences and HindIII/SacI-Csp6I restriction sites were mapped to the human genome (hg18) using BLAST (mask: low complexity; expect value: 0.1). In all, 755 clones were sequenced for SAMD4A (144, 182, 172, 161, 48, and 48 for 0, 10, 30, 60 min, 0 min+BAY, and 30 min+BAY libraries, respectively), 745 clones for EXT1 (183, 140, 181, 139, 52, and 50 for 0, 10, 30, 60 min, 0 min+BAY, and 30 min+BAY libraries, respectively), and 374 clones for ETS2 (148 and 226, for 0 and 60 min, respectively). Numbers of clones analysed at a particular time are pooled from two independent experiments; Supplementary Figure S7 illustrates reproducibility obtained with different experiments/libraries. In all, <1% sequences in each library were not flanked by appropriate sequence motifs and were not analysed. 4C showed high reproducibility when ∼80 inserts or more were read by conventional sequencing, e.g., in the ETS2 libraries prepared 60 min after stimulation using HindIII ∼75% sequences were shared between two replicates (not including self-ligated inserts; Supplementary Figure S7A). Reproducibility was adequate even when different enzymes were used to generate libraries (e.g., ∼39% sequences were shared between SAMD4A libraries prepared using SacI and HindIII, 10 min after stimulation; Supplementary Figure S7A). For next-generation sequencing, >10 ng DNA was processed as per manufacturer’s instructions (Illumina). Briefly, amplimers were re-cut with HindIII and Csp6I, electrophoretic profiles evaluated using Bioanalyzer (Agilent Technologies), ends repaired and linkers ligated, products separated on agarose gels and fragments of 300±100 bp re-purified. From each library, ∼15 × 106 36-bp single-end reads were obtained, ∼70% of which contained one or other restriction site. After eliminating the first 5 bases containing the restriction-enzyme motif, ∼97% resulting 31-base reads were successfully aligned to 175 (0 min), 269 (10 min), 723 (30 min), and 258 (60 min) unique loci in the human genome (hg18) allowing <2 mismatches (multiple mappings and mappings to repeat elements disallowed). Reads within 5 kbp (the average HindIII restriction fragment size) of a RefSeq gene were categorized as genic, and ranked according to the number of unique mapping sites (all such genic contacts are listed in Supplementary Table S2; typical genome browser view in Supplementary Figure S8A). To focus on frequent contacts, we selected only those represented by ⩾2 sites of ⩾10 identical reads mapping to one RefSeq gene. The number of reads mapping to one site varied greatly (i.e., from 1 to 2.5 × 106). When contacts were compared to those seen by conventional sequencing an overlap of ∼25% was observed (Supplementary Figure S7B). 4C-seq data are available in the Sequence Read Archive (NCBI) under accession numbers SRX045413.2, SRX045414.2, SRX045412.3, and SRX045415.3.
Chromatin interaction analysis with paired-end tag sequencing
ChIA-PET was as described (Li et al, 2010). Briefly, confluent HUVECs were serum-starved (16 h), treated (0–30 min) with TNFα, crosslinked using 10 mM ethyl-glycol-bis-succinimidylsuccinate (EGS; Thermo Scientific) in 50% glacial acetic acid (45 min) and then in 1% paraformaldehyde (20 min; TAAB), quenched (5 min) in 2.5 M glycine, harvested, sonicated (Branson), and ChIP performed using magnetic beads (Invitrogen) and the Pd75C9 antibody directed against phospho-Ser2/-Ser5 within the heptad repeat at the C-terminus of the largest catalytic subunit of RNA polymerase II (a gift of H Kimura). Chromatin captured on magnetic beads was trimmed to create blunt ends, phosphate groups added to 5’ ends, ends ligated to each other via biotinylated half-linkers, and complexes eluted. Crosslinks were then reversed, DNA purified and digested with MmeI (whose binding site is encoded by the linker). After immobilization on M-280 Streptavidin Dynabeads (Invitrogen), adaptors were ligated, and the efficiency of library production evaluated by PCR. Finally, di-tags were prepared, sequenced on a GAII analyzer (Illumina), and resulting paired-end tags (PETs) analysed. Libraries yielded ∼35 × 106 20-bp paired-end reads each, from which 10.8 × 106 and 8.8 × 106 were successfully aligned to the genome (hg19) for the 0- and 30-min samples, respectively. For stringency, two or more reads having the same sequences, or mapping within 2 bp of the left and right ends of another read were classified as one PET; as a result most PETs were represented by many reads (for typical genome browser views, see Supplementary Figure S8B). PETs displayed some overlap to those seen by 4C as the two approaches carry different biases resulting from pull-down and amplification, respectively (e.g., in SAMD4A libraries ∼13% of contacts are shared; Supplementary Figure S5B). For Figure 5C, genes were selected as follows. Sequences encoding 852 of the 1523 miRNAs in the database lie within 731 different RefSeq transcripts and 714 distinct genomic regions; PETs involving 701 and 703 of these regions (which we call ‘miRNA genes’) were seen in 0- and 30-min libraries. PETs/contacts seen between these miRNA genes, and between these miRNA genes and (i) an equal number of randomly selected genes, (ii) 916 and 921 genes at 0 and 30 min, respectively, that are the most highly expressed but non-induced (<1.5-fold), and (iii) 900 highly induced 30 min after stimulation (>1.9-fold) were then counted, and interaction frequencies (i.e., the number of PETs divided by the number of possible pairwise combinations expressed as a percentage) calculated.
Assessment of responsiveness to TNFα and SMAD binding
Responsiveness to TNFα was assessed using publicly available HUVEC microarray data ( http://sbmdb.genome.rcast.u-tokyo.ac.jp/huvecdb/main_search.jsp); a gene was considered as responsive if mRNA levels increased ⩾1.5-fold between 0 and 1 or 4 h after stimulation (average of all relevant probes in the database; 1 h after stimulation there are ∼500 responsive genes according to these criteria). SMAD binding was assessed using ChIP-chip and ChIP-seq data for SMAD1/5, and 2/3 from human keratinocytes (Koinuma et al, 2009a, 2009b).
Analysis of miRNA targets
To assess functional relevance of miRNAs encoded by co-associating genes (by 4C and ChIA-PET), we selected from the HUVEC database the 100 most downregulated genes 1 or 4 h after TNFα stimulation; mRNA levels of these genes fell to between 2 and 42% of the 0-h level. Using default settings in the miRWalk database (Dweep et al, 2011), we queried how many mRNAs encoded by these 2 × 100 genes possessed (both predicted and experimentally validated) target sequences for the 24 miRNAs in their 3’ untranslated regions. miR-711, -761, -3169, -3170, and -4293 were not included in the database, so a list of putative targets was generated using TargetScan (Lewis et al, 2005). Results are shown in Supplementary Table S4.
RNA FISH
RNA FISH was performed as described (Papantonis et al, 2010). Probes for SAMD4A intron 1, EXT1 intron 1, and EDN1 intron 2 were sets of five 50-mers; in each roughly every tenth thymine was tagged with Alexa 488 or 555. Probes were purified and labelled with an efficiency of 3.5–4.5 fluors/50-mer; RCOR1 intron 1 was targeted by a set of 24 20-mers carrying one fluor each (Papantonis et al, 2010). Sets of multiplexed probes were four 55-mers (five internal amino-C6-modified thymines each, to which an average of four fluors was attached; IBA, Germany) targeting a <450 bp intronic region of seven genes shown to contact SAMD4A: MYH9 (on HSA 22), TNFAIP2, FBXO34, GCH1 (on 14), LARP1B (on 4), ZNF608 (on 5), and IL4R (on 16). Precursors of miR-15a and -17 (on HSA 13), -155 (on 21), -105, -504, and -767 (on X), -191 and -425 (on 3), -1203 (on 17), and -1289-2 (on 5) were targeted by single 55-mers. These probes permit detection of single transcripts, which—when tested singly—never gave more than two foci (see also Wada et al, 2009 and Larkin et al, 2012), consistent with detection of nascent transcripts at both alleles. If additional (still-unspliced) transcripts were being detected, then we would expect to see additional foci. Note that the half-lives of segments within SAMD4A intron 1 are ∼5 min (e.g., Larkin et al, 2012). Note also that pairwise combinations of these probes yield colocalization frequencies comparable to those seen by others for co-transcribed genes (e.g., Brown et al, 2008; Schoenfelder et al, 2010). After DAPI staining, images were collected using an Axioplan 2 microscope (Zeiss) with a CoolSNAPHQ camera (Photometrics) via MetaMorph v. 7.1 (Molecular Devices). A focus is defined as ⩾4 contiguous 90-nm pixels that contain signal above background (defined as the average intensity of a 50 pixel line-scan across the focus). Typically, FISH foci were 10±3 pixels in size and deemed to colocalize if ⩾25% red and green pixels overlapped.
Measurements of separations below the diffraction limit
The separation between peak intensities of overlapping foci was determined after identifying the position of each peak with 22-nm precision. Foci were identified manually and verified by a custom algorithm checking for: Gaussian shape and intensity above mean nuclear intensity plus 1 s.d. Next, the location of the emitter was estimated using the Joint Distribution algorithm (JD; Larkin and Cook, 2012). Pixel shift between fluorescence channels was corrected using 0.1-μm TetraSpeck beads (Molecular Probes) fluorescing at relevant wavelengths. Residual differences in alignment were accounted for along with spot intensity, shape, and signal-to-noise ratio to calculate uncertainty. Individual separation measurements and corresponding uncertainties were used to infer a probability distribution, which was compared to the model of a transcription factory (i.e., a 35-nm shell around a 90-nm core; Papantonis et al, 2010). All calculations were performed in MATLAB (MathWorks) using custom software routines.
Accession codes
ChIA-PET data are available in the Gene Expression Omnibus repository under accession number GSE41553.
Supplementary Material
Acknowledgments
We thank Hiroshi Kimura for the Pd75C9 and Marc Vigneron for the 7C2 antibodies, Jon Bartlett for technical assistance, Shogo Yamamoto, Kaori Shiina, and Akashi Taguchi for help with deep sequencing. This work was supported by the Biotechnology and Biological Sciences Research Council and the ERASysBio+ initiative under the FP7/ERA-NET Plus scheme (AP), the Wellcome Trust (AP, JDL), the Felix Scholarship Trust of Oxford University (SB), the Medical Research Council (BD), the Japanese Society for the Promotion of Science through the Funding Program for World-Leading Innovative R&D on Science and Technology (TK), Grants-in-Aid for Scientific Research (B)22310117 (YW, TK) and Exploratory Research 23659050 (YW) from the Japanese Society for the Promotion of Science, and Scientific Research on Innovative Areas 23125503 (YW) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author contributions: AP, YW, TK, YR, and PRC conceived and designed experiments; AP prepared 4C libraries and did conventional sequencing, 3C, qRT–PCR, ChIP, blots, and RNA FISH; SB performed immuno-FISH and immunofluorescence; JDL developed and applied algorithms for high-resolution and NN analyses; BD isolated factory fractions; YK, MK, HA, and YW did ChIP-seq, ‘deep’-sequenced 4C libraries, and prepared ChIA-PET templates; GL, HMP, XR, and YR performed ChIA-PET and analysed the resulting data; ST, TK, and GL generated high-throughput data sets and mined ChIA-PET data; ST provided help with bioinformatics and generated Circos diagrams; PS did the statistical analysis for the number of NFκB factories; AP, YW, TK, YR, GL, and PRC analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Apostolou E, Thanos D (2008) Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134: 85–96 [DOI] [PubMed] [Google Scholar]
- Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand DA, Sée V, White MR (2009) Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science 324: 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ (2008) Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol 182: 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T (2006) SATB1 packages densely looped transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Chakalova L, Fraser P (2010) Organization of transcription. Cold Spring Harb Perspect Biol 2: a000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Liverpool TB (2010) Bursts and pulses: insights from single cell studies into transcriptional mechanisms. Curr Opin Genet Dev 20: 478–484 [DOI] [PubMed] [Google Scholar]
- Cook PR (2010) A model for all genomes; the role of transcription factories. J Mol Biol 395: 1–10 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- de Wit E, de Laat W (2012) A decade of 3C technologies: insights into nuclear organization. Genes Dev 26: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MTT (2010) Chromosome conformation capture of transcriptional interactions between cytochrome c oxidase genes and genes of glutamatergic synaptic transmission in neurons. J Neurochem 115: 676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk - Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44: 839–847 [DOI] [PubMed] [Google Scholar]
- Eskiw CH, Rapp A, Carter DR, Cook PR (2008) RNA polymerase II activity is located on the surface of ∼87 nm protein-rich transcription factories. J Cell Sci 121: 1999–2007 [DOI] [PubMed] [Google Scholar]
- Ethier SD, Miura H, Dostie J (2012) Discovering genome regulation with 3C and 3C-related technologies. Biochim Biophys Acta 1819: 401–410 [DOI] [PubMed] [Google Scholar]
- Ferrai C, Xie SQ, Luraghi P, Munari D, Ramirez F, Branco MR, Pombo A, Crippa MP (2010) Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol 8: e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY et al. (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göndör A, Ohlsson R (2009) Chromosome crosstalk in three dimensions. Nature 461: 212–217 [DOI] [PubMed] [Google Scholar]
- Göndör A, Rougier C, Ohlsson R (2008) High-resolution circular chromosome conformation capture assay. Nat Protoc 3: 303–313 [DOI] [PubMed] [Google Scholar]
- Grøntved L, Hager GL (2012) Impact of chromatin structure on PR signaling: Transition from local to global analysis. Mol Cell Endocrinol 357: 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB (2006) Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Imamura T, Aburatani H, Miyazono K (2009a) Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci 100: 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H (2009b) Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol Cell Biol 29: 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolovos P, Knoch TA, Grosveld FG, Cook PR, Papantonis A (2012) Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JD, Cook PR (2012) Maximum precision closed-form solution for localizing diffraction-limited spots in noisy images. Opt Express 20: 18478–18493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JD, Cook PR, Papantonis A (2012) Dynamic reconfiguration of long human genes during one transcription cycle. Mol Cell Biol 32: 2738–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing often flanked by adenosines indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, Wei CL, Ruan Y, Sung WK (2010) ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G et al. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R (2006) Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413 [DOI] [PubMed] [Google Scholar]
- Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR (2011) The proteomes of transcription factories containing RNA polymerases I II or III. Nat Methods 8: 962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeeste E, Ten Dijke P (2011) The dynamic roles of TGF-β in cancer. J Pathol 223: 205–218 [DOI] [PubMed] [Google Scholar]
- Mitchell JA, Fraser P (2008) Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev 22: 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ (2008) Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol 15: 902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K (2008) Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 7: 1539–1544 [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D (2011a) The dynamic architecture of Hox gene clusters. Science 334: 222–225 [DOI] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W (2011b) Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol 13: 944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR (2010) Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol 8: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA (2008) Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol 182: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME (1997) Novel inhibitors of cytokine-induced IkappaB-alpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem 272: 21096–21103 [DOI] [PubMed] [Google Scholar]
- Pombo A, Cook PR (1996) The localization of sites containing nascent RNA and splicing factors. Exp Cell Res 229: 201–203 [DOI] [PubMed] [Google Scholar]
- Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR (1999) Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J 18: 2241–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Friedli M, Gehrig C, Arcangeli M, Marin M, Guipponi M, Farinelli L, Barde I, Verp S, Trono D, Antonarakis SE (2011) Chromosome conformation capture uncovers potential genome-wide interactions between human conserved non-coding sequences. PLoS ONE 6: e17634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P (2010) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip 4C. Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Smale ST (2010) Selective transcription in response to an inflammatory stimulus. Cell 140: 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F (2010) The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez Y, Fernández-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC (2008) Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA 105: 14082–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez Y, Wang C, Manes TD, Pober JS (2010) Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol 184: 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DE, Ferris M, Nguyen H, Abboud E, Brody AR (2009) TNF-alpha induces TGF-beta1 expression in lung fibroblasts at the transcriptional level via AP-1 activation. J Cell Mol Med 13: 1866–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, Izumi A, Kobayashi M, Meguro H, Kanki Y, Mimura I, Yamamoto K, Mataki C, Hamakubo T, Shirahige K, Aburatani H et al. (2009) A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA 106: 18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele H, Chartrand P (2006) Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res 14: 477–495 [DOI] [PubMed] [Google Scholar]
- Xu M, Cook PR (2008) Similar active genes cluster in specialized transcription factories. J Cell Biol 181: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A (2011) Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 43: 1059–1065 [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R (2006) Circular chromosome conformation capture 4C uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.