Abstract
Long non-coding RNAs (lncRNAs) transcribed extensively from the genome have been proposed to be key regulators of diverse biological processes. However, little is known about the role of lncRNAs in regulation of the cell-cycle G1/S checkpoint following DNA damage, a key step in the maintenance of genomic fidelity. Here we show that growth-arrested DNA damage-inducible gene 7 (gadd7), a DNA damage-inducible lncRNA, regulates the G1/S checkpoint in response to UV irradiation. Interestingly, UV-induced gadd7 directly binds to TAR DNA-binding protein (TDP-43) and interferes with the interaction between TDP-43 and cyclin-dependent kinase 6 (Cdk6) mRNA, resulting in Cdk6 mRNA degradation. These findings demonstrate a role for gadd7 in controlling cell-cycle progression and define a novel mechanism by which lncRNAs modulate mRNA expression at the post-transcriptional level by altering mRNA stability.
Keywords: cell-cycle progression, Cdk6, gadd7, lncRNA, TDP-43
Introduction
Mammalian genomes transcribe a larger number of non-coding RNAs than previously expected (Mattick, 2009). In addition to small regulatory RNAs (e.g., microRNAs, small interfering RNAs and others), surprisingly numerous long non-coding RNAs (lncRNAs) have been identified (Mattick and Makunin, 2006; Guttman et al, 2009). LncRNAs are generally considered as non-protein-coding transcripts varying in length from several hundred bases to tens of kb. Similar to protein-coding genes, lncRNAs have been implicated in diverse biological processes, including invasiveness and metastasis (Gupta et al, 2010), apoptosis (Huarte et al, 2010), dosage compensation (Deng and Meller, 2006) and embryogenesis (Pauli et al, 2011). Furthermore, lncRNAs have been reported to regulate gene expression through a wide range of mechanisms (Mercer et al, 2009). For example, lncRNAs can serve as a repressor or an activator to regulate the process of transcription. LincRNA-p21, a lncRNA downstream of p53, acts as a repressor in p53-dependent transcriptional responses by physically associating with heterogeneous nuclear ribonucleoprotein (hnRNP) K and modulating its localization (Huarte et al, 2010). In addition, the ability of lncRNAs to recognize complementary sequences also allows lncRNAs to regulate various steps in the post-transcriptional processing of mRNAs, as in the case of the Zeb2 antisense RNA, which complements an intron in the 5′-untranslated region (5′UTR) of Zeb2 and inhibits its splicing (Beltran et al, 2008). Recently, a handful of studies have implicated lncRNAs in a variety of disease states, especially in neurodegenerative diseases and cancers (Huarte and Rinn, 2010; Qureshi et al, 2010; Wapinski and Chang, 2011). Although lncRNAs have been recognized as a critical functional expression in complex organisms, there are still huge gaps in our understanding of lncRNAs.
gadd7 (growth-arrested DNA damage-inducible gene 7) is a 754-nt polyadenylated lncRNA that was isolated from Chinese hamster ovary (CHO) cells on the basis of increased expression following UV irradiation (Fornace et al, 1988; Hollander et al, 1996). Induction of gadd7 is also observed after several other types of DNA damage and growth arrest signals (Fornace et al, 1988; Hollander et al, 1996). In addition, overexpression of gadd7 leads to suppression of cell growth (Hollander et al, 1996), suggesting that gadd7 may be involved in cell-cycle control. More recently, it has been reported that gadd7 is also a regulator of lipid-induced oxidative and endoplasmic reticulum stress (Brookheart et al, 2009).
Here we further demonstrate that gadd7 is induced by both growth arrest and DNA damage signals, and plays a key role in regulation of the G1/S checkpoint after DNA damage. Moreover, we find that gadd7 regulates Cdk6 mRNA decay by dissociating TAR DNA-binding protein (TDP-43) from cyclin-dependent kinase 6 (Cdk6) mRNA upon UV irradiation. Collectively, this study describes gadd7 as a cell-cycle regulator, and explores the underlying mechanism by which gadd7 regulates the cell-cycle checkpoint in response to DNA damage, which provides novel insights into the understanding of the biological functions of lncRNAs.
Results
gadd7 is induced in response to genotoxic and non-genotoxic stresses
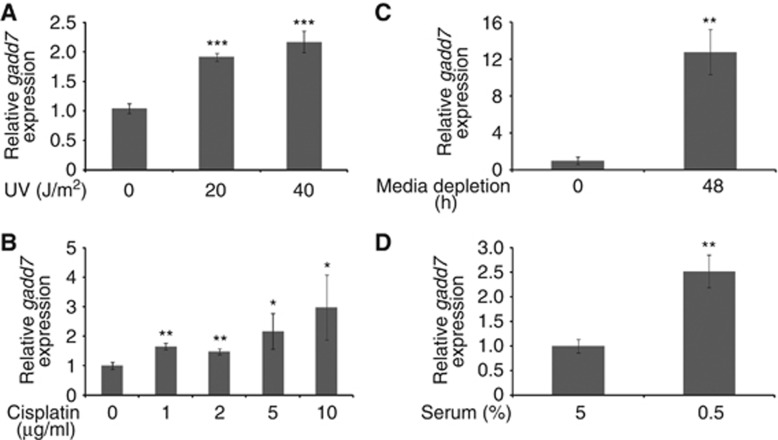
It has been reported that gadd7 is a DNA damage-inducible transcript (Fornace et al, 1988; Hollander et al, 1996). We here further investigated the induction of gadd7 by DNA damage and growth arrest signals. The results demonstrated that expression of gadd7 RNA was substantially induced in a dose-dependent manner following CHO-K1 cells’ exposure to UV irradiation (Figure 1A; Supplementary Figure S1A). Furthermore, cisplatin, a chemotherapeutic drug that causes DNA damage, also induced gadd7 expression in a dose-dependent manner (Figure 1B; Supplementary Figure S1B). Similar to the induction by DNA damage, increased gadd7 expression was seen following treatment of CHO-K1 cells with growth arrest signals such as media depletion (Figure 1C; Supplementary Figure S1C) and serum reduction (Figure 1D; Supplementary Figure S1D). In these assays, Gadd45a, a well-known DNA damage-inducible transcript (Rosemary Siafakas and Richardson, 2009), was included as positive control. These results further confirm that gadd7 is induced upon various types of genotoxic and non-genotoxic stresses that elicit growth arrest.
Figure 1.
Induction of gadd7 following DNA damage and growth arrest signals in CHO-K1 cells. (A, B) Induction of gadd7 expression upon DNA damage. Exponentially growing CHO-K1 cells were treated with different doses of UV (A) or cisplatin (B). The expression of gadd7 was detected by qRT–PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are from one of three independent experiments and are represented as mean±s.d. with n=3. *P<0.05, **P<0.01 or ***P<0.001 compared with the untreated groups. (C, D) Induction of gadd7 expression upon growth arrest signals. CHO-K1 cells were treated with the two growth arrest signals, media depletion (C) and serum reduction (D). The expression of gadd7 was detected and analysed as in (A, B). **P<0.01 compared with the untreated groups.
gadd7 plays a role in regulation of the G1/S checkpoint following DNA damage
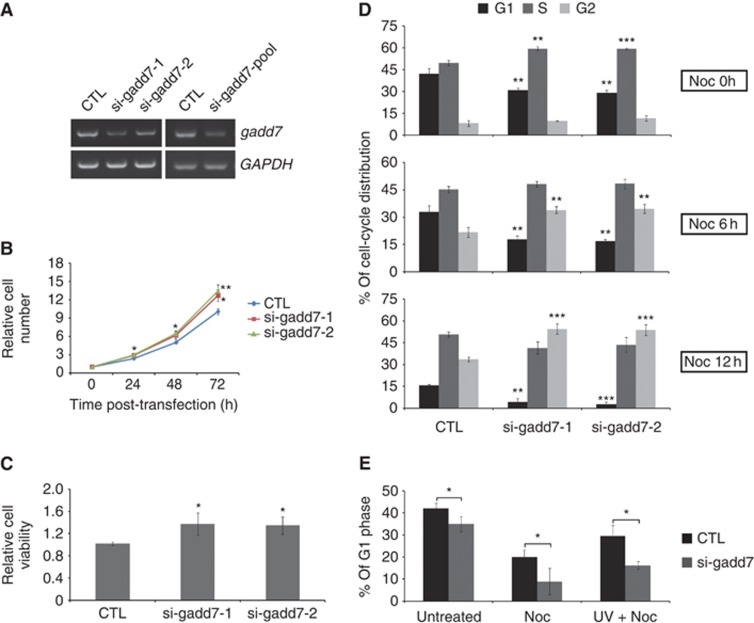
Growth arrest is a well-known and critical cellular event after DNA damage. We aimed to determine the physiological role of gadd7 in this process. First, the approach of RNA interference (RNAi) was employed to knock down endogenous gadd7 in CHO-K1 cells, and significant decrease of gadd7 expression was achieved (Figure 2A). We found that depletion of gadd7 resulted in increased cell proliferation compared with cells treated with non-specific siRNA control as measured by growth curves and MTT assay (Figure 2B and C), suggesting that gadd7 plays a negative role in the control of cell growth.
Figure 2.
Defect in the G1/S checkpoint in gadd7-depleted cells. (A) RNAi-mediated knockdown of gadd7 in CHO-K1 cells. CHO-K1 cells were transfected with control siRNA (CTL) or two individual siRNAs targeting gadd7 separately or combined. The expression of gadd7 was detected by semi-quantitative RT–PCR after transfection for 48 h. GAPDH was used as internal reference. (B, C) Knockdown of gadd7 increases cell growth. The relative numbers of CHO-K1 cells transfected with control siRNA or two individual siRNAs against gadd7 were counted daily for 3 days (B), and the relative cell viability was determined by MTT assay after transfection for 72 h (C). Data are represented as mean±s.d. from three independent experiments. *P<0.05 or **P<0.01 compared with control siRNA. (D) Depletion of gadd7 causes a rapid G1/S transition. CHO-K1 cells transfected with the indicated siRNAs for 48 h were treated with nocodazole (Noc) for 0 h (top panel), 6 h (middle panel) or 12 h (bottom panel). Cell-cycle distribution was measured by PI staining followed by flow cytometry. The percentage of cells in G1, S or G2 phase transfected with control siRNA is defined as control. Data are represented as mean±s.d. from three independent experiments. **P<0.01, ***P<0.001. (E) gadd7 knockdown results in G1/S checkpoint defect following UV irradiation. CHO-K1 cells transfected with control siRNA or gadd7 siRNA pool for 48 h were treated with nocodazole alone or in combination with UV irradiation just prior to the addition of nocodazole. After 12 h, the distribution of cell cycle was analysed as in (D). Data are represented as mean±s.d. from three independent experiments. *P<0.05.
Next we examined whether gadd7 regulates cell-cycle progression. We observed that knockdown of gadd7 led to the changes in cell-cycle distribution. The cell population in G1 phase was reduced, but that in S phase was increased after depletion of gadd7 (Figure 2D, top; Supplementary Figure S2A), suggesting that gadd7 may affect G1/S transition. To better understand the gadd7-regulatory function in G1/S transition, cell-cycle distribution analyses were further conducted in the presence of nocodazole, which blocks cells in mitosis (Zieve et al, 1980). After treatment with nocodazole for 6 h, a remarkable reduction in the G1 population and an accumulation of G2/M phase were observed in gadd7-depleted cells compared with siRNA control (Figure 2D, middle; Supplementary Figure S2A). At 12 h, nearly all gadd7-knockdown cells progressed through G1 phase and entered S and G2/M phase, while many of the cells treated with control siRNA were still in G1 phase (Figure 2D, bottom; Supplementary Figure S2A). Thus, gadd7 might be essential for the control of an orderly G1/S transition.
Since gadd7 is induced by various types of DNA damage signals, we examined the effect of gadd7 on the G1/S checkpoint in response to DNA damage. In the control cells, UV irradiation resulted in a large accumulation of the G1 population in the presence of nocodazole treatment, indicating that there is an intact G1/S checkpoint after DNA damage. In contrast, the accumulation of G1 phase was substantially reduced in cells silenced for gadd7 after UV irradiation (Figure 2E; Supplementary Figure S2B). These observations demonstrate that gadd7 is a key regulator of the G1/S checkpoint after UV irradiation. In addition, we also investigated the effect of gadd7 overexpression on G1 arrest in the absence of UV treatment. However, overexpression of gadd7 alone appeared not to generate a G1 arrest (Supplementary Figure S3A and B), suggesting that other factors induced by UV irradiation may act in concert with gadd7 to regulate the G1/S checkpoint.
gadd7 specifically binds to TDP-43
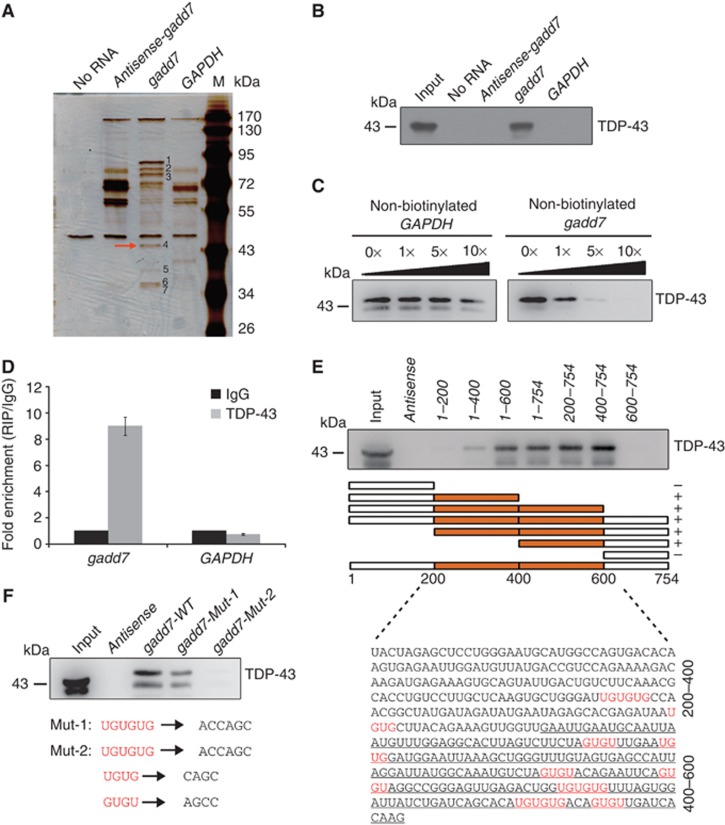
Recent studies have shown that lncRNAs perform their functions through interaction with proteins (Rinn et al, 2007; Huarte et al, 2010). Therefore, we assumed that the role of gadd7 in cell-cycle control might be possibly mediated by such a mechanism. To identify the proteins that are associated with gadd7, biotin RNA pull-down assay was performed. The biotinylated gadd7 and the two negative control transcripts, biotinylated antisense-gadd7 and biotinylated GAPDH, were transcribed in vitro (Supplementary Figure S4A) and incubated with whole CHO-K1 cell lysates. Biotin-labelled transcripts and their associated cellular proteins were captured by streptavidin beads and then subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis. The results of biotin RNA pull-down assay are presented in Figure 3A. Compared with antisense-gadd7 (line 2) and GAPDH (line 4) RNAs, there were seven bands that were specifically associated with gadd7 (line 3). The protein bands specific to gadd7 were then extracted, digested with trypsin and subjected to mass spectrometry analysis. Interestingly, this analysis showed that the majority of these proteins binding to the gadd7 transcript were hnRNPs (Supplementary Table S1). To focus on the role of gadd7 in the control of cell cycle, TAR DNA-binding protein (TDP-43) (Figure 3A, red arrow) was chosen for further studies given its function in regulating cell-cycle distribution (Ayala et al, 2008). First, the association of gadd7 with TDP-43 was validated by immunoblotting assay and the results showed that TDP-43 was clearly detected in gadd7–pull-down protein complexes, but not in the complexes associated with antisense-gadd7 or GAPDH (Figure 3B). To further confirm the specificity of this interaction, competition assay was performed by adding different amounts of non-biotinylated gadd7 or GAPDH. The interaction between gadd7 and TDP-43 was competed by non-biotinylated gadd7 in a dose-dependent manner, but not by GAPDH (Figure 3C).
Figure 3.
gadd7 directly interacts with TDP-43. (A) Identification of cellular proteins associated with gadd7 in vitro. Proteins from CHO-K1 cell extracts were pulled down with the biotin-RNAs, subjected to SDS–PAGE and visualized by silver staining. The bands specific to gadd7, as pointed out by the numbers (bands 1–7), were subjected to mass spectrometry. The TDP-43 (band 4) is indicated by the red arrow. (B) Immunoblotting analysis of the specific interaction of gadd7 with TDP-43. Proteins pulled down with the biotin-RNAs as in (A) were analysed by immunoblotting with TDP-43 antibody. (C) Specific association of gadd7 with TDP-43 is confirmed by competition assay. Various amounts of unlabelled gadd7 were added to compete with biotin-labelled gadd7 in interacting with TDP-43 (right panel). Unlabelled GAPDH RNA was used as negative control (left panel). TDP-43 associated with the biotin-gadd7 was analysed by immunoblotting. (D) gadd7 interacts with TDP-43 in vivo. TDP-43 was immunoprecipitated from CHO-K1 cells and co-precipitated RNAs were detected by qRT–PCR using primers for gadd7 and GAPDH (as negative control). IP enrichment is determined as the amount of RNA associated to TDP-43 IP relative to IgG control. Data are from one of three independent experiments and are represented as mean±s.d. with n=3. (E) Two regions in the middle of gadd7 are necessary to associate with TDP-43. A series of gadd7 deletion mutants and gadd7 antisense were used to perform RNA pull-down assay as in (A), and followed by immunoblotting with TDP-43 antibody. The regions bound by TDP-43 are indicated using the boxes shown in orange. UG/GU repeats in gadd7 transcript are shown in red. The ranges of gadd7 sequence are indicated on the right with 400–600 nt underlined. (+) binding; (−) unbinding. (F) The interaction of gadd7 with TDP-43 is GU/UG-repeat dependent. Two point mutants of gadd7 were treated as in (A) and associated TDP-43 was detected by immunoblotting. Mut-1 denotes three UGUGUG elements mutated to ACCAGC. Mut-2 denotes UGUG and GUGU repeats mutated to CAGC and AGCC, respectively, in addition to UGUGUG elements mutation. WT, wild type. Figure source data can be found with the Supplementary data.
To confirm the interaction between gadd7 and TDP-43 in vivo, RNA immunoprecipitation (RIP) was performed. An antibody against TDP-43 was incubated with CHO-K1 cell lysates, and co-precipitated RNAs were then analysed by RT–polymerase chain reaction (RT–PCR) or qRT–PCR using primers for gadd7 or GAPDH (negative control). As expected, we observed an enrichment of gadd7, but not GAPDH, with TDP-43 IP when compared with non-specific IgG IP (Figure 3D; Supplementary Figure S4B).
We then aimed to map the regions of gadd7 that are required for the interaction. For this purpose, a series of gadd7 deletion mutants, as displayed in Figure 3E, were synthesized by in vitro transcription and labelled with biotin (Supplementary Figure S4C). RNA pull-down assay was carried out and followed by immunoblotting assay to measure the interactions. We found that deletion mutants that contain the regions from nucleotides 200 to 400 or 400 to 600 were able to bind to TDP-43 (Figure 3E). Interestingly, UG/GU (UG or GU) repeats, the canonical TDP-43 target sequences (Polymenidou et al, 2011; Tollervey et al, 2011), are present in the regions from nucleotides 200 to 400 and especially in 400–600, indicating that these two regions with UG/GU repeats might be necessary for gadd7’s association with TDP-43.
To confirm the requirement of these UG/GU repeats for binding to TDP-43, two point-mutant versions, namely gadd7-Mut-1 and gadd7-Mut-2, in which three UGUGUG elements or all UG/GU repeats were mutated, respectively (Figure 3F), were transcribed in vitro for RNA pull-down experiments (Supplementary Figure S4D). The results showed that mutation of three UGUGUG elements in gadd7 RNA substantially reduced its interaction with TDP-43, and additional mutation of UGUG and GUGU sites almost completely abolished such interaction (Figure 3F), indicating that UG/GU repeats are responsible for the interaction between gadd7 and TDP-43. Collectively, all these observations indicate that gadd7 directly interacts with TDP-43 through UG/GU repeats.
The interaction between gadd7 and TDP-43 is increased following UV irradiation
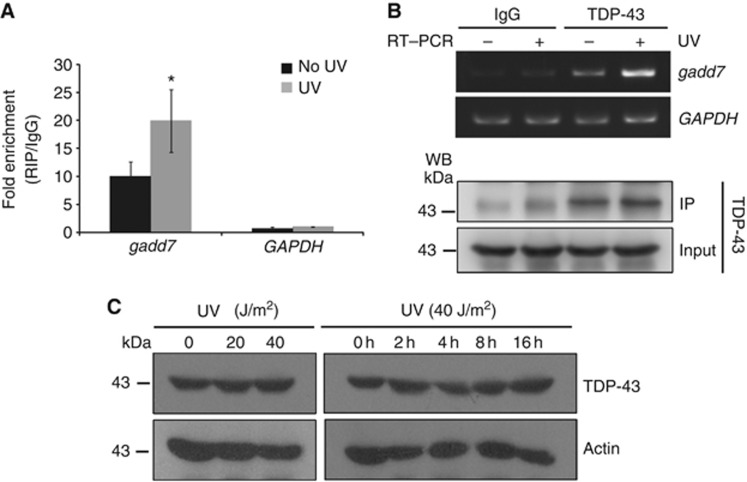
Based on the induction of gadd7 following UV irradiation, we examined the interaction between gadd7 and TDP-43 in response to UV irradiation. To this end, RIP with an antibody against TDP-43 was performed in CHO-K1 cells subjected to UV irradiation, followed by qRT–PCR or RT–PCR analysis. As expected, UV irradiation led to increased interaction between gadd7 and TDP-43, while the interaction between GAPDH and TDP-43 showed no significant change (Figure 4A and B). To exclude the possibility that increased interaction between gadd7 and TDP-43 may be the result of changed levels of TDP-43 upon UV irradiation, the expression of TDP-43 was examined. We observed that the expression of TDP-43 was unchanged in response to UV irradiation either in a dose-dependent manner or in a time course-dependent manner (Figure 4C). Together with the observation that gadd7 expression is increased after UV irradiation, these findings suggest that upon UV irradiation gadd7 is substantially induced and then binds to TDP-43.
Figure 4.
UV irradiation results in increased interaction between gadd7 and TDP-43. (A, B) Exponentially growing CHO-K1 cells subjected to UV irradiation (40 J/m2) were cultured for additional 4 h and then harvested for RIP assay with TDP-43 antibody and IgG. RNAs present in the immunoprecipitates were detected by qRT–PCR (A) or semi-quantitative RT–PCR (B, upper panel) using primers for gadd7 or GAPDH (as negative control). IP of TDP-43 was confirmed by immunoblotting using the cognate antibody (B, lower panel). IP enrichment is determined as the amount of RNA associated to TDP-43 IP relative to the corresponding IgG control. Data are from one of two independent experiments and are represented as mean±s.d. with n=3. *P<0.05 compared with the no UV-treated group. (C) The expression of TDP-43 is unchanged following UV irradiation. Exponentially growing CHO-K1 cells were treated with different doses of UV, and then harvested at the indicated times. The expression of TDP-43 and actin was detected by immunoblotting with the corresponding antibodies. Actin was used as loading control. Figure source data can be found with the Supplementary data
gadd7 and TDP-43 co-regulate Cdk6 mRNA expression
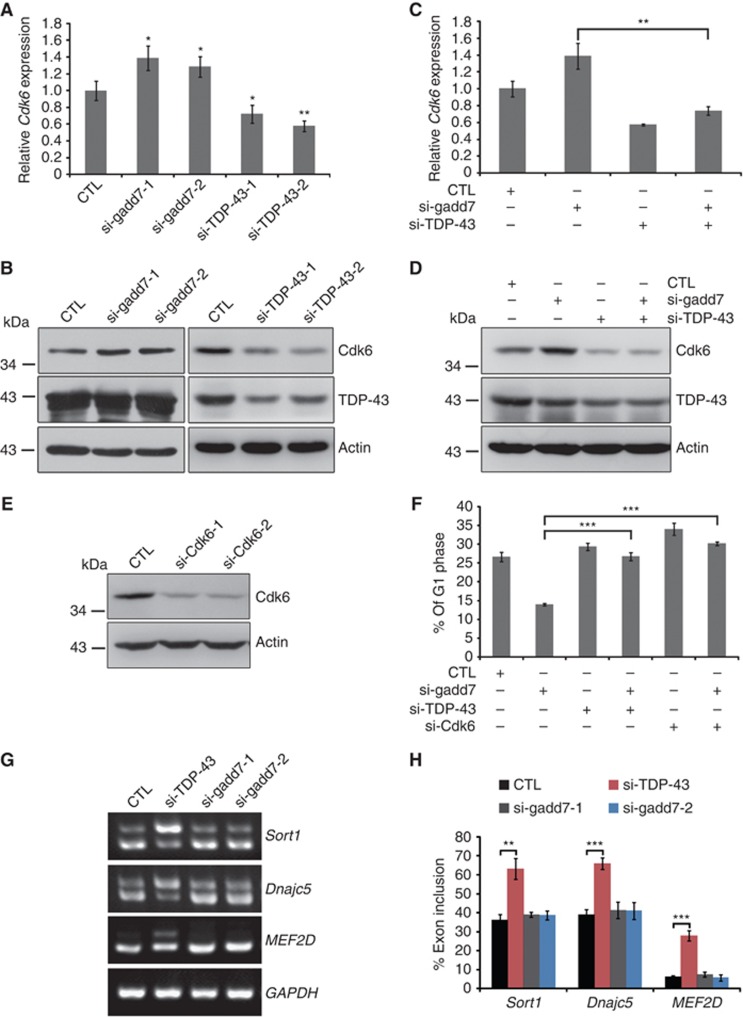
We next investigated the functional relevance of the interaction between gadd7 and TDP-43. Recent studies by Ayala et al (2008) demonstrate that TDP-43 inhibits Cdk6 expression through its recruitment to the UG-rich transcript of Cdk6 in HeLa cells. Cdk6 is known to associate with Cyclin D and regulate G1/S transition of the cell cycle (Lee and Yang, 2003). Thus, we speculated that gadd7 might be involved in the regulation of Cdk6 expression. Indeed, we observed upregulation of Cdk6 transcript and protein levels upon knockdown of gadd7 in CHO-K1 cells (Figure 5A and B). However, to our surprise, knockdown of TDP-43 in CHO-K1 cells significantly decreased the expression of Cdk6 in both RNA and protein levels (Figure 5A and B), which was not consistent with the findings of Ayala et al (2008). To further confirm the effect of TDP-43 on Cdk6 expression, we introduced the expression vector of human full-length TDP-43 into CHO-K1 cells and then examined the expression of Cdk6. Consistent with the observation that silencing of TDP-43 in CHO-K1 cells downregulates Cdk6, overexpression of TDP-43 resulted in upregulated expression of Cdk6 (Supplementary Figure S5A and B). These findings indicate that TDP-43 might act as an activator rather than a repressor in the expression of Cdk6 in CHO-K1 cells. Furthermore, simultaneous knockdown of gadd7 and TDP-43 substantially attenuated the induction of Cdk6 by gadd7 depletion at both RNA and protein levels (Figure 5C and D). Given the direct interaction of gadd7 with TDP-43 and loss of Cdk6 induction with simultaneous knockdown of gadd7 and TDP-43, we conclude that gadd7 downregulates the expression of Cdk6 mainly through its inhibition of TDP-43.
Figure 5.
The expression of Cdk6 is regulated by gadd7 and TDP-43. (A, B) Effect of knockdown of gadd7 or TDP-43 on Cdk6 mRNA and protein levels. Control siRNA (CTL) or the indicated siRNAs were transfected into CHO-K1 cells. Forty-eight hours after transfection, Cdk6 mRNA level (A) was detected by qRT–PCR and normalized to GAPDH. The protein level (B) was detected by immunoblotting and Actin was used as loading control. Data are from one of three independent experiments and are represented as mean±s.d. with n=3. *P<0.05 or **P<0.01 compared with control siRNA. (C, D) Induction of Cdk6 by gadd7 depletion was attenuated when TDP-43 was simultaneously knocked down. CHO-K1 cells were transfected with control siRNA or an siRNA pool targeting gadd7 or TDP-43 (either alone or in combination). After 48 h, Cdk6 mRNA level (C) and protein level (D) were detected as in (A) and (B), respectively. Data are from one of three independent experiments and are represented as mean±s.d. with n=3. **P<0.01. (E) Knockdown of Cdk6 in CHO-K1 by RNAi. Cdk6 expression was examined by immunoblotting. (F) Co-transfection of gadd7 siRNA with Cdk6 or TDP-43 siRNA rescues the defective G1/S checkpoint. The G1/S checkpoint of CHO-K1 cells treated with siRNAs against gadd7, Cdk6 or TDP-43, singly or together, was analysed as in Figure 2E. Data are represented as mean±s.d. from three independent experiments. ***P<0.001. (G) Effects of TDP-43 and gadd7 on alternative splicing. The indicated siRNAs were co-transfected into CHO-K1 cells. Forty-eight hours after transfection, total RNA was extracted, and splicing patterns were examined by semi-quantitative RT–PCR. (H) Quantification of inclusion rates from three independent experiments by measuring the intensity of each band shown in (G). **P<0.01, ***P<0.001. Figure source data can be found with the Supplementary data.
Since gadd7 is involved in regulation of the G1/S checkpoint, we hypothesized that this effect might be mediated through its interaction with TDP-43 and inhibition of Cdk6. To this end, Cdk6 was knocked down by RNAi in CHO-K1 cells (Figure 5E), and the effect on the G1/S checkpoint of CHO-K1 cells upon co-transfection of gadd7 siRNA with either TDP-43 or Cdk6 siRNA was analysed. Clearly, defect in the G1/S checkpoint in gadd7-depleted cells was rescued by TDP-43 or Cdk6 depletion (Figure 5F).
gadd7 interferes with the interaction between TDP-43 andCdk6 mRNA
TDP-43 plays an important role in RNA processing, especially in pre-mRNA splicing, as target sequences (UG-rich sequences) are preferentially localized in long intronic regions and near splice site acceptors (Polymenidou et al, 2011; Sephton et al, 2011; Tollervey et al, 2011). Hence, several selected targets of TDP-43, such as MEF2D (Tollervey et al, 2011), Sort1 and Dnajc5 (Polymenidou et al, 2011), were used to investigate whether gadd7 is also involved in the splicing event. We found that knockdown of TDP-43 led to more inclusion of exons in MEF2D, Sort1 and Dnajc5 compared with control siRNA, while depletion of gadd7 had no effect on alternative splicing of these genes (Figure 5G and H). Since the function of TDP-43 in CFTR exon 9 alternative splicing is well characterized (Buratti and Baralle, 2001; Buratti et al, 2001), we subsequently analysed the role of gadd7 in this important event. CHO-K1 cells were co-transfected with TG13T5 minigene and siRNAs targeting TDP-43 or gadd7. Similar to what was observed in MEF2D, Sort1 and Dnajc5, knockdown of TDP-43, but not gadd7, led to considerably increased inclusion of CFTR exon 9 (Supplementary Figure S6A–C). These data demonstrate that TDP-43 is indeed a powerful splicing modulator in CHO-K1 cells, but that gadd7 has no role in regulating this process.
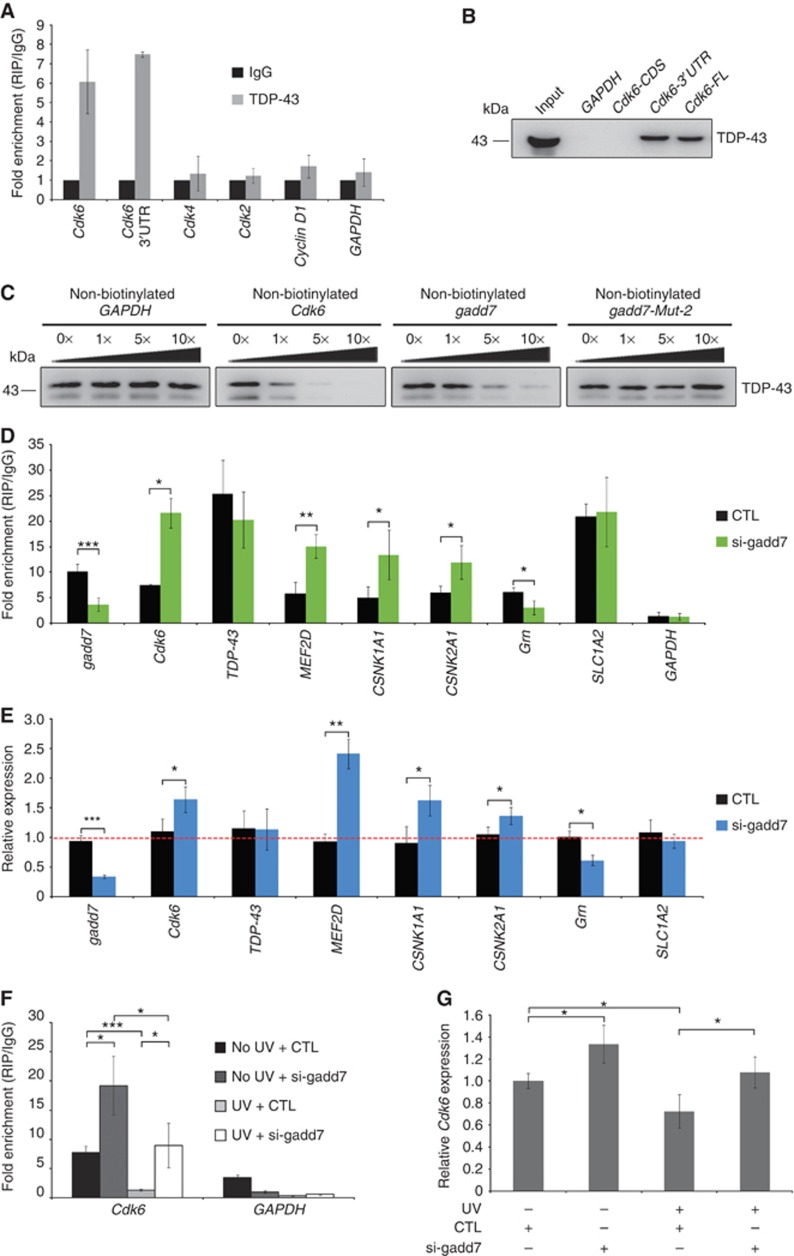
In addition to participation in pre-mRNA splicing, TDP-43 has also been implicated in mRNA decay because of its binding to target 3′UTRs (Colombrita et al, 2012). Several investigations have shown that TDP-43 stabilizes the human neurofilament (hNFL) mRNA by binding to UG motifs present on the stem loops of the 3′UTR (Strong et al, 2007; Volkening et al, 2009). UG-rich sequences are also present in the Cdk6 3′UTR and are bound by TDP-43, as determined by individual-nucleotide resolution CLIP (iCLIP)-seq (Tollervey et al, 2011). To test the hypothesis that TDP-43 modulates Cdk6 expression in a similar manner in CHO-K1 cells, we first performed RIP assay to analyse the interaction between TDP-43 and Cdk6 mRNA. TDP-43 IP specifically retrieved Cdk6 mRNA but not GAPDH or other G1 phase-related genes, such as Cdk4, Cdk2 and Cyclin D1, when compared with non-specific IgG IP (Figure 6A), indicating that TDP-43 binds to Cdk6 mRNA in CHO-K1 cells. Furthermore, to validate whether the Cdk6 3′UTR is responsible for its binding to TDP-43, RNA pull-down assay was carried out with different regions of Cdk6 mRNA: coding sequence (CDS), 3′UTR or full length (including both CDS and 3′UTR). Clearly, both 3′UTR and full length of Cdk6 were able to bind to TDP-43 strongly, but the CDS region failed to bind to TDP-43 (Figure 6B). The interaction specificity was examined by competition experiment. The binding of TDP-43 to Cdk6 mRNA was efficiently competed by increasing amounts of non-biotinylated Cdk6 mRNA (Figure 6C, second panel), but not GAPDH (Figure 6C, first panel). Taken together, these observations demonstrate that TDP-43 specifically associates with Cdk6 3′UTR in CHO-K1 cells.
Figure 6.
gadd7 abrogates the interaction between TDP-43 and Cdk6 mRNA. (A) TDP-43 binds to Cdk6 mRNA in vivo. RIP with TDP-43 antibody was performed using CHO-K1 lysates and subsequently subjected to qRT–PCR analysis for the indicated RNAs. IP enrichment is determined as the amount of RNA associated to TDP-43 IP relative to IgG control. Data are from one of three independent experiments and are represented as mean±s.d. with n=3. (B) Cdk6 3′UTR is responsible for its binding to TDP-43. RNA pull-down assay was performed with different regions of Cdk6 mRNA. FL, full length. (C) Competition assay analysis of the binding of TDP-43 to Cdk6 mRNA in the presence of various amounts of unlabelled GAPDH (first panel), Cdk6 (second panel), gadd7 (third panel) or gadd7-Mut-2 (fourth panel). (D) The binding of TDP-43 to its 3′UTR targets is affected by gadd7 loss. CHO-K1 cells transfected with control siRNA (CTL) or siRNA pool targeting gadd7 for 48 h were harvested for RIP assay with TDP-43 antibody and IgG. Binding of TDP-43 to the indicated RNA targets was determined by qRT–PCR with primers against their 3′UTRs. IP enrichment is determined as the amount of RNA associated to TDP-43 IP relative to the corresponding IgG control. Data are represented as mean±s.d. from three independent experiments. *P<0.05, **P<0.01, ***P<0.001. (E) The expression of indicated RNA targets as in (D) is changed upon gadd7 depletion as analysed by qRT–PCR. Data are from one of three independent experiments and are represented as mean±s.d. with n=3. *P<0.05, **P<0.01, ***P<0.001. (F) RIP analysis of the interaction between TDP-43 and Cdk6 mRNA upon treatment with control siRNA or siRNA pool targeting gadd7 in the presence or absence of UV irradiation (40 J/m2). Binding of TDP-43 to Cdk6 mRNA was determined by qRT–PCR. IP enrichment is determined as the amount of RNA associated to TDP-43 IP relative to the corresponding IgG control. Data are represented as mean±s.d. from three independent experiments. *P<0.05, ***P<0.001. (G) qRT–PCR quantification of Cdk6 mRNA expression in CHO-K1 cells as treated in (F). Data are from one of three independent experiments and are represented as mean±s.d. with n=3. *P<0.05. Figure source data can be found with the Supplementary data.
We then sought to determine whether gadd7 represses Cdk6 expression by affecting the binding of TDP-43 to Cdk6 mRNA. To this end, we first conducted RIP analysis in gadd7-depleted cells and observed that gadd7 depletion led to increased interaction between TDP-43 and Cdk6 mRNA (Figure 6D). The increased interaction with Cdk6 mRNA was not the result of increased TDP-43 levels in gadd7-knockdown cells (Figure 5B), suggesting that gadd7 antagonizes the ability of TDP-43 to bind to the Cdk6 mRNA. The gadd7 inhibitory effect is further supported by competition experiment (Figure 6C). Non-biotinylated gadd7 RNA significantly reduced the association of TDP-43 with Cdk6 mRNA in a dose-dependent manner, while gadd7-Mut-2 RNA with mutated TDP-43 binding sites failed to interfere with the binding even at a high dose.
Moreover, since large sets of putative TDP-43 RNA targets, including 3′UTR targets, have been identified recently through RIP-seq or cross-linking and immunoprecipitation (CLIP)-seq, we also selected several 3′UTR targets, such as its own 3′UTR (Ayala et al, 2011; Polymenidou et al, 2011), MEF2D 3′UTR, CSNK1A1 3′UTR, CSNK2A1 3′UTR, SLC1A2 3′UTR (Tollervey et al, 2011) and Grn 3′UTR (Colombrita et al, 2012), to examine whether gadd7 affects the binding of TDP-43 to other 3′UTRs. We observed that gadd7 depletion led to an increase in the binding of TDP-43 to most of these targets (Figure 6D), coincident with the changed expression of these targets on gadd7 depletion (Figure 6E), although the binding to its own 3′UTR or SLC1A2 3′UTR was not changed and the binding to Grn 3′UTR was reduced (Figure 6D). To avoid the interference from changed expression and re-association of mRNAs in native RIP, RIP was carried out in UV cross-linked CHO-K1 cells and normalized to transcript levels. Similar to what was observed in native RIP, depletion of gadd7 was sufficient to increase the binding of TDP-43 to most of these 3′UTRs, including Grn 3′UTR, but not to its own 3′UTR or SLC1A2 3′UTR (Supplementary Figure S7). Since RIP with UV cross-linking could avoid the re-association of mRNAs, the binding of TDP-43 to Grn should have been increased in vivo when gadd7 was knocked down.
Additionally, treatment with UV irradiation resulted in dismissal of TDP-43 from Cdk6 mRNA, whereas this dismissal was significantly attenuated after depletion of gadd7 (Figure 6F). These results correlate with the finding that the expression of Cdk6 was downregulated upon UV irradiation (Figure 6G; Supplementary Figure S8A and B), but depletion of gadd7 attenuated the reduction of Cdk6 by UV (Figure 6G). Taken together, these findings indicate that the interaction between TDP-43 and Cdk6 mRNA is disrupted by gadd7, which at least partially contributes to downregulated expression of Cdk6 upon UV irradiation.
gadd7 modulates Cdk6 mRNA decay through TDP-43
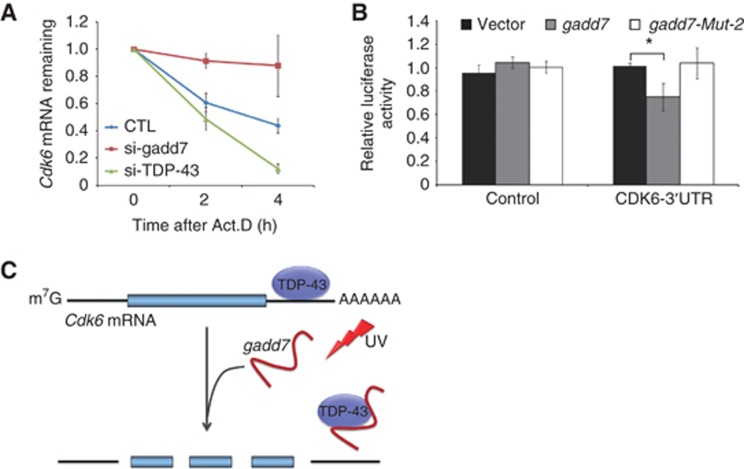
As the interaction between TDP-43 and Cdk6 3′UTR was disrupted by gadd7, we next aimed to determine whether the stability of Cdk6 mRNA is regulated by gadd7. We examined the decay of Cdk6 mRNA following treatment of CHO-K1 cells with actinomycin D, a RNA Pol II inhibitor. Depletion of TDP-43 accelerated the degradation of Cdk6 mRNA in the presence of actinomycin D relative to negative control. In contrast, depletion of gadd7 showed a pattern of much slower degradation of Cdk6 mRNA upon actinomycin D treatment (Figure 7A), indicating that gadd7 is involved in regulation of Cdk6 mRNA decay.
Figure 7.
gadd7 modulates Cdk6 mRNA decay via TDP-43. (A) Knockdown of gadd7 or TDP-43 affects Cdk6 mRNA decay. CHO-K1 cells were transfected with siRNA control (CTL) or siRNA pool targeting gadd7 or TDP-43. Forty-eight hours after transfection, cells were treated with actinomycin D (Act.D) for 0, 2 and 4 h. Total RNA was prepared and Cdk6 mRNA expression was analysed by qRT–PCR and normalized to GAPDH. The transcript remaining is defined as relative expression at the indicated times compared with the expression level at 0 h. Data are from one of three independent experiments and are represented as mean±s.d. with n=3. (B) Relative luciferase activity of the reporter containing the Cdk6 3′UTR is reduced in cells overexpressing gadd7 wild type but not mutant compared with the control vector. Relative luciferase activity is the ratio of firefly luciferase to Renilla luciferase. Data are represented as mean±s.d. from three independent experiments. *P<0.05. (C) Proposed model for the function of gadd7 in regulating Cdk6 mRNA stability. DNA damage-induced gadd7 binds to TDP-43, and inhibits TDP-43 occupancy to Cdk6 mRNA, thus leading to the instability of Cdk6 mRNA.
We further conducted luciferase reporter assay to address the role of gadd7 in the Cdk6 mRNA stability. A 2026-bp fragment of Cdk6 3′UTR containing TG repeats was cloned into the region downstream of the firefly luciferase gene of pGL3-Control vector. Both pGL3-Control vector and pGL3-Cdk6-3′UTR vector were transfected into CHO-K1 cells along with gadd7 wild-type vector (pcDNA3.1-gadd7) or mutant vector (pcDNA3.1-gadd7-Mut-2). The results showed that overexpression of gadd7 resulted in a reduction of luciferase activity, while the gadd7 mutant did not have any influence on the activity relative to the control empty vector (Figure 7B). These findings demonstrate that the effect of gadd7 on Cdk6 mRNA stability is dependent on TDP-43.
Discussion
Although lncRNAs are transcribed pervasively from mammalian genomes, the current understanding on the functions of lncRNAs is largely limited. Here we have identified a novel mechanism by which gadd7, a DNA damage-inducible lncRNA, plays an important role in regulation of the cell-cycle G1/S checkpoint in response to DNA damage. We demonstrate that following UV irradiation gadd7 is greatly induced and interacts with TDP-43, which in turn destabilizes Cdk6 mRNA and promotes the degradation of Cdk6 mRNA, thus leading to the inhibition of cell-cycle progression.
The cell-cycle G1/S checkpoint following DNA damage is an important event that maintains genomic integrity, and so disruption of the G1/S checkpoint may result in instability of the genome (Bartek and Lukas, 2001). To our knowledge, the G1/S checkpoint in the mammalian is governed by a series of protein factors, and there is little understanding on the functions of lncRNAs in the control of such an important event. In this study we find that as the lncRNA gadd7 is implicated in regulation of the G1/S checkpoint. Following CHO-K1 cells’ exposure to genotoxic stress such as UV irradiation or cisplatin treatment gadd7 is rapidly induced, while depletion of gadd7 via the RNAi approach substantially abrogates cell-cycle G1 arrest after UV treatment, indicating that gadd7 is required for regulation of the G1/S checkpoint after DNA damage. However, overexpression of gadd7 alone in the absence of UV treatment appears not to induce a G1 arrest, suggesting that the well-established G1/S checkpoint may involve not only gadd7, but also other factors that are activated by DNA damage. This observation is consistent with the previous demonstration that gadd7 overexpression alone does not increase cellular ROS levels or induce ER stress (Brookheart et al, 2009).
The mechanism by which gadd7 regulates the G1/S checkpoint after DNA damage is investigated in this study. It has been shown that lncRNAs bind to and modulate the functions of proteins (Rinn et al, 2007; Huarte et al, 2010). Through RNA pull-down assay and mass spectrometry analysis, TDP-43, an hnRNP protein, was identified to associate with gadd7. This interaction is further supported by RIP, competition assay, deletion-mapping experiment and point-mutation experiment. We find that UG/GU repeats located at the two regions in the middle of gadd7 are required for the interaction with TDP-43. We also investigated the interaction between gadd7 and TDP-43 following UV treatment. As expected, in response to UV irradiation, the interaction of gadd7 with TDP-43 was increased.
TDP-43 belongs to the family of hnRNPs, which is a multifunctional RNA/DNA-binding protein involved in transcription, splicing, mRNA transport and stability (Buratti and Baralle, 2008; Cohen et al, 2011). More importantly, recent studies have shown that dominant mutations in the TDP-43 cause a series of neurodegenerative diseases, including frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS) (Kabashi et al, 2008; Sreedharan et al, 2008). Both the capacity of RNA binding to UG repeats and the interaction with other hnRNPs through the C-terminal region are critical for the function of TDP-43 (Buratti et al, 2005; D'Ambrogio et al, 2009; Kuo et al, 2009). Until now, many targets of TDP-43 have been identified, among which Cdk6 is reported to be repressed by TDP-43 through recruitment to the UG-rich Cdk6 transcript in HeLa cells (Ayala et al, 2008). Cdk6 is a well-known member of the CDK family that is important for G1-phase progression (Lee and Yang, 2003). However, we here find that the expression of Cdk6 is activated by TDP-43, which is not consistent with the previous report. The reason for this discrepancy may be that the mechanisms involved in control of TDP-43-mediated Cdk6 expression are various in cell lines of different species and backgrounds. As is known, HeLa is a HPV-positive cancer cell line derived from human cervical cancer cells, but CHO-K1 is an immortalized cell line derived from CHO. Therefore, the mechanisms by which TDP-43 regulates Cdk6 expression may be different in the two cell lines, thus leading to different outcomes. Indeed, as reported by Ayala et al (2008), the loss of TDP-43 has no effect on Cdk6 protein levels in chicken cells (DF-1), since chicken Cdk6 lacks TG repeats completely. Although all of human, rat, mouse and hamster Cdk6 have the TG repeats, the distribution and amounts of TG repeats are quite different. Until now, to our knowledge, the regulatory effect of TDP-43 on Cdk6 expression has not been well studied in other cell lines, although the binding of TDP-43 to Cdk6 has been observed, as measured by RIP-seq or iCLIP-seq (Sephton et al, 2011; Tollervey et al, 2011). The positive role of TDP-43 in Cdk6 expression in CHO-K1 cells may be mediated by stabilizing its mRNA rather than inhibiting pre-mRNA splicing. Indeed, as proof of principle, TDP-43 binds to Cdk6 3′UTR, which leads to increase in its mRNA half-life, while gadd7 decreases the Cdk6 mRNA abundance by affecting the binding of TDP-43 to Cdk6 mRNA. Of note, the in vitro binding affinity of TDP-43 to Cdk6 is larger than to gadd7 in competition experiments, although the in vivo binding of TDP-43 to Cdk6 mRNA is weaker than to gadd7 RNA in RIP assay. The reason for this discrepancy may be due to the differences between these two kinds of experiments, and also because RNA secondary structures probably mediate the association between TDP-43 and these targets. The secondary structures of biotin-labelled RNA transcripts produced in vitro may be different from those of in vivo transcripts. Furthermore, following UV irradiation, gadd7 is substantially induced and more gadd7 binds to TDP-43, thus leading to decreased interaction between TDP-43 and Cdk6 mRNA and thus downregulation of Cdk6 expression.
In addition to Cdk6 mRNA, gadd7 could also reduce the binding of TDP-43 to some other 3′UTR targets, such as MEF2D, CSNK1A1, CSNK2A1 and Grn. However, gadd7 did not affect the interaction between TDP-43 and another 3′UTR target, SLC1A2. Furthermore, gadd7 is also not sufficient to affect the binding of TDP-43 to its own 3′UTR. TDP-43 has been reported to autoregulate its own protein level by directly binding and enhancing splicing of intron 7 in its own 3′UTR, thereby triggering degradation (Polymenidou et al, 2011; Avendano-Vazquez et al, 2012). Hence, splicing of intron 7 in its own 3′UTR is a critical event for TDP-43 autoregulation. gadd7, however, seems to have no effect on alternative splicing of TDP-43 targets as measured by splicing assay and CFTR reporter assay, which may be the reason for the unchanged binding to its own 3′UTR and expression level upon gadd7 depletion. Collectively, we conclude that gadd7 is able to affect the interactions between TDP-43 and some of its 3′UTR targets. One of the possible mechanisms by which gadd7 affects the interaction between TDP-43 and its RNA targets is by directly binding to and sequestering TDP-43 from its 3′UTR targets. Furthermore, other indirect effects may also exist. TDP-43 has extensive interactions with RNA-binding proteins (Freibaum et al, 2010; Sephton et al, 2011). The changed expression of these RNA-binding proteins or interaction with TDP-43 upon gadd7 depletion may also affect the binding of TDP-43 to its targets. Moreover, under stress, TDP-43 is localized to stress granules (SGs) (Colombrita et al, 2009; Freibaum et al, 2010; McDonald et al, 2011), which are dynamic triage centres for mRNA storage, decay or re-initiation during stress conditions (Anderson and Kedersha, 2008); thus, the localization of TDP-43 to SGs may likely contribute to the changed interaction with its RNA targets upon gadd7 depletion.
Like other lncRNAs, such as Xist and rncs-1, gadd7 is poorly conserved in sequence, since sequence searches have not revealed homologues in human or mouse species (Pang et al, 2006; Hellwig and Bass, 2008). In fact, the secondary structure of lncRNAs is more highly conserved than the nucleotide sequence (Washietl et al, 2005; Torarinsson et al, 2006). Thus, orthologues with similar secondary structure or functional elements to gadd7 may exist in humans or mice and exert their function by modulating TDP-43 function. Identification of gadd7 orthologues in humans will be critical for exploring the pathogenesis of FTLD and ALS that is correlated with TDP-43.
In summary, we demonstrate that gadd7, as the first DNA damage-inducible lncRNA, is implicated in regulation of the G1/S checkpoint. Hereby, we propose a model in which UV-induced gadd7 binds to TDP-43 and disrupts the interaction of TDP-43 with Cdk6 mRNA, thus resulting in the instability of cdk6 mRNA (Figure 7C). Our findings indicate that lncRNAs may serve as regulators by dissociating the protein–RNA interaction, thereby modulating the mRNA expression at the post-transcriptional level.
Materials and methods
Plasmids
The full-length gadd7 sequence lacking a poly A tail (on the basis of the gadd7 sequence reported as L40430 in GenBank) was amplified by PCR on the complementary DNA (cDNA) of CHO-K1 cells, and cloned into the HindIII/XbaI sites of the mammalian expression vector pcDNA3.1(+) (Invitrogen) to generate pcDNA3.1-gadd7 vector. pcDNA3.1-antisense-gadd7 was constructed by subcloning the antisense of gadd7 into XbaI/HindIII sites of the pcDNA3.1(–) vector (Invitrogen). Two mutant versions of gadd7 cDNA with HindIII/XbaI sites and three mutated TGTGTG elements or all mutated TG/GT repeats, respectively, were chemically synthesized by Invitrogen and inserted into pcDNA3.1(+) to create pcDNA3.1-gadd7-Mut-1 and pcDNA3.1-gadd7-Mut-2. A 2026-bp fragment of Cdk6 3′UTR containing TG repeats (on the basis of the Cdk6 mRNA sequence reported as XM_003496965.1 in GenBank) was PCR amplified from CHO-K1 cells’ cDNA, and cloned into the XbaI site of pGL3-Control vector (Promega) to generate pGL3-Cdk6-3′UTR vector.
Cell culture and treatment
CHO-K1 cells were purchased from the Cell Culture Center, Chinese Academy of Medical Sciences (Beijing, China). CHO-K1 cells were grown in Dulbecco’s modified Eagle’s medium/Ham’s F-12 (DMEM/F12, 1:1) medium supplemented with 5% fetal bovine serum (Gibco) and 2 mM L-glutamine at 37°C with 5% CO2. For UV treatment, exponentially growing CHO-K1 cells were rinsed with phosphate-buffered saline (PBS) and irradiated with UV radiation at different doses. After cells’ exposure to UV irradiation, fresh medium was added and the cells were cultured for 4 or 6 h till harvest. For cisplatin treatment, exponentially growing CHO-K1 cells were grown in medium containing cisplatin at different concentrations for 4 h till harvested. For media depletion, CHO-K1 cells were grown to confluence, then held without refeeding for an additional 48 h. For serum reduction, exponentially growing CHO-K1 cells were cultured with fresh DMEM/F12 media with 5% or 0.5% serum for 24 h. For actinomycin D (Serva) treatment, exponentially growing CHO-K1 cells were treated with 500 ng/ml actinomycin D for 0, 2 and 4 h. For nocodazole (Sigma) treatment, exponentially growing CHO-K1 cells were treated with 200 ng/ml nocodazole for 6 or 12 h.
RNA interference
In all, 20–30% confluent CHO-K1 cells were transfected with 50 nM of siRNAs using Lipofectamine 2000 (Invitrogen) following the manufacturer’s direction. All siRNAs were obtained from Invitrogen (25-mer duplex Stealth siRNAs for gadd7 and TDP-43; 21-mer duplex siRNA for Cdk6). The target sequences are the following: siRNA-1, 5′-GAGCACGAGAUAAUGUGCUUACAGA-3′ and siRNA-2, 5′-CCAGUGACACAAGUGAGAAUUGGAU-3′ for gadd7; siRNA-1, 5′-UGGUUCAGGUCAAGAAAGAUCUUAA-3′ and siRNA-2, 5′-CAGCGUGCAUAUAUCCAAUGCUGAA-3′ for TDP-43; siRNA-1, 5′-CCUAAAGCCACAGAACAUUTT-3′ and siRNA-2, 5′-GCAAGAGUGAUUGCAGCUUTT-3′ for Cdk6.
Viability and cell-cycle analysis
CHO-K1 cells grown in 24-well plates were transfected with 50 nM of control siRNA or siRNAs targeting gadd7. At 24, 48 and 72 h after transfection, cell numbers were counted and plotted for the growth curves. MTT absorbance of CHO-K1 cells was detected by using a microplate reader at 570 nm 72 h post transfection. For cell-cycle analysis, 48 h after transfection, cells grown in six-well plates were harvested, washed with PBS and fixed with ice-cold 70% ethanol at −20 °C overnight. Samples were then washed with PBS and stained with propidium iodide (PI) (Sigma) containing RNase A (Sigma) for 30 min at 37°C. Cell-cycle distribution was measured by flow cytometry (Becton Dickinson).
Luciferase reporter assay
CHO-K1 cells were transfected by Lipofectamine 2000 (Invitrogen) in 96-well plates with 100 ng pGL3-Control vector or pGL3-Cdk6-3′UTR vector, 2 ng pRL-CMV control vector and 100 ng pcDNA3.1-gadd7 or pcDNA3.1-gadd7-Mut-2. Forty-eight hours after transfection, cells were harvested and analysed using a dual-luciferase assay kit (Promega). Firefly activity was normalized to Renilla luciferase activity.
RNA isolation, real-time RT–PCR and semi-quantitative RT–PCR
Total RNA from CHO-K1 cells was extracted with TRIzol (Invitrogen). First-strand cDNA was synthesized by using the Superscript II-reverse transcriptase kit (Invitrogen) according to the manufacturer’s instructions. Real-time PCR (qPCR) was conducted using SYBR Premix Ex Taq (Takara) on an ABI 7300 Real-Time PCR System (Applied Biosystems). Gene-specific qPCR primer pairs are provided in Supplementary Table S2. All samples were normalized to GAPDH.
Semi-quantitative RT–PCR was conducted using the Taq polymerase (Takara). The gene-specific PCR primer pairs used are as follows: gadd7, forward 5′-GGGAAGCTGAGGTTTTTCC-3′, reverse 5′-CACACCAGTCTCAACTCCC-3′; GAPDH, forward 5′-GCTGAGAACGGGAAGCTTGT-3′, reverse 5′-GCCAGGGGTGCTAAGCAGTT-3′; Cdk6, forward 5′-GCTGAGGCACCTGGAGACCT-3′, reverse 5′-GTCCAGACCTCGGAGAAGCTG-3′. GAPDH was used as the internal control.
Immunoblotting
Cell protein lysates were prepared using 1 × PBS supplemented with 1% Nonidet P-40, 1 × protease inhibitors cocktail (Roche) and 50 μg/ml phenylmethylsulphonyl fluoride. Proteins were resolved by 10% SDS–PAGE and transferred to polyvinylidene difluoride membrane. Membranes were incubated with the indicated primary antibodies and anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (HRP). After addition of HRP substrate, the chemiluminescence signal was detected with X-ray film or Luminescent Image Analyzer LAS-4000 (Fujifilm). Antibodies used for immunoblotting are as follows: rabbit anti-TDP-43 (Proteintech Group, 10782-2-AP, 1:1000), mouse anti-Cdk6 (Santa Cruz Biotechnologies, sc-56282, 1:300), rabbit anti-Cyclin D1 (Santa Cruz Biotechnologies, sc-753, 1:500) and mouse anti-Actin (Sigma-Aldrich, A1978).
RNA pull-down assay
RNA pull-down assay was performed as described previously, with some modifications (Rinn et al, 2007). The plasmids, pcDNA3.1-gadd7/pcDNA3.1-gadd7-Mut-1/pcDNA3.1-gadd7-Mut-2/pcDNA3.1-antisense-gadd7, were linearized with the corresponding restriction enzymes used to clone the vector at the 3′, in order to prepare the template DNAs for in vitro transcription of gadd7 wild type, point mutants or antisense. Template DNAs for in vitro transcription of GAPDH, various deletion mutants of gadd7 or different fragments of Cdk6 mRNA were PCR amplified from CHO-K1 cells’ cDNA using the primers containing T7 promoter sequence in the sense primer. The primers used are listed in Supplementary Table S3.
All biotin-labelled RNA transcripts were produced in vitro using the MEGAscript T7 Kit (Ambion) with biotin-16-UTP (Ambion), and purified by MEGAclear Kit (Ambion) according to the protocol provided by the manufacturer. Ten picomoles of biotinylated RNAs was heated to 56°C for 5 min and 37°C for 10 min, and then slowly cooled to 4°C. RNAs were mixed with 500 μg of pre-cleared CHO-K1 cell extracts in binding buffer (10 mM HEPES pH 7.0, 50 mM KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100) supplemented with tRNA (0.1 μg/μl), heparin (0.5 μg/μl) and RNasin (1 unit), and incubated at room temperature for 30 min. Fifty microlitres of washed streptavidin agarose beads (Invitrogen) was added to each binding reaction and further incubated at room temperature for 15 min. Beads were washed five times with the binding buffer, and boiled in 1 × loading buffer for 10 min. The retrieved proteins were subjected to SDS–PAGE, and further visualized by silver staining or immunoblotting assay. Protein bands were excised and identified by in-gel trypsin digestion and analysed by mass spectrometry.
Native RIP and UV cross-linking RIP
Native RIP experiment was carried out with the EZ-Magna RIP Kit (Millipore) according to the manufacturer’s protocol using 5 μg of rabbit anti-TDP-43 antibody (Proteintech Group, 10782-2-AP) or rabbit IgG (Millipore). The co-precipitated RNAs were extracted with phenol:chloroform:isoamyl alcohol and detected by semi-quantitative RT–PCR or qRT–PCR. Proteins isolated from the beads were detected by immunoblotting. For UV cross-linking RIP, cells were irradiated with 100 mJ/cm2 UV light before performing RIP assay as described previously (Anko et al, 2012).
Statistical analysis
Data are presented as mean±s.d. Statistical analyses were carried out with Student’s t-tests. Differences were considered statistically significant at P<0.05.
Supplementary Material
Acknowledgments
We thank Professor Xuemin Zhang (National Center of Biomedical Analysis, Beijing, China) for mass spectrometry analysis. We also thank Professor Hongyu Hu (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) for providing the plasmids of pCMVtag2B and pCMVtag2B-TDP-43, and Professors Emanuele Buratti and Francisco E Baralle (International Center for Genetic Engineering and Biotechnology, Trieste, Italy) for the TG13T5 minigene. This work was supported by funding from the 973 National Fundamental Research Program of China (2009CB521801, 2010CB912800) and the National Natural Science Foundation of China (81021061).
Author contributions: XL, MG and QZ designed the experiments. XL, DL and WZ performed the experiments and analysed the data. XL and QZ wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson P, Kedersha N (2008) Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM (2012) The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendano-Vazquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, Baralle FE (2012) Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev 26: 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, Baralle FE (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J 30: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Misteli T, Baralle FE (2008) TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA 105: 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2001) Pathways governing G1/S transition and their response to DNA damage. FEBS Lett 490: 117–122 [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookheart RT, Michel CI, Listenberger LL, Ory DS, Schaffer JE (2009) The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J Biol Chem 284: 7446–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci 13: 867–878 [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem 280: 37572–37584 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20: 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Lee VM, Trojanowski JQ (2011) TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol Med 17: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A (2012) TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem 287: 15635–15647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem 111: 1051–1061 [DOI] [PubMed] [Google Scholar]
- D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE (2009) Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res 37: 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Meller VH (2006) Non-coding RNA in fly dosage compensation. Trends Biochem Sci 31: 526–532 [DOI] [PubMed] [Google Scholar]
- Fornace AJ Jr, Alamo I Jr., Hollander MC (1988) DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci USA 85: 8800–8804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res 9: 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, Bass BL (2008) A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci USA 105: 12897–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Alamo I, Fornace AJ Jr (1996) A novel DNA damage-inducible transcript, gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Res 24: 1589–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Rinn JL (2010) Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19: R152–R161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40: 572–574 [DOI] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS (2009) Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37: 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Yang HY (2003) Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev 22: 435–449 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5: e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 1: R17–R29 [DOI] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet 20: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS (2006) Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14: 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF (2010) Long non-coding RNAs in nervous system function and disease. Brain Res 1338: 20–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemary Siafakas A, Richardson DR (2009) Growth arrest and DNA damage-45 alpha (GADD45alpha). Int J Biochem Cell Biol 41: 986–989 [DOI] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G (2011) Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem 286: 1204–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319: 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C (2007) TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci 35: 320–327 [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J (2006) Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res 16: 885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ (2009) Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res 1305: 168–182 [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361 [DOI] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF (2005) Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390 [DOI] [PubMed] [Google Scholar]
- Zieve GW, Turnbull D, Mullins JM, McIntosh JR (1980) Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp Cell Res 126: 397–405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.