Abstract
The positioning of the nucleosome by ATP-dependent remodellers provides the fundamental chromatin environment for the regulation of diverse cellular processes acting on the underlying DNA. Recently, genome-wide nucleosome mapping has revealed more detailed information on the chromatin-remodelling factors. Here, we report that the Schizosaccharomyces pombe CHD remodeller, Hrp3, is a global regulator that drives proper nucleosome positioning and nucleosome stability. The loss of Hrp3 resulted in nucleosome perturbation across the chromosome, and the production of antisense transcripts in the hrp3Δ cells emphasized the importance of nucleosome architecture for proper transcription. Notably, perturbation of the nucleosome in hrp3 deletion mutant was also associated with destabilization of the DNA–histone interaction and cell cycle-dependent alleviation of heterochromatin silencing. Furthermore, the effect of Hrp3 in the pericentric region was found to be accomplished via a physical interaction with Swi6, and appeared to cooperate with other heterochromatin factors for gene silencing. Taken together, our data indicate that a well-positioned nucleosome by Hrp3 is important for the spatial-temporal control of transcription-associated processes.
Keywords: chromatin remodeller, heterochromatin silencing, Hrp3, non-coding transcription, nucleosome positioning
Introduction
The modulation of nucleosome positioning plays an important role in regulating gene expression by altering the access of the transcriptional machinery and histone-modifying enzymes (Wyrick et al, 1999; Jenuwein and Allis, 2001). The structure and dynamics of chromatin can be modulated through nucleosome positioning; this is mediated by ATP-dependent chromatin remodellers (Flaus and Owen-Hughes, 2001; Narlikar et al, 2002), such as members of the SNF2 family of nucleosome-stimulated ATPases (Gorbalenya and Koonin, 1993; Eisen et al, 1995; Clapier and Cairns, 2009). In Saccharomyces cerevisiae, the ISWI and CHD families, which are members of the SWI2/SNF2 class of chromatin-remodelling complexes, have been shown to regulate nucleosome positioning both in vitro (Tsukiyama et al, 1999; Lusser et al, 2005; Stockdale et al, 2006) and in vivo (Gkikopoulos et al, 2011). The gene expression of PHO5 in budding yeast is a representative example showing the regulation of gene expression by nucleosome positioning (Straka and Hörz, 1991; Martinez-Campa et al, 2004), and yChd1 was shown to be directly involved in the activation of PHO5 gene expression via remodelling of promoter nucleosomes in vivo (Ehrensberger and Kornberg, 2011). Despite similar results in in vivo and in vitro studies, mapping studies of genome-wide nucleosome positioning using chd1, isw1, and isw2 deletion strains have indicated that members of the ISWI and CHD families of chromatin-remodelling complexes possess distinct in vivo functions in terms of regulating global nucleosome positioning (Whitehouse et al, 2007; Gkikopoulos et al, 2011). However, the mechanisms that control the distinct and overlapping contributions of each remodeller to global nucleosome positioning in budding yeast are still not fully understood. Interestingly, the fission yeast Schizosaccharomyces pombe lacks members of the ISWI family but has two CHD homologues, Hrp1 and Hrp3 (Yoo et al, 2000; Jae Yoo et al, 2002). While the two remodellers show functional overlaps in sister-chromatid cohesion and mat2/3 silencing due to their sequence similarities, they also play distinct roles in chromosome segregation and heterochromatin silencing. In heterochromatic regions, Hrp1 specifically targets the centromere-associated histone H3 variant, CENP-A, to the central core region and contributes to silencing in this region (Yoo et al, 2000; Jae Yoo et al, 2002; Walfridsson et al, 2005). However, the mechanism by which Hrp3 is involved in heterochromatin silencing is poorly understood. Furthermore, although genome-wide ChIP-chip data on the roles of Hrp1 and Hrp3 are available (Walfridsson et al, 2007), global nucleosome positioning by these remodellers has not been mapped in detail.
Consistent with the relationship between nucleosome positioning and gene expression, several lines of evidence suggest that CHD is involved in regulating transcription-related processes. For instance, Drosophila dCHD1 is associated with actively transcribed regions (interband puffs) (Stokes et al, 1996); S. cerevisiae yChd1 interacts with members of the Paf1 complex, which is known to associate with RNAPII in actively transcribed regions (Simic et al, 2003); and mammalian CHD1 maintains open chromatin and pluripotency in mouse embryonic stem cells, suggesting that it is involved in transcriptional regulation (Gaspar-Maia et al, 2009). The regulation of chromatin dynamics during transcription is important for preventing aberrant transcription initiation from hidden promoters, as well as for regulating classical gene expression (Kaplan et al, 2003). Indeed, loss of Isw2 in budding yeast resulted in the production of non-coding antisense transcripts, which were generated when the nucleosome shifted to upstream of the promoter NFRs (nucleosome-free regions) (Whitehouse et al, 2007).
Here, we use genome-wide sequencing to show that the fission yeast CHD protein, Hrp3, is a global regulator of nucleosome positioning throughout the genome. Hrp3-mediated nucleosome positioning is linked to nucleosome stability and loss of Hrp3 caused perturbation of nucleosome structure and inappropriate transcription at the centromeric region and within the transcribed regions. Moreover, we found that Hrp3 appears to act directly on heterochromatin via a physical interaction with heterochromatin protein, Swi6 (known as HP1 in mammals). Since transcription of pericentric repeats during S phase of cell cycle is essential for the nucleation of heterochromatin assembly (Hall et al, 2002; Volpe et al, 2002; Chen et al, 2008), our findings indicate that Hrp3-mediated transcription-associated modulation of nucleosome structure may be required for heterochromatin silencing. Collectively, our data suggest that Hrp3 controls global chromatin structure in fission yeast, leading to gene regulation in euchromatin and heterochromatin.
Results
Hrp3 organizes nucleosome positioning within transcribed regions
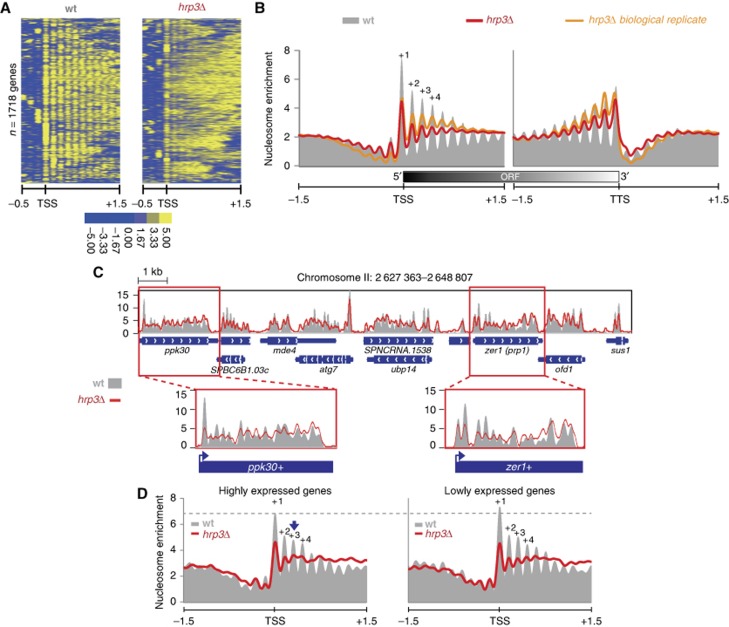
Ekwall’s group previously showed that Hrp1 and Hrp3 affect nucleosome density by cooperatively interacting with the histone chaperone, Nap1 (Walfridsson et al, 2007). While the Mi-2 type remodeller, Mit1, has been shown to be required for regular nucleosome spacing in fission yeast (Lantermann et al, 2010), the effect of Hrp1 and Hrp3 on nucleosome positioning has not been fully elucidated. To characterize Hrp1/Hrp3-mediated nucleosome positioning, we explored the nucleosome occupancy profiles of deletion mutants using MNase-seq (micrococcal nuclease digestion followed by sequencing). After MNase digestion, nucleosomal DNA fragments were isolated and subjected to paired-end sequencing. The size distributions of the sequenced fragments between samples were similar, suggesting that sequencing libraries are constructed reliably (Supplementary Figure S1). Nucleosome occupancy was estimated using the nucleR program (Flores and Orozco, 2011). The total reads in each sample were normalized using the unit called RPM (reads per million). The positions of the first and last nucleosomes within the open reading frame (ORF) of each gene were determined based on annotation of the transcription start site (TSS) and the transcription termination site (TTS), respectively (Lantermann et al, 2010). To examine global nucleosome changes, we generated a heatmap illustrating the nucleosome occupancy profiles of non-overlapping regions around 1718 TSSs, and performed manual investigations of several loci. For this analysis, we included the positioning data from mit1Δ in addition to hrp1Δ and hrp3Δ. Interestingly, the nucleosome positions in hrp3Δ were highly disrupted within genic regions, compared to hrp1Δ and mit1Δ (Figure 1A; Supplementary Figure S2A). These data suggest that Hrp3 plays an important role in determining the global nucleosome structure of fission yeast. While clear periodic enrichments of nucleosomal dyads on 3-kb regions around the TSSs and TTSs were observed in the wild-type (wt), this pattern was substantially perturbed in hrp3Δ (Figure 1B). Nucleosome occupancies around the TSSs in the mutant were reduced from the +1 nucleosome and gradually increased across the 3′ region of the TTSs, indicating that there was perturbation of the downstream nucleosomal array within transcribed regions. This indicates that Hrp3 is a key factor involved in nucleosome positioning within transcribed regions. These results were further confirmed by investigating the nucleosome positions of each mutant at the individual loci, ppk30+, zer1+, SPBC543.02C and pek1+ (Figure 1C; Supplementary Figure S2B). Consistent with the genome-wide data of nucleosome positioning, hrp3Δ also caused perturbation of positioned nucleosomes within those regions. Since changes of nucleosome structure within the ORF could be involved in transcription-coupled processes, we also checked the distribution of nucleosomal occupancy according to transcription strength (Figure 1D). In hrp3Δ, the periodicity around the TSS regions of highly expressed genes was perturbed to a higher degree than those of lower expressed genes, suggesting an involvement of Hrp3 in RNAPII-associated transcription.
Figure 1.
Loss of Hrp3 disrupts nucleosome positioning within transcribed regions. (A) A heatmap shows patterns of nucleosome positions around 2-kb regions containing 1718 non-overlapping TSSs. A colour bar represents nucleosome density. (B) A nucleosome density graph illustrates nucleosome arrangements within 3-kb regions containing the TSSs and TTSs. The y axis indicates the level of normalized nucleosome enrichment (reads per million; RPM). This result was confirmed by using identical analysis with a biological replicate (right panel). (C) The nucleosome patterns at a 21-kb region in wild type and hrp3Δ. (D) Based on the expression profiling analysis (see Materials and methods), the top 200 and bottom 200 genes were regarded as highly expressed and lower expressed genes, respectively. Nucleosomes were aligned to 3-kb regions around the TSSs of the given gene set. These figures represent a single experiment.
Hrp3 suppresses antisense RNA transcription
In S. cerevisiae, up to 85% of the genome is transcribed by RNAPII (David et al, 2006), and pervasive transcription produces extensive non-coding transcripts arising from intronic and intergenic regions (Stolc et al, 2004; Dutrow et al, 2008; Nagalakshmi et al, 2008). Nucleosomes perturbed during transcription elongation should be reorganized by chromatin-related factors (e.g., histone chaperones and nucleosome-spacing enzymes) in order to prevent transcription from cryptic promoters within transcribed region (Whitehouse et al, 2007; Cheung et al, 2008; Imbeault et al, 2008; Anderson et al, 2009). To test whether Hrp3-mediated nucleosome positioning modulates the initiation of cryptic transcripts within transcribed region, we monitored genome-wide transcript levels from both strands in hrp3Δ using the customized microarray containing 43 987 probes as described previously (Zofall et al, 2009; Yamane et al, 2011; Zhang et al, 2011). This genome-wide antisense profiling revealed that the hrp3 deletion mutant caused a significant increase in antisense transcripts at many euchromatic loci, but no detectable change in the sense transcripts (Figure 2A). Strand-specific reverse-transcription (RT)–PCR analysis confirmed the appearance of antisense RNAs at zer1+ of hrp3Δ (Figure 2B) but not hrp1Δ or mit1Δ (Supplementary Figure S3). Although Hrp1 and Hrp3 physically interact in vivo, they have distinct roles in chromosome segregation and heterochromatin silencing in central core region (Walfridsson et al, 2005). These results prompted us to investigate the genetic and biochemical differences between Hrp1 and Hrp3. Since a large number of factors that regulate transcription are known to suppress cryptic transcription (Cheung et al, 2008), we tested the effect of 6-azauracil (6-AU; a transcriptional elongation inhibitor) on hrp1Δ and hrp3Δ cells. The hrp3Δ cells exhibited hypersensitivity to 6-AU compared to hrp1Δ and wt cells, suggesting that Hrp3 is involved in transcriptional elongation (Supplementary Figure S4). To further investigate the biochemical distinction between Hrp1 and Hrp3, we purified Hrp3 proteins from double-tagged strains expressing Hrp3-TAP and Hrp1-3XFLAG (Supplementary Figure S5A). Mass spectrometry showed that only TAP-tagged Hrp3 was isolated from the separated fractions (Supplementary Figure S5B), suggesting that Hrp3 is physically and functionally separated from Hrp1. Taken together, these findings suggest that Hrp3 may control proper chromatin architecture to prevent aberrant transcription during transcriptional elongation in fission yeast.
Figure 2.
Deletion of hrp3 produces antisense transcripts. (A) Expression profiling revealed that the deletion of hrp3 caused the generation of antisense transcripts. The signal intensity (cy5/cy3) of antisense transcripts was measured using microarrays. A total of 840 genes in chromosome II were examined, and the relative positions of the genes were preserved (x axis). A colour bar represents expression levels of antisense transcripts. (B) Strand-specific RT–PCR of RNAs isolated from wild-type and hrp3Δ cells. Single primers complementary to either forward or reverse transcripts were used to synthesize the cDNAs of zer1+ and act1+ (control). (C) Mutants were clustered according to the similarity of their antisense profiles (Pearson’s correlation and average linkage method). A set of microarray data for various genes (ago1Δ, clr4Δ, clr6Δ, pht1Δ, rrp6Δ, set2Δ, and swi6Δ) was obtained from the literature (Zofall et al, 2009; Zhang et al, 2011). A colour bar represents the Pearson’s correlation coefficient. (D) Several genes in the euchromatic region (chr2:1459011–1485370) show antisense transcripts in mutants (log2 of cy5/cy3). Red arrows indicate the genes that produced antisense transcripts. A colour bar represents expression levels of antisense transcripts. (E) The levels of antisense transcripts from convergent and non-convergent genes were measured. The average signal intensity (log2 of cy5/cy3 from both sense and antisense transcripts) per gene was calculated and converted to z-scores. P-values for paired t-tests (two-tailed) comparing mutants and wild-type (wt) antisense levels are indicated (*P<0.1, **P<0.01). Microarray results were generated in a single experiment. Figure source data can be found with the Supplementary data.
Hrp3 cooperates with Set2 and Clr6 HDAC complex II to suppress the transcription of antisense RNAs at euchromatic regions
Histone deacetylation by Clr6 HDAC complex II coupled with Set2-dependent histone H3 methylation is the best characterized pathway known to suppress spurious transcription by RNAPII (Nicolas et al, 2007). More recently, heterochromatin factors such as Clr4 (known as SUV39H in mammals), Ago1, and H2A.Z have also been implicated in suppressing antisense RNAs at euchromatic loci (Nicolas et al, 2007; Zofall et al, 2009; Zhang et al, 2011). Here, we compared the distribution of antisense transcripts in the different mutants by performing hierarchical clustering according to similarities (Pearson correlation coefficients) in the genome-wide antisense profiles. As shown in Figure 2C, the hrp3Δ, set2Δ, and Clr6 HDAC complex II mutants displayed remarkable similarities in their patterns of antisense transcription. The deletions of heterochromatin factors and exosome subunit rrp6, in contrast, yielded distinctly different clusters. The findings from various individual loci (mug93, git5-1, SPBC32H8.08c, SPBC32H8.09 and mok12) in Figure 2D further supported the above observation that antisense transcripts in the hrp3 mutant were upregulated in set2Δ and Clr6 HDAC complex subunit, alp13Δ but not in rrp6Δ and clr4Δ.
Notably, while H2A.Z, Clr4, and Rrp6 prevent the generation of read-through transcripts at convergent genes (Zofall et al, 2009), Clr6 HDAC complex II prevents transcription from cryptic promoters in transcribed regions (Nicolas et al, 2007). These results were confirmed by comparing the antisense expression levels at convergent and non-convergent genes in each single mutant (Figure 2E). Consistent with previous data, pht1Δ, the gene encoding H2A.Z, and rrp6Δ showed relatively higher antisense signals at convergent genes, whereas hrp3Δ, alp13Δ, and set2Δ showed increased antisense levels at both convergent and non-convergent genes. These results suggest that Hrp3 is not responsible for transcription termination, but instead is involved in suppressing cryptic antisense generation during pervasive transcription.
To further dissect the role of Hrp3 in the suppression of antisense transcription at euchromatic loci, we combined the hrp3Δ strain with the set2Δ and alp13Δ mutants. The hrp3Δalp13Δ double mutant showed a synergistic increase in antisense RNA levels and the percentage of genes with upregulated antisense transcripts (wt (0.9%) versus hrp3Δset2Δ (21%)) (Figure 3A). The hrp3Δset2Δ double mutant also showed a synergistic increase in antisense transcripts. These findings were confirmed by showing the cumulative increase of antisense transcripts in both double mutants using strand-specific RT–qPCR for zer1+ (Figure 3B). Together, our observations indicate that Hrp3 acts in concert with the Set2 and Clr6 HDAC complex II pathway to control antisense RNA transcription. Next, we examined whether Hrp3 also affects the histone acetylation controlled by the Clr6 HDAC complex II by using chromatin immunoprecipitation (ChIP) to measure the acetylation levels of histone H3K9 and H3K14 at several loci of zer1+ (Figure 3C). In the strain lacking Hrp3, histone H3 acetylation was slightly increased in the coding region but not in the promoter. However, combining hrp3Δ with set2Δ resulted in cumulative increases in histone H3 acetylation, concomitant with elevations in the levels of antisense RNA, whereas hrp3Δalp13Δ double mutant did not show any synergistic increase. The loss of Hrp3 did not affect the occupancies of Clr6-associated HDAC complex I (Sds3) or II (Cph1) (Supplementary Figure S6). These results suggest that Hrp3, chromatin-remodelling enzyme, and Set2 and Clr6 HDAC complex II synergistically act to suppress cryptic transcription at euchromatic regions.
Figure 3.
Hrp3 suppresses antisense transcription along with Set2 and Clr6 HDAC complex II. (A) The genome-wide levels of antisense transcripts in wild-type (wt), hrp3Δ, alp13Δ, and hrp3Δalp13Δ were calculated around the ORF regions (left panel). The genes showing significant expression of antisense transcripts (average log2 ratio >1.5) were counted in each sample (right panel). (B) The expression levels of sense (left panel) or antisense transcripts (right panel) corresponding to zer1+ in the indicated strains were analysed by RT–qPCR. All values were normalized to the expression level of act1+. The graphs show the amount of transcripts in each strain relative to that in the wild type. (C) ChIP analysis was performed with antibodies against H3, H3 Lys9 acetylation (H3K9Ac), and H3 Lys14 acetylation (H3K14Ac). The precipitated chromatin was amplified by qPCR using primers encompassing the promoter region, the middle region and the 3′ region of zer1+. The signals of H3K9Ac and H3K14Ac were normalized to that of H3. The graphs in the left and right panels show the occupancies of H3K9Ac/H3 and H3K14Ac/H3, respectively, in the mutants relative to those in wild type. Error bars in (B) and (C) represent the standard deviations from three independent experiments.
Hrp3 maintains proper chromatin organization and nucleosome stability as a spacing enzyme
To ascertain the direct link between the enzymatic activity of Hrp3 and the prevention of non-coding transcription, we generated an ATPase-defective mutant of Hrp3 (hrp3K406A) harbouring a lysine-to-alanine substitution at amino acid 406 in the Walker A motif of Hrp3. The loss of ATPase activity in hrp3K406A was observed in ATPase assay (Supplementary Figure S7). As expected, the antisense RNA levels for zer1+ and sod2+ were increased in hrp3K406A (Figure 4A, right panel) relative to the sense transcripts (Figure 4A, left panel). These data suggest that Hrp3-mediated chromatin-remodelling activity is required for the prevention of aberrant transcription. We further used a micrococcal nuclease (MNase) sensitivity assay to check that the lack of Hrp3 causes defects in nucleosome organization. The bulk nucleosome ladders in hrp3Δ exhibited increased smear patterns and more rapid production of small fragments compared to wt cells (Supplementary Figure S8A). Similar phenotypes were also observed in an ATPase-defective mutant of Hrp3 (Supplementary Figure S8B), and in Southern blotting of the sod2+ gene (Supplementary Figure S9A and B).
Figure 4.
The ATPase activity of Hrp3 is required for nucleosome stability. (A) The remodelling activity of Hrp3 is important for suppressing antisense transcripts. Strand-specific RT–qPCR of the zer1+ and sod2+ genes was performed in Hrp3-3XFLAG and hrp3K406A-3XFLAG strains, as described in Figure 3B. The graphs show the amount of transcripts relative to those of Hrp3-3XFLAG. (B) Hrp3 does not affect the level of H3 occupancy in zer1+ (left panel) and sod2+ (right panel). ChIP analysis was performed with antibodies to H3 in wild type and hrp3Δ. The data represent the average of three independent experiments, and the error bars show the standard deviations. (C, D) Hrp3 affects the sensitivity of chromatin to salt-dependent disruption. Equal amounts of nuclei extracted from wild type and hrp3Δ (C), or Hrp3-3XFLAG and hrp3K406A-3XFLAG strains (D) were pelleted and resuspended in solutions of increasing NaCl concentration. The amount of insoluble H3 obtained following each salt wash was analysed by western blotting using α-H3 antibody. The graph depicts the amount of pelleted H3 normalized with respect to the amount of H3 obtained following the 0.2 M NaCl wash. Error bars indicate the standard deviations from three independent experiments. Figure source data can be found with the Supplementary data.
Sensitivity for MNase is due to either histone loss or an alteration of chromatin structure (Kaplan et al, 2003). Our ChIP experiments revealed that the MNase sensitivity in hrp3Δ was not caused by histone loss (Figure 4B), suggesting that the loss of Hrp3 may weaken the histone–DNA (or histone–histone) interactions and destabilize the nucleosome on the chromatin. To test the effect of Hrp3 on nucleosome stability, we prepared chromatin from wt and mutant cells and treated the chromatin pellets with buffers containing increasing NaCl concentrations (Chandrasekharan et al, 2009). To check the degree of salt-dependent nucleosome disruption, we measured the amount of H3 in pellet fractions resuspended with the indicated salt concentrations (0.2–2.0 M NaCl). The hrp3Δ mutant showed a dramatic loss of histone between 0.4 and 0.6 M NaCl, whereas the wt showed a distinguished loss between 0.8 and 1 M (Figure 4C, upper panel, compare lanes 1–7 between wt and hrp3Δ; also see bottom panel). These data suggest that Hrp3 plays an important role in regulating chromatin configuration by affecting nucleosome stability. As seen for hrp3Δ, the hrp3K406A mutant also showed decreased nucleosome stability (Figure 4D, upper panel, compare lanes 1–7 between wt and hrp3K406A; also see bottom panel), indicating that the ATP-dependent chromatin-remodelling activity of Hrp3 is involved in the stability of chromatin architecture.
Hrp3 controls the transcription of centromeric dg/dh repeats through an RNAi-independent pathway
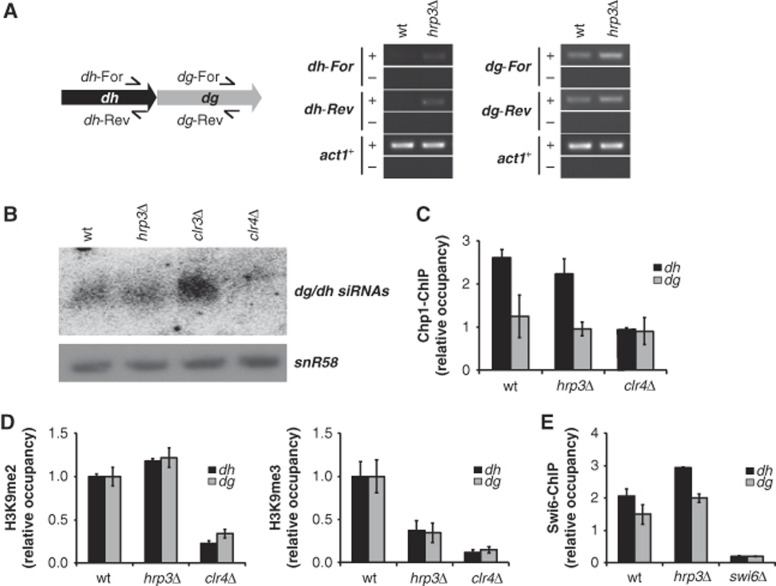
The importance of Hrp3 for stable chromatin structure and the regulation of non-coding transcription prompted us to investigate the influence of Hrp3 on heterochromatin silencing, which is another target of the Clr6 HDAC complex II in fission yeast (Nicolas et al, 2007). A previous study showed that hrp3Δ is hypersensitive to the microtubule-destabilizing drug, TBZ, providing additional evidence that Hrp3 is involved in centromeric silencing (Walfridsson et al, 2007). To examine whether Hrp3 might be involved in centromeric repeat sense/antisense transcription, the expression levels of forward and reverse dg/dh element transcripts in the hrp3 mutant were analysed by strand-specific RT–PCR. Interestingly, the hrp3 deletion mutation caused the accumulation of transcripts from both strands, whereas these transcripts were hardly detectable in wt cells (Figure 5A). In the wt strain, transcripts from pericentromeric repeats are processed into siRNAs by the RNAi machinery, which in turns target RITS and heterochromatin proteins such as Clr4 to the centromeres (Noma et al, 2004; Verdel et al, 2004; Cam et al, 2005). Accordingly, we examined whether the increase of dg/dh transcripts in the hrp3Δ mutant affects RNAi-mediated heterochromatin assembly, and found that there was no obvious defect in the siRNA production of corresponding dg/dh repeat elements in the hrp3Δ mutant (Figure 5B). This result was correlated with normal localization of the RITS Chp1 subunit at the heterochromatin (Figure 5C), suggesting that Hrp3 controls the transcription of centromeric repeats in an RNAi-independent manner. Furthermore, we performed chromatin IP against H3K9 methylation and Swi6 in hrp3Δ to check for defects in the H3K9me-HP1 platform assembly. Although hrp3Δ showed deregulation of pericentromeric transcription, there was no change in the level of H3 Lys9 di-methylation (H3K9me2). In contrast, the level of H3 Lys9 tri-methylation (H3K9me3) was significantly decreased compared to the wt (Figure 5D). H3K9me3 is enriched almost exclusively at pericentromeric regions (Peters et al, 2003; Rice et al, 2003), and reflects the significant silencing of heterochromatin genes in mouse cells (Wang et al, 2003; Yamada et al, 2005). However, since the biological meaning of H3K9me3 is not yet well understood in fission yeast, the reduction of H3K9me3 levels in hrp3Δ will warrant further study. Because HP1 family proteins such as Swi6 bind to heterochromatin by recognizing methylated H3K9 (Nakayama et al, 2001), we also tested whether Swi6 occupancy was altered in the hrp3 mutant. The occupancy of Swi6, however, appeared similar in hrp3Δ and wt cells (Figure 5E), suggesting that Hrp3 acts downstream of the recruitment of Swi6 to heterochromatin.
Figure 5.
Hrp3 controls centromeric transcripts in an RNAi-independent manner. (A) The hrp3Δ mutant showed an increase in dh/dg transcripts. Transcripts derived from the forward (dh/dg-For) or reverse (dh/dg-Rev) strand of centromeric dg and dh were determined by RT–PCR, with actin (act1+) used as a loading control. ‘+’ indicates DNA generated through reverse transcription and ‘−’ indicates control omitting the RT step. (B) Deletion of hrp3 does not decrease the siRNAs corresponding to dg and dh repeats. siRNAs from each strain were analysed by northern blot analysis with a probe specific for dg/dh sequences. (C) hrp3Δ does not affect the localization of the RITS subunit, Chp1, at dh/dg regions. ChIP experiments were performed using antibodies against Chp1. The graph shows the relative occupancy of Chp1 normalized with respect to the results obtained in the clr4Δ mutant. (D) H3K9 tri-methylation is significantly decreased in the hrp3Δ mutant. H3K9me2 and H3K9me3 levels at dh/dg regions in wild-type and hrp3Δ strains were examined by ChIP using antibodies specific for H3K9me2 and H3K9me3. The clr4Δ strain was used as a negative control. The graphs show the occupancies of H3K9me2 (left panel) and H3K9me3 (right panel) in the mutant relative to those in wild-type cells. (E) hrp3Δ does not affect the localization of Swi6 at dh/dg regions. ChIP experiments were performed using antibodies against the Swi6. The relative occupancy of Swi6 was normalized with respect to the results obtained in swi6Δ cells. The error bars in (C–E) represent the standard deviations from three independent experiments. Figure source data can be found with the Supplementary data.
Hrp3 is physically associated with heterochromatin via Swi6
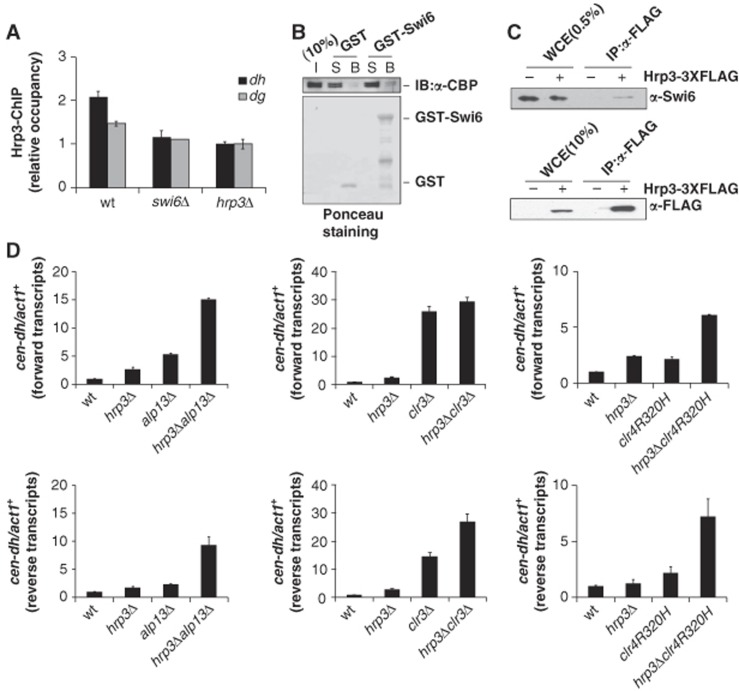
The HP1 family protein, Swi6, functions in both RNAi-mediated and RNAi-independent pathways by providing a platform for the recruitment of many proteins to heterochromatic loci, including SHREC, Clr6 HDAC complex II (Yamada et al, 2005; Nicolas et al, 2007; Sugiyama et al, 2007), and the anti-silencing factor, Epe1 (Zofall and Grewal, 2006; Isaac et al, 2007). Thus, we next investigated whether Hrp3 directly or indirectly regulates transcription of dg/dh repeat elements, and if so, whether this occurs through the Swi6 protein. First, we checked whether the localization of Hrp3 at the centromeric region was affected by the Swi6 protein. ChIP analysis against Hrp3 showed that the occupancy of Hrp3 at centromeric regions was significantly reduced in the absence of the Swi6 protein (Figure 6A). To test whether Hrp3 interacts with Swi6, we carried out GST pull-down and co-immunoprecipitation assay using Hrp3 and Swi6. To verify the interaction in vitro, TAP-tagged purified Hrp3 was mixed with GST-Swi6 or GST, and the associated proteins were subjected to western blot analysis using α-CBP antibody against known calmodulin-binding peptide (Figure 6B). Compared to background, a weak but reproducible interaction between Hrp3 and Swi6 was observed. In addition, we performed co-immunoprecipitation experiment using α-FLAG agarose resin in cells expressing FLAG-tagged Hrp3 and immunoprecipitates were western blotted with an antibody against Swi6 (Figure 6C). Consistent with these results, previous report has shown that Hrp3 copurified with Swi6 by tandem mass spectrometry (Motamedi et al, 2008). Our data collectively showed that Hrp3 binds to Swi6 in vitro and in vivo.
Figure 6.
Hrp3 acts together with other heterochromatin factors. (A) The occupancy of Hrp3 at dh/dg regions is affected by Swi6. ChIP experiments were performed using antibodies against Hrp3. The relative occupancy of Hrp3 was normalized with respect to the results obtained in hrp3Δ cells. The error bars represent the standard deviations from three independent repeats. (B) Hrp3 physically interacts with Swi6. GST-Swi6 or GST alone was incubated with TAP-purified Hrp3, and TAP-tagged Hrp3 was detected by western blotting using α-CBP antibody. The input lane (I) contains 10% of the amount of Hrp3 protein used in the binding assay. Lanes labelled (S) contain 10% of the unbound material recovered after incubation of the TAP-tagged Hrp3 with GST-Swi6 or GST. Lanes labelled (B) show the bound TAP-tagged Hrp3. Ponceau staining is shown in the bottom panel. (C) Hrp3 interacts with Swi6 in vivo. Cells carrying Hrp3-3XFLAG were extracted and precipitated with α-FLAG M2 agarose resin, and immunoprecipitated fractions were analysed by western blotting with α-Swi6 antibody. The lane labelled ‘WCE for Swi6 detection’ contained the equivalent of 0.5% of the input protein, while ‘WCE for Hrp3-3XFLAG detection’ contained 10% of the input. (D) Hrp3 acts cooperatively with other TGS factors and Clr4 to control centromeric repeat transcription. Transcripts derived from the forward (upper) or reverse (lower) strands of centromeric dh were determined with RT–qPCR. All values were normalized with respect to the expression of act1+. The error bars indicate standard deviations from three independent experiments. Figure source data can be found with the Supplementary data.
Hrp3 acts together with other heterochromatin factors to promote heterochromatin silencing
The above results suggest that Hrp3 functions directly in heterochromatin silencing through a physical interaction with Swi6. To further elucidate the influence of Hrp3 on the heterochromatin silencing pathway, we made double-deletion mutants of Hrp3 and other heterochromatin factors, and checked the centromeric repeat transcript levels in each mutant (Figure 6D). As seen in euchromatic regions, the double mutant of hrp3Δ combined with alp13Δ, a component of Clr6 HDAC complex II, showed a cumulative increase in the level of dh transcripts. In addition, the increase of dh reverse transcripts in hrp3Δclr3Δ double mutant was also synergistic compared to those of the single mutants. However, we could not detect a synergistic increase of dh forward transcripts indicating both redundant and non-redundant pathway between the two factors. Previously, the TGS (transcriptional gene-silencing) factors, Clr6 HDAC complex II, SHREC subunit Clr3, and histone H3-K9 methyltransferase Clr4 were shown to synergistically promote heterochromatin silencing (Nakayama et al, 2001; Chen et al, 2008). Here, we propose that there is also a functional interaction between Hrp3 and the Clr4 pathway in pericentromeric silencing. The double-mutant strain containing hrp3Δ and clr4R320H (partial loss-of-function mutation of clr4; Nakayama et al, 2001) showed an additive increase of dh transcript levels compared to those seen in the single-deletion mutants (Figure 6D).
To confirm the defects of heterochromatin silencing in hrp3Δ, we used a ura4+ reporter assay (harbouring the ura4+ gene in the otr1 region of the centromere) in double mutants of the hrp3 deletion mutant combined with either clr4R320H or alp13Δ. Although the hrp3Δ mutation alone had relatively little effect on the otr1R::ura4+ reporter, hrp3Δclr4R320H double mutant displayed a synergistic defect in heterochromatic silencing (Supplementary Figure S10, upper panel). Similar result was also observed in hrp3Δalp13Δ double mutants (Supplementary Figure S10, bottom panel). These data suggest that Hrp3 is required for heterochromatin silencing and the regulation of dg/dh transcription.
Hrp3 affects cell cycle-dependent transcription of centromeric repeat
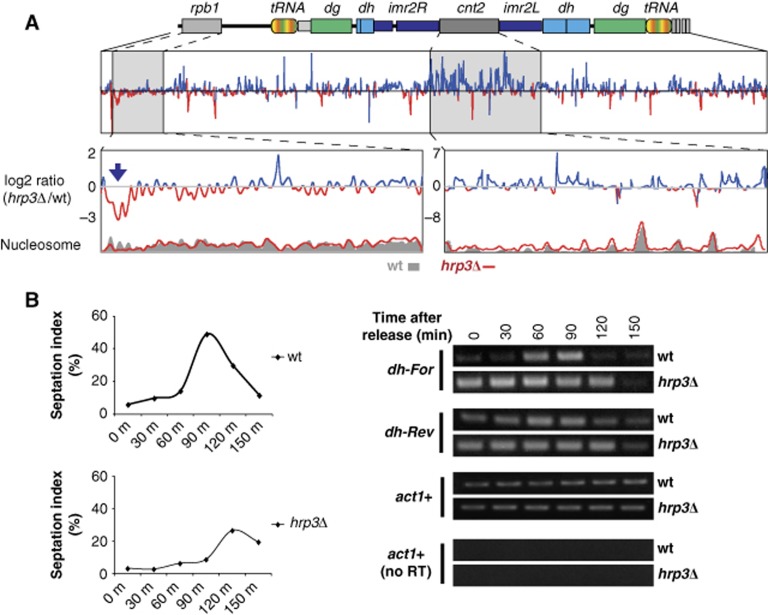
Transcription from centromeric repeats by RNAPII, which is essential for heterochromatin assembly, is dynamically regulated in cell cycle-dependent manner (Chen et al, 2008). Based on our observations that Hrp3 is involved in RNAPII-associated transcriptional elongation, including the regulation of dg/dh transcription, we investigated whether nucleosome instability induced by defects in Hrp3 affects dh transcription by RNAPII limited during S phase. The increase of dg/dh repeat transcripts in the inactive hrp3 mutant (hrp3K406A) supports our contention that the chromatin-remodelling activity of Hrp3 is required for heterochromatin silencing (Supplementary Figure S11). To investigate nucleosomes at heterochromatic regions in the absence of hrp3, we examined the centromere of chromosome II (Figure 7A; Supplementary Figure S12). Although we did not detect a drastic change of nucleosome distribution at the outer repeat (otr) region of the hrp3 deletion mutant, there were obvious changes at the innermost repeat (imr) and central core (cnt) regions (Supplementary Figure S12, bottom graph, log2(hrp3Δ/wt)). Consistent with the above findings, the nucleosome at euchromatic region adjacent to the pericentric heterochromatin, rdp1, was also perturbed in hrp3Δ. However, it is possible that we failed to detect any change at the pericentric region due to the technical limitations of nucleosome mapping in the repeat-rich sequences of the otr region. Therefore, we cannot exclude the possibility that Hrp3 may play a role in nucleosome positioning at the otr region. We also studied whether the Hrp3-induced changes in nucleosome architecture at heterochromatic regions could affect cell cycle-dependent transcription of dh elements. In wt cells, forward transcription of dh occurs through recruited RNAPII during the S phase (Chen et al, 2008); in contrast, reverse-strand transcription of centromeric repeats is not influenced by heterochromatin. We checked the expression levels of dh elements in temperature-sensitive cdc25-22 mutant cells, which are arrested at the G2/M-phase boundary under the non-permissive temperature. The progression of cell cycle after release from synchronization was confirmed by the septation index (the percentage of cells with a septum) (Kim and Huberman, 2001; Figure 7B, left panel). We found that transcripts derived from dh elements in wt cells were accumulated during the S and G2 phases. Loss of Hrp3 in the cdc25-22 background, however, resulted in persistent transcription of dh repeats throughout the cell cycle (Figure 7B, right panel), suggesting that hrp3Δ disturbs the cell-cycle control of centromeric repeat transcription. cdc25-22 hrp1Δ, however, did not cause apparent increase in transcript levels of dh repeats during cell cycle compare to wt cells (Supplementary Figures S13).
Figure 7.
hrp3Δ perturbs nucleosome position at centromeric regions, and alleviates cell cycle-dependent heterochromatin silencing. (A) The absence of Hrp3 causes nucleosome perturbation on both heterochromatic and euchromatic regions near the centromere on chromosome II. The blue arrow indicates a region showing lower nucleosome occupancy in hrp3Δ compared to wild type. The relative level of nucleosome density (log2 ratio) between hrp3Δ and wild type (wt) was calculated using a sliding window approach (50 bp). High and low occupancies of nucleosomes in hrp3Δ compared to wild type are indicated by the blue and red lines, respectively. (B) Centromeric dh repeats are constitutively transcribed in hrp3Δ. RNAs were isolated from synchronized cdc25-22 and cdc25-22 hrp3Δ cells, and transcripts corresponding to the forward (cen For) or reverse (cen Rev) strand of the centromeric dh repeat were assayed using strand-specific RT–PCR. Actin (act1+) was used as a loading control. The septation index (right panel) was used to monitor cell-cycle progression. Figure source data can be found with the Supplementary data.
Discussion
In this study, genome-wide nucleosome positioning and biochemical experiments in hrp3Δ showed that Hrp3 is responsible for maintaining overall genome-wide nucleosome positioning and stability via its ATP-dependent chromatin-remodelling activity. From the viewpoint of chromatin dynamics during transcription, the increase of non-coding RNAs in the ATPase-defective mutant of Hrp3 suggests that it might suppress cryptic transcription by re-organizing perturbed nucleosomes and governing nucleosome stability during RNAPII progression. Previous reports have shown that the histone chaperones such as Spt6 and Spt16 (Kaplan et al, 2003) and histone-modifying enzymes including Set2 and Clr6 HDAC complex II (Carrozza et al, 2005) are required to re-organize the nucleosome structure and prevent cryptic antisense transcription within intragenic regions. Our results indicate that Hrp3-mediated chromatin remodelling suppresses cryptic transcription from cryptic promoters, independent of the Set2 and Clr6 HDAC complex II pathway. Most importantly, the hrp3 deletion mutant showed perturbation of the nucleosome array throughout the genome, especially in transcribed regions, and this was correlated with the production of non-coding transcripts from ORFs. These data confirm the importance of Hrp3-mediated nucleosome positioning within transcribed regions during transcription elongation.
Surprisingly, the influence of Hrp3 on euchromatic regions extended to the heterochromatin, where Hrp3 suppressed the inappropriate transcription of centromeric repeats that serve as the centre of heterochromatin nucleation (Grewal and Klar, 1997; Hall et al, 2002). In addition to RNAi-mediated PTGS, TGS is also required for heterochromatin silencing (Volpe et al, 2002; Noma et al, 2004; Verdel et al, 2004). HP1 proteins regulate heterochromatin formation by providing a platform for the recruitment of the TGS factors, SHREC (Yamada et al, 2005; Sugiyama et al, 2007) and Clr6 HDAC complex II (Nicolas et al, 2007; Fischer et al, 2009), and the anti-silencing factor, Epe1 (Zofall and Grewal, 2006; Isaac et al, 2007; Fischer et al, 2009). In particular, the nucleosomal positioning activity of the SHREC complex (containing Clr3 and Mit1) affects the heterochromatic TGS by leading to the formation of condensed heterochromatin (Sugiyama et al, 2007). These data indicate that proper positioning of the nucleosome by chromatin remodellers is crucial to heterochromatin silencing. In addition, RNAPII-associated transcription in the pericentric region might also require re-organization of nucleosome following the wake of RNAPII transcription. Therefore, we speculate that in this process, Hrp3 creates an environment for silencing at heterochromatin by regulating chromatin architecture. The localization of Hrp3 to the pericentromeric region is maintained by a physical interaction with Swi6. Furthermore, the role of Hrp3 differs from those of other TGS factors (e.g., SHREC and Clr6 HDAC complex II, which inhibit the access of RNA polymerase II to heterochromatic repeats; Fischer et al, 2009) and the histone-modifying proteins (e.g., Set2, Alp13, and Clr4). Instead, our findings suggest that the ATP-dependent remodelling activity of Hrp3 plays a distinct role in the regulation of non-coding transcription in vivo. Taken together, the results from our study of Hrp3 provide mechanistic insights into the importance of chromatin-remodelling activity for the proper transcription of euchromatic and heterochromatic regions.
Materials and methods
Yeast strains
All yeast strains used in this study are listed in Supplementary Table S1. All deletion strains and tagged strains were constructed using standard PCR-based methods. To construct the Hrp3K406A-3XFLAG allele, a hrp3+ gene fragment containing 3XFLAG and KANMX6 was cloned into the pGEM-T Easy vector (Promega). The cloned vector was used as the template for site-directed mutagenesis by Pfu polymerase (Stratagene). The mutant allele was sequenced, PCR amplified, and introduced into the wt strain by transformation.
Expression profiling and data analysis
DNase-treated mRNA was amplified and labelled with Agilent’s Low RNA Input Linear Amplification kit PLUS. Labelled samples from mutant (Cy5) and wt (Cy3) were mixed and hybridized to the customized microarray (Agilent Custom Gene Expression 4 × 44K), as previously described (Zofall et al, 2009). The microarray contained 43 987 probes of 60-base oligonucleotides, alternately representing the plus and minus strands. The raw microarray data were extracted with the Agilent Feature Extraction Software and processed using the GeneSpring program. Probes with non-significant P-values (P-value log ratio ≥0.05) were set to 1, as previously described (Zofall et al, 2009). For comparison, a set of microarray data was obtained from the literature, representing genes such as ago1Δ (GSM432549), clr4Δ (GSM432548), clr6Δ (GSM432567), pht1Δ (GSM432542), rrp6Δ (GSM432554), set2Δ (GSM432566), and swi6Δ (GSM432559). To produce a density graph for antisense transcripts around the ORF regions (Figure 3A), each gene locus was divided into 20 bins (5 upstream, 5 downstream, and 10 for the ORF). Then, the average signal intensity (log2 of cy5/cy3) of the probes in each bin was calculated and plotted. The microarray data have been deposited in the GEO database (accession number GSE37697).
RNA extraction and strand-specific RT–PCR
RNA was purified using the previously described hot-phenol method (Schmitt et al, 1990), treated with DNase (rDNase I, Ambion), and subjected to reverse transcription (ImProm-II reverse transcription system, Promega) using primers complementary to either forward or reverse transcripts. Reverse-transcribed cDNAs were amplified by PCR or real-time PCR using gene-specific primers. A full list of the utilized primer is provided in Supplementary Table S2.
Northern blot analysis
For siRNA detection, 25 μg of total RNA was resolved on a 12.5% denaturing acrylamide gel, electrotransferred onto Hybond-N+ membranes (Amersham), and UV crosslinked. siRNAs (21 nt) were probed with 32P-labelled oligos homologous to the dg/dh repeats. As the loading control, we used an oligonucleotide homologous to a snoRNA. siRNAs corresponding to centromeric dg/dh repeats were visualized by using a Phospho-Image plate (Fuji). Loading control was visualized by exposing to Kodak film.
Chromatin immunoprecipitation
ChIP was carried out as previously described (Strahl-Bolsinger et al, 1997), with modifications. Lysates were sonicated six times (20 s on, 180 s off) using a Fisher Scientific sonic Dismembrator Model 500 sonicator at 35% output. The samples were immunoprecipitated, and then subjected to protease treatment and crosslink reversal. The DNA was then extracted with phenol/chloroform and precipitated with ethanol.
Antibodies
The α-H3K14Ac (Millipore 07-353), α-H3K9me2 (Abcam ab1220), α-H3K9me3 (Abcam ab 8898), and α-Chp1 (Abcam ab 18191) antibodies were purchased as indicated. The rabbit polyclonal α-H3, α-H3K9Ac, α-Myc, α-Swi6, α-CBP, and α-Hrp3 antibodies were produced in-house as previously described (Oh et al, 2010).
Quantitative real-time PCR
To analyse the ChIP DNA and cDNA samples, real-time qPCR analysis was performed using the CFX™96 Real-Time System (Bio-Rad).
GST pull-down and immunoprecipitation
For GST pull-down experiments, TAP-tagged Hrp3 proteins were incubated with GST or GST-Swi6. After pull-down with glutathione-sepharose (GE Healthcare), the eluted proteins were resolved by 10% SDS–PAGE and examined by western blotting using α-CBP antibody. For immunoprecipitation experiments, whole-cell lysates of FLAG-tagged Hrp3 strains were incubated with anti-FLAG M2 agarose (Sigma) at 4°C for 3 h, washed three times with NP-40 buffer, resolved by 10% SDS–PAGE, and detected using α-Swi6 antibody. FLAG-tagged Hrp3 proteins were also detected by western blotting using α-FLAG antibody (Sigma).
TAP purification
Six litres of yeast cell cultures expressing TAP-tagged Hrp3 were grown in YES medium at 30°C to an optical density of ∼1 at 600 nm. The cell pellet was resuspended in PDB 150 buffer (5 mM HEPES pH 7.4, 0.5% Triton X-100, 10% glycerol, 1 mM EDTA, and proteinase inhibitors) and disrupted in a bead beater (Biospec). The resulting crude whole-cell extracts were clarified by ultracentrifugation and directly applied to IgG sepharose resin (GE Healthcare). After a 150 mM salt wash, the resin was resuspended in 2 ml of TEV protease buffer (10 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.1% Nonidet P-40, 1 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 1 mM dithiotreitol) containing TEV protease and incubated overnight at 4°C. The cleaved, eluted Hrp3 was mixed with 8 ml calmodulin binding buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM MgCl2, 1 mM immidazole, 2 mM CaCl2, 10 mM β-mercaptoethanol, 10% glycerol), 100 μl of 1 M CaCl2, and 400 μl of calmodulin beads and incubating on rotating platform at 4°C for 3 h. Bound protein was eluted by adding calmodulin elution buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM MgCl2, 1 mM immidazole, 3 mM EGTA, 10% glycerol).
Chromatin isolation and MNase digestion
Crude chromatin was isolated from cells as previously described (Bernardi et al, 1991), with slight modifications. The pelleted chromatin was resuspended in buffer A (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 1 mM PMSF) containing 5 mM CaCl2 and 500 U MNase (NEB) at 37°C for varying durations. The samples were separated on 1.5% agarose gels and transferred onto a Nylon membrane (Amersham). The membranes were sequentially probed with random-primed fragments corresponding to the ORF region of the sod2+ gene.
Nucleosomal DNA preparation and data processing
Cells were suspended in 200 ml of culture medium at a density of 2 × 107 cells/ml, and then crosslinked with formaldehyde (final concentration of 1% v/v) for 15 min at room temperature. The fixed cells were subjected to chromatin isolation and MNase digestion as described above (Bernardi et al, 1991). Pelleted nuclei were digested with 600 U of MNase (NEB) at 37°C for 20 min, and samples were subjected to proteinase K treatment followed by overnight de-crosslinking at 65°C. Naked DNA was prepared from genomic DNA digested with 100 U of MNase. Mono-nucleosomal fragments were gel purified from 1.5% agarose gels run in TAE, and DNA was extracted with QIAGEN gel extraction kits and subject to paired-end sequencing using the Illumina platform. The reads (sequenced tags) were aligned with respect to the S. pombe genome sequence (EF2 assembly, http://fungi.ensembl.org/), using the Bowtie alignment program (version 0.12.7) (Langmead et al, 2009). The aligned reads from each sample were further normalized with respect to the RPM (real per million), and the data were smoothed using the Fast Fourier Transform algorithm of the nucleR program (version 1.0.0) (Flores and Orozco, 2011). The outputs were visualized using the IGV (integrative genomics viewer, www.broadinstitute.org/software/igv/). The nucleosome sequencing data have been deposited in the GEO database (accession number GSE40451).
Differential salt solubility assay
Nuclei were isolated as described above (in the chromatin isolation and MNase digestion section), and a differential salt assay was performed according to the previously published protocol (Bernardi et al, 1991; Chandrasekharan et al, 2009). Equal volumes of nuclei obtained from the different salt washes of wt or mutant yeast strains were subjected to western blotting using an α-H3 antibody. Immunoprecipitated proteins were detected by an ECL reagent (Millipore) and analysed with LAS-3000 (Fuji Film). Relative band intensity was quantified using the Image J-software.
Supplementary Material
Acknowledgments
This work was supported by grants from the Stem Cell Research Program (2012M 3A9B 4027953) and the Research Program for New Drug Target Discovery (2007-0052983), Ministry of Education, Science and Technology, South Korea. This work was also funded by the KRIBB/KRCF Research Initiative Program (NAP).
Author contributions: YSS and YC performed the experimental analyses using biological materials. KK undertook the genome-wide analysis. SO purified the proteins using TAP purification. KC analysed the purified proteins by mass spectrometry. JL undertook computational analysis. SISG provided the strains. YSS and YC wrote the manuscript. DL supervised the study. SISG and DL edited the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK (2009) The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol Cell Biol 29: 5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F, Koller T, Thoma F (1991) The ade6 gene of the fission yeast Schizosaccharomyces pombe has the same chromatin structure in the chromosome and in plasmids. Yeast 7: 547–558 [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS (2005) Comprehensive analysis of heterochromatin-and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chandrasekharan M, Huang F, Sun Z (2009) Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA 106: 16686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F (2008) Chromatin-and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR (2008) Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet 40: 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrensberger AH, Kornberg RD (2011) Isolation of an activator-dependent, promoter-specific chromatin remodeling factor. Proc Natl Acad Sci USA 108: 10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SIS (2009) Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA 106: 8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2001) Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev 11: 148–154 [DOI] [PubMed] [Google Scholar]
- Flores O, Orozco M (2011) nucleR: a package for non-parametric nucleosome positioning. Bioinformatics 27: 2149–2150 [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason M, Heidersbach A, Ramalho-Santos J, McManus M, Plath K, Meshorer E (2009) Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T (2011) A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333: 1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol 3: 419–429 [Google Scholar]
- Grewal SIS, Klar AJS (1997) A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146: 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SIS (2002) Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Imbeault D, Gamar L, Rufiange A, Paquet E, Nourani A (2008) The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J Biol Chem 283: 27350. [DOI] [PubMed] [Google Scholar]
- Isaac S, Walfridsson J, Zohar T, Lazar D, Kahan T, Ekwall K, Cohen A (2007) Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 175: 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae Yoo E, Kyu Jang Y, Ae Lee M, Bjerling P, Bum Kim J, Ekwall K, Hyun Seong R, Dai Park S (2002) Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem Biophys Res Commun 295: 970–974 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096. [DOI] [PubMed] [Google Scholar]
- Kim SM, Huberman JA (2001) Regulation of replication timing in fission yeast. EMBO J 20: 6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Strålfors A, Yuan GC, Ekwall K, Korber P (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT (2005) Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166 [DOI] [PubMed] [Google Scholar]
- Martinez-Campa C, Politis P, Moreau JL, Kent N, Goodall J, Mellor J, Goding CR (2004) Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol Cell 15: 69–81 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJE, Li X, Gerber S, Denison C, Gygi S, Moazed D (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SIS (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, FitzGerald PC, Kobayashi R, Grewal SIS (2007) Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 14: 372–380 [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SIS (2004) RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Oh S, Jeong K, Kim H, Kwon CS, Lee D (2010) A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun 399: 512–517 [DOI] [PubMed] [Google Scholar]
- Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Schmitt M, Brown T, Trumpower B (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM (2003) Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T (2006) Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem 281: 16279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes DG, Tartof KD, Perry RP (1996) CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci USA 93: 7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE (2004) A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306: 655–660 [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83. [DOI] [PubMed] [Google Scholar]
- Straka C, Hörz W (1991) A functional role for nucleosomes in the repression of a yeast promoter. EMBO J 10: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal S (2007) SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C (1999) Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev 13: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Dai Park S, Ekwall K (2005) The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res 33: 2868–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K (2007) A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J 26: 2868–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y (2003) mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 12: 475–487 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T (2007) Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FCP, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [DOI] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185 [DOI] [PubMed] [Google Scholar]
- Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SIS (2011) Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell 41: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo EJ, Jin YH, Jang YK, Bjerling P, Tabish M, Hong SH, Ekwall K, Dai Park S (2000) Fission yeast hrp1, a chromodomain ATPase, is required for proper chromosome segregation and its overexpression interferes with chromatin condensation. Nucleic Acids Res 28: 2004–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SIS (2011) Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331: 1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SIS (2009) Histone H2A. Z c. Nature 461: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SIS (2006) Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell 22: 681–692 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.