Abstract
ChaC1 is a mammalian proapoptic protein of unknown function induced during endoplasmic reticulum stress. We show using in vivo studies and novel in vitro assays that the ChaC family of proteins function as γ-glutamyl cyclotransferases acting specifically to degrade glutathione but not other γ-glutamyl peptides. The overexpression of these proteins (but not the catalytically dead E>Q mutants) led to glutathione depletion and enhanced apoptosis in yeast. The ChaC family is conversed across all phyla and represents a new pathway for glutathione degradation in living cells, and the first cytosolic pathway for glutathione degradation in mammalian cells.
Keywords: apoptosis, ChaC1, γ-glutamyl cyclotransferases, glutathione, 5-oxoproline
Introduction
Apoptosis, or programmed cell death, is an evolutionarily conserved process that is essential for normal development and senescence of living cells. A variety of different stimuli have been shown to trigger apoptosis. For many of these stimuli, glutathione (γ-glutamyl-cysteinyl-glycine, GSH), the redox buffer of living cells, and reactive oxygen species (ROS) have a crucial role [1]. Overexpression of glutathione biosynthetic enzymes (and higher intracellular glutathione levels) protect against apoptosis induced by different stimuli and have been associated with apoptotic-resistant phenotypes in several models of apoptosis and cancerous cells [2, 3]. Conversely, glutathione depletion is found as an early hallmark of apoptosis [4]. Apoptosis execution by glutathione depletion occurs as a result of ROS generation and the consequent downstream cascade, but can also occur in an ROS-independent manner [5, 6].
Endoplasmic reticulum (ER) stress is a cellular response to disturbances in ER function and is critical for cellular survival. However, chronic or persistent ER stress moves from a prosurvival phase to a propapoptotic phase, the latter leading to cell death via apoptosis, and is an important factor in many neurodegenerative diseases, as well as diabetes, atherosclerosis and renal disease. Although the exact mechanisms of ER stress-induced apoptosis are still not clear, it is found that both the IREI- and PERK (IRI/ASK1/JNK and PERK/ATF4/CHOP)-initiated signalling cascades of the unfolded protein response (UPR) are involved in apoptosis induction [7]. However, evidences suggest that upregulation of the transcription factor CHOP (C/EBP homologue protein) during ER stress (mediated by ATF4) is a main link between ER stress and apoptosis [8]. CHOP overexpression has been associated with upregulation of Bim (a proapoptotic protein) and downregulation of Bcl2, a major antiapoptotic protein [9, 10]. CHOP overexpression also leads to ERO1 induction and consequently to hyperoxidation of the ER as well as release of calcium stores [11, 12]. Cytoplasmic hyperoxidation and glutathione depletion have also been observed as a consequence of ER stress and is another potential trigger of apoptosis [10]. Glutathione depletion in the cell principally occurs by the oxidation of GSH to GSSG, the conjugation of GSH to electrophiles or by plasma membrane efflux of GSH [13–15]. However, the exact mechanism of CHOP-mediated glutathione depletion is not known [10]. Surprisingly, cytosolic glutathione degradation, a potential arm of glutathione depletion, has never been found to deplete glutathione.
Recently, a novel proapoptotic protein ChaC1, of unknown function, was identified in mammalian cells. ChaC1 localized to the cytosol, and was under the regulation of CHOP during the UPR of ER stress [16]. The ChaC1 protein was also identified as a prognostic marker in certain types of cancer although in these cells a role in apoptosis could not be established [17]. Homologues of the protein are found across phyla. In Escherichia coli, chaC is a gene of the cha operon (Ca++/H+ antiporter) and is assumed to be a possible regulator of this operon [18]. The homologue in Saccharomyces cerevisiae, YER163c, is also listed as a protein of unknown function.
In this manuscript, we demonstrate that members of the ChaC family contain the BtrG/γ-glutamyl cyclotransferase (γ-GCT) fold and belong to the family of γ-GCTs. Using in vivo growth assays in yeast, and novel in vitro assays developed to investigate glutathione degradation, the preferential activity of ChaC proteins towards glutathione was revealed, which involved cleaving of the γ-glutamyl bond of glutathione to yield 5-oxoproline and cys-gly. Overpexpression of mouse wild-type (WT) ChaC1 but not its catalytically inactive mutant (E>Q) led to glutathione depletion and enhanced apoptosis in yeasts. The studies delineate the molecular function of ChaC1, and explain the proapoptotic nature of these proteins. The findings also reveal a novel pathway for glutathione degradation in living cells, and the first cytosolic pathway for glutathione degradation in mammalian systems.
Results and Discussion
The ChaC proteins belong to the γ-GCT superfamily
The ChaC1 protein and its yeast homologue YER63c that were modelled by the PHYRE server indicated the presence of a BtrG/γ-GCT fold (as also indicated in the Conserved Domain Database, supplementary Fig S1 online) that is characterized by unique beta barrels surrounded by alpha helices in these proteins [19]. When the homology models of these proteins were superimposed on the γ-GCT (C7orf24) structure they revealed very high structural similarity, despite no sequence similarity in their primary sequences (Fig 1A; supplementary Fig S2A online). Moreover, 39YGSL42 and E116 residues of ChaC1, and 13YGSL16 and E115 residues of YER163c are equivalent to the critical 22YGSN25 and E98 residues of γ-GCT that are involved in substrate binding and catalysis and superimposed very well on the corresponding residues of γ-GCT (Fig 1B; supplementary Fig S2B online).
Figure 1.
The ChaC family proteins have structure (BtrG/γ-GCT fold) and active site similar to γ-GCT/γ-GACT. The protein ChaC1 was modelled using PHYRE server. The homology model obtained was superimposed on the crystal structure of γ-GCT (C7orf24, PDB code 2PN7 and 2RBH) with the help of PyMOL molecular graphics software. (A) Superimposition of homology-modelled ChaC1 structure (cyan colour) on γ-GCT structure (yellow colour). (B) Putative active site residues of ChaC1 (39YGSL42 and E116, cyan in colour) were superimposed on corresponding active site residues of γ-GCT (22YGSN25 and E98, yellow in colour). γ-GCT residues are labelled in yellow colour. γ-GACT, γ-glutamylamine cyclotransferase; γ-GCT, γ-glutamyl cyclotransferase.
Multiple sequence alignment of the ChaC1 homologues from different phyla revealed that all the members contained the signature motif and catalytic glutamate residue confirming that the ChaC family of proteins belongs to the family of γ-GCTs (supplementary Fig S3 online). This family includes γ-GCT that acts on various γ-glutamyl amino acids to yield 5-oxoproline and amino acids [19]. γ-glutamylamine cyclotransferase (γ-GACT), another enzyme found in mammalian cells, acts on γ-glutamyl amine to yield 5-oxoproline and the amine [20] (Table 1). BtrG, a bacterial protein, acts on γ-glutamyl butirosin, an intermediate in butirosin biosynthesis, and cleaves the γ-glutamyl moiety to yield 5-oxoproline and butirosin [21]. These different members do not show any significant sequence similarity among themselves despite having a common BtrG/γ-GCT fold. However, they all produce 5-oxoproline from different γ-glutamyl compounds.
Table 1. Comparison of the BtrG/γ-GCT-fold proteins.
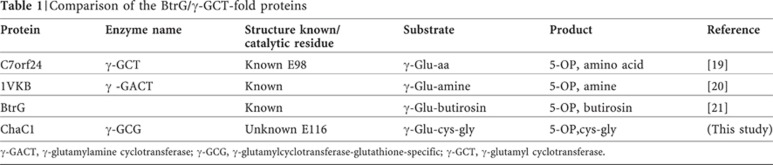
γ-GACT, γ-glutamylamine cyclotransferase; γ-GCG, γ-glutamylcyclotransferase-glutathione-specific; γ-GCT, γ-glutamyl cyclotransferase.
ChaC proteins allow utilization of glutathione in yeasts
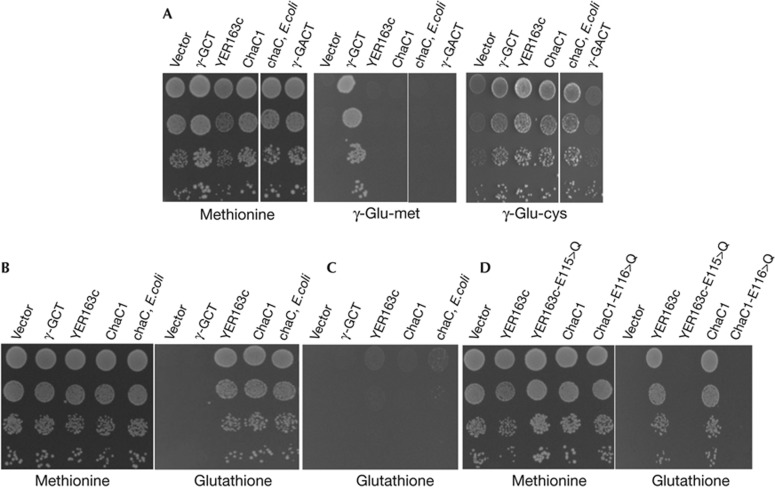
To examine if a possible γ-GCT activity might be associated with ChaC family proteins, it was necessary to identify the γ-glutamyl compounds that might be potential substrates against which this activity would be displayed. Towards this goal, we cloned ChaC1 (mouse), YER163c (S.cerevisiae) and E. coli chaC genes along with known BtrG/γ-GCT-fold proteins, γ-GCT (C7orf24) and γ-GACT genes of mouse in the yeast expression vector p426GPD. These constructs were transformed into the S.cerevisiae strain met15Δ ecm38Δ dug3Δ. This strain is an organic sulphur auxotroph owing to a met15Δ. The strain also carries a deletion in ECM38 (which encodes for γ-glutamyl transpeptidase, γ-GT) and also carries a deletion in DUG3 (which degrades glutathione by the alternative DUG pathway [22]). As a consequence of the absence of the two enzymes capable of cleaving the γ-glutamyl bond, the strain is deficient in glutathione and γ-glutamyl amino-acid utilization. This strain was used for in vivo assays to evaluate the γ-glutamyl degrading ability of the ChaC family proteins. We examined two different sulphur-containing γ-glutamyl amino acids, γ-glu-cys and γ-glu-met. As γ-GCT (C7orf24) is a known γ-GCT, transformants carrying this open reading frame could grow on both γ-glu-cys and γ glu-met. In contrast, γ-GACT failed to act on either of the γ-glutamyl dipeptides as it is specific for γ-glutamylamine (expression was confirmed by an haemagglutinin-tagged protein, data not shown), while members of the ChaC family proteins cleaved γ-glu-cys but were completely inactive against γ-glu-met (Fig 2A). Glutathione is a γ-glutamyl-tripeptide (γ-glu-cys-gly) and we examined whether glutathione was a substrate of the ChaC family using this assay. Interestingly, it was observed that strains expressing ChaC family proteins were able to grow very well on glutathione (Fig 2B). Furthermore, this was also dependant on the cys-gly peptidase (Dug1p) suggesting also that the release of cys-gly from glutathione by the action of ChaC family proteins (Fig 2C). In contrast, γ-GCT expression (C7orf24) did not lead to growth when glutathione was used as the sulphur source, (Fig 2B). The result suggests that the ChaC family proteins alone display γ-GCT activity towards glutathione.
Figure 2.
The ChaC proteins expressed in yeast act on glutathione to enable glutathione utilization in a cys-gly peptidase (Dug1p)-dependent manner. Other γ-GCTs are inactive against glutathione. ChaC family proteins encoding gene from S.cerevisiae (YER163c), mouse (ChaC1) and E. coli (chaC) along with mouse γ-GCT (C7orf24) and mouse γ-GACT were cloned in the yeast expression vector p426GPD and transformed in yeast strain ABC1723 (met15Δ ecm38Δ dug3Δ, that is organic sulphur auxotroph and deficient in glutathione and γ-glu-amino-acids utilization). The transformants were overnight grown and harvested, washed and resuspended in water and serially diluted to give 0.2, 0.02, 0.002 and 0.0002 OD600 nm of cells. A 10-μl aliquot of each dilution was spotted on minimal medium containing (A) 0.07 mg/ml methionine, 300 μM γ-glu-met and 300 μM γ-glu-cys (methionine is used as a positive control), (B) 300 μM Glutathione. (C) To examine Dug1p dependence, transformants of yeast strain ABC2295 (met15Δ, ecm38Δ dug3Δ dug1Δ that cannot cleave cys-gly as they carry a dug1Δ) were spotted on 300 μM glutathione, and failed to grow on glutathione control methionine plates not shown. (D) Putative catalytic glutamate residues of YER163c (E115) and ChaC1 (E116) were changed into Glutamine (Q) using site-directed mutagenesis. Functionality of the mutant genes was checked using glutathione as a substrate, as done above. Methionine plates are used as growth assay controls. (Photographs were taken after 3 days of growth.) γ-GACT, γ-glutamylamine cyclotransferase; γ-GCT, γ-glutamyl cyclotransferase.
To further confirm this γ-GCT activity towards glutathione, we mutated the putative catalytic glutamate residue of YER163c (E115) and ChaC1 (E116) to glutamine (E>Q). These mutations led to loss in activity against glutathione (Fig 2D) although protein expression was not altered (data not shown), demonstrating the involvement of this residue in catalysis. This suggests that the γ-GCT, γ-GACT and ChaC family proteins share similar cyclotransferase reaction mechanisms and structure, despite the differences in their protein sequences and substrate preferences.
Purified ChaC proteins have γ-GCT activity in vitro
To confirm the in vivo activities of the ChaC family proteins, recombinant ChaC1 and YER163c proteins were purified from E. coli using His-tags and Ni-NTA affinity chromatography (supplementary Fig S4 online). To identify the products of these ChaC family proteins on different γ-glutamyl amino acids (γ-glu-ala, γ-glu-cys, γ-glu-met, γ-glu-his and γ-glu-lys) and glutathione, products of the reaction mixture were analyzed by high-performance liquid chromatography (HPLC) as described in methods. The results showed that although YER163c showed some activity towards γ-glu-ala, none of the other γ-glu-amino acids examined were substrates as no new peak corresponding to 5-oxoproline appeared with these γ-glu-amino acids (supplementary Fig S5A,B online). However, when these proteins were incubated with glutathione, the peak corresponding to glutathione disappeared and new peaks corresponding to 5-oxoproline and cys-gly appeared (Fig 3). The generation of 5-oxoproline from glutathione confirmed the γ-GCT activity of the ChaC family proteins. These results revealed that ChaC proteins cleave glutathione according to the following reaction scheme (supplementary Fig S6 online),
 |
Figure 3.
The ChaC proteins exhibit γ-glutamyl cyclotransferase activity specifically towards glutathione producing 5-oxoproline and cys-gly, but do not act on γ-glutamyl amino acids to any significant extent. Ten micrograms of proteins (B) ChaC1 and (C) YER163c were incubated with 10 mM glutathione and different γ-glutamyl amino acids (γ-glu-ala, γ-glu-cys, γ-glu-met, γ-glu-his and γ-glu-lys) for 60 min. Twenty microlitres of the terminated reaction mix was analyzed using high-performance liquid chromatography system on a C18 column (250 × 4.6 mm2, Phenomenex) and a mobile phase of 2% (v/v) aqueous perchloric acid at 1.0 ml/min. Data are shown for glutathione only and data for γ-glutamyl amino acids are shown in supplementary Fig 5 online. Substrate and products of reaction mix were monitored at 210 nm. Cys-gly was detected at 5.9 min, 5-oxoproline was detected at 9 min and glutathione was detected at 11.6 min (oxidized cys-gly was detected at 7.1 min). Substrate and products were identified with authentic standards. (D) ChaC activity against glutathione and different γ-glutamyl amino acids as seen by the 5-oxoprolinase-coupled Amplex Red-based glutamate detection method. ChaC proteins of 2.5 μg were incubated with 10 mM of different γ-glu-amino acids or glutathione for 60 min. 5-oxoprolinase enzyme of 5 μgwas then added to the heat-inactivated, above reaction mix and incubated for a further 60 more minutes. This was followed by 50 μl of Amplex Red-based glutamate/glutamate oxidase assay kit solution and a further incubation for 30 more minutes. Fluorescence was measured using 544 and 584 nm excitation and emission wavelengths, respectively. Experiments were performed in triplicates. Data are means±s.e.m. Panel A refers to the Control sample (without added ChaC proteins) with glutathione as substrate.
To quantitate the relative activities of the ChaC proteins towards different γ-glu-peptides and glutathione, we measured their γ-GCT activity by 5-oxoproline (a common product of BtrG/γ-GCT-fold proteins acting on various γ-glutamyl compounds, Table 1) release. In a novel assay, the 5-oxoproline released was acted on by 5-oxoprolinase (purified from yeast) to yield glutamate that was estimated by the sensitive Amplex Red fluorometric method [23]. We observed that ChaC1 and YER163c showed barely any activity against γ-glu-dipeptides, while, in contrast, glutathione was the most preferred substrate for both the ChaC family proteins (Fig 3D). The absence of any activity on γ-glu-cys in vitro (unlike what we observed in vivo) could be owing to the fact that the growth experiments are measured over a long time and might pick up residual activity.
We also developed an alternative assay that could determine the kinetic parameters with glutathione as a substrate. This assay exploits the cys-gly generation capability of ChaC family proteins from glutathione. The cys-gly generated from glutathione by ChaC family proteins is converted into free cysteine and glycine with the help of Dug1p (a cys-gly peptidase from S.cerevisiae), followed by estimation of cysteine [24]. Using this method, we again found that glutathione but not γ-glu-cys was acted on by ChaC family proteins (supplementary Fig S7 online). The kinetic parameters revealed similar Km for both the proteins ChaC1 (3.13+0.40 mM) and YER163c (1.52+0.18 mM) although the Vmax were significantly different, ChaC1, 980+50, and YER163c, 110+7 μmoles h−1 mg per protein, respectively (supplementary Fig S8 online). More importantly, the Km for glutathione in the range between 1.5 and 3 mM reveals that the enzymatic action on glutathione is physiologically relevant.
ChaC overexpression leads to glutathione depletion
Our results revealed that glutathione is the preferred substrate of ChaC proteins. Glutathione depletion is an important factor for apoptosis initiation and execution. Moreover, ChaC1 has been shown as a proapototic factor in mammals [1, 16]. To examine if the ChaC family proteins caused glutathione depletion in vivo and lead to apoptosis, the WT and E>Q mutants were expressed in the S.cerevisiae strain met15Δ, ecm38Δ and dug3Δ. Cells expressing WT ChaC1 and YER163c led to significant depletion in glutathione despite ongoing glutathione synthesis in these cells. However, cells expressing the catalytically dead mutants ChaC1-E>Q and YER163c-E>Q did not show any glutathione depletion (Fig 4A). As these strains had ongoing glutathione synthesis, we also examined the effects in the yeast strain met15Δ, ecm38Δ, dug3Δ, gsh1Δ that is unable to synthesize glutathione owing to a deletion in GSH1. Only the mutants ChaC1-E>Q or YER163c-E>Q expressing cells of the above strain were able to grow on the plate containing 10 μM glutathione, while WT ChaC1 and YER163c expression showed highly compromised growth (Fig 4B). Addition of extracellular glutathione rescued these cells from this effect (Fig 4C). These data confirm the ability of these proteins to deplete glutathione. We subsequently evaluated the ability of ChaC1 WT and E>Q mutant to induce apoptosis in these cells. We observed a significant increase in apoptosis caused by the WT clone as compared with the mutant as seen both by Annexin V assays (Fig 4D) and TdT-mediated dUTP nick end labelling assays (Fig 4E). Furthermore, the enhanced apoptosis in yeasts could be reversed by addition of glutathione (Fig 4D,E).
Figure 4.
The ChaC proteins, but not the catalytically dead E>Q mutants, cause glutathione depletion and enhance apoptosis in yeast. (A) Glutathione was estimated in the yeast strain ABC1723 expressing ChaC family proteins (ChaC1 and YER163c) and their catalytically dead mutants (ChaC1-E>Q and YER163c-E>Q). Glutathione estimation was performed using the DTNB-glutathione reductase method. The experiment was done in triplicates. Data are mean±s.e.m. Expression of ChaC1 and YER163c leads to ∼50% and ∼40% glutathione depletion, respectively, in comparison with vector control. (B,C) The ChaC proteins cause growth retardation in a gsh1Δ background that can be reversed by addition of glutathione. Chac and their mutants were expressed in yeast strain ABC2712 (met15Δ ecm38Δ dug3Δ gsh1Δ that is a glutathione auxotroph because of the deletion in GSH1). The transformants were spotted on minimal medium containing (B) 10 μM glutathione and (C) 300 μM glutathione. (D,E) The ChaC proteins but not the E>Q mutants cause enhanced apoptosis. Yeast strain ABC2712 expressing ChaC1 and mutant ChaC1-E>Q were grown in minimal medium, with and without glutathione as indicated in the figure. Cells showing apoptosis were stained with (D) FITC-labelled annexin V for detection of exposed phosphatidylserine and analysed by flow cytometry or (E) FITC-labelled anti-5-bromodeoxyuridine Antibody Solution (TdT-mediated dUTP nick end labelling assay) in the dark for 120 min at room temperature as described in methods. Hydrogen peroxide treatment at 100 mM for 1 h was used as a positive control. FITC, fluorescein isothiocyanate.
In summary, we have characterized a new group of γ-GCTs that function specifically to degrade glutathione. The yeast homologue has been renamed as GCG1 (γ-GCT acting on glutathione). The mammalian homologue, ChaC1, is a known proapoptotic protein that is highly induced in UPR stress under the CHOP cascade [16]. As CHOP overexpression also leads to glutathione depletion and ROS generation [10], our finding fills an important gap in this cascade as it provides an explanation how expression of CHOP leads to glutathione depletion, and assists in bringing about the permissive condition for apoptosis execution. The studies described here represent a novel pathway for glutathione degradation, and the only known pathway of cytosolic glutathione degradation in higher eukaryotes, that is conserved from bacteria to mammals (supplementary Fig S9 online). As the ChaC1 protein is also significantly induced in certain cancers and have been suggested as a prognostic marker [17], they are likely to have a significant role in the physiology and redox biology of both normal and malignant cells. These findings, and the sensitive and convenient assays developed for assaying these activities will greatly facilitate our understanding of these processes in living organisms.
Methods
HPLC analysis. To study the γ-GCT activity of the ChaC proteins, 10 μg of either YER163c or ChaC1 was incubated with 10 mM γ-glutamyl-containing dipeptide (γ-glu-ala, γ-glu-cys, γ-glu-met, γ-glu-his or γ-glu-lys) or glutathione in an 100-μl reaction mix containing 50 mM Tris–HCl buffer (pH 8). The reaction mix was incubated for 1 h at 37 °C. The reactions were terminated by heat denaturation at 95 °C for 5 min. Samples were centrifuged for 30 min to remove the inactivated proteins before HPLC analysis. To analyze the products of the reaction, 20 μl of the reaction mixes were subjected to HPLC analysis using C18 HPLC column (250 × 4.6 mm, Phenomenex) with a 2% (v/v) aqueous solution of perchloric acid used as the mobile phase with 1 ml/min flow rate. Peaks of different γ-glu-dipeptides, cys-gly, glutathione and 5-oxoproline were monitored at 210 nM. Substrate and products were identified with authentic standards. Reaction product peaks corresponding to 5-oxoproline and cys-gly appeared at 9 min and 5.9 min, respectively. A third peak (between 5-oxoproline and cys-gly peaks), at 7.1 min appeared in reaction mix, was identified as oxidized cys-gly, and appeared even if standard cys-gly was incubated in Tris buffer (pH 8) for prolonged periods.
5-oxoprolinase-coupled γ-GCT assay for ChaC family proteins. As 5-oxoproline appeared as a common product of the ChaC protein’s action on γ-glutamyl compounds, a 5-oxoprolinase-coupled, extremely sensitive, 5-oxoproline detection-based γ-GCT assay was developed. Ten millimolar of γ-glutamyl-containing dipeptide (γ-glu-ala, γ-glu-cys, γ-glu-met, γ-glu-his or γ-glu-lys) or glutathione was incubated with 2.5 μg of YER163c or ChaC1 protein for 60 min at 37 °C in 50 μl of reaction mixture containing 50 mM Tris–HCl (pH 8). The reaction mixture was then placed at 95 °C for 5 min to inactivate the enzyme. After inactivation, 10 μl of the 5-oxoprolinase reaction mix was added. This mix typically consists of 5 mM ATP, 10 mM MgCl2, 150 mM KCl and 5 μg of yeast 5-oxoprolinase, and enables the conversion of released 5-oxoproline to glutamate. Incubation was carried out for 1 h at 37 °C. After this, 50 μl of the glutamate/glutamate oxidase Amplex Red assay kit solution (Invitrogen) was added to the reaction mixture and incubated for 30 more minutes at 37 °C. The reaction mixture was then transferred to 96-well plates and the fluorescence was measured using a fluorescence microplate reader with an excitation at 544 nm and an emission at 584 nm. Controls lacking the ChaC proteins were treated similarly. The assay was performed in triplicates and data are means±s.e.m.
Cys-gly peptidase (Dug1p)-coupled γ-GCT assay for ChaC family proteins Cys-gly is a product when glutathione is acted on by the ChaC proteins. As 5-oxoprolinase purified from yeast is very labile, a cys-gly peptidase-coupled, sensitive and convenient spectrophotometric assay was also developed to study the kinetic parameters of the ChaC proteins. In this assay, 2.5 μg of YER163c or ChaC1 protein was incubated with glutathione for 30 min at 37 °C in 50 μl of reaction mixture containing 50 mM Tris–HCl (pH 8). The reaction mixture was then placed at 95 °C for 5 min to inactivate the enzyme. After inactivation, 10 μl of cys-gly peptidase reaction mixture, that will convert cys-gly dipeptide into free cysteine and glycine, was added. The reaction mixture consisted of 20 μM MnCl2 and 5 μg of Dug1p, a cys-gly peptidase. This reaction mixture was further incubated for 1 h at 37 °C. The free cysteine generated was measured by acidic ninhydrin. In brief, after the incubation, 40 μl of water was added to the above reaction mixture to make up the volume to 100 μl. Thereafter, 100 μl acetic acid and 100 μl of acidic ninhydrin solution (20 g ninhydrin dissolved in acetic acid and HCl that are mixed in 3:2 ratio) were added to the reaction mixture and the reaction mixture boiled in a water bath for 10 min to develop the pink colour. Two hundred microlitres coloured solutions were transferred in 96-well enzyme-linked immunosorbent assay (ELISA) plates and the OD was taken at 560 nm using ELISA reader. Controls containing everything except the ChaC protein were treated similarly. Cysteine liberated from the γ-glu-cys by ChaC family proteins activity was measured by acidic ninhydrin-based cysteine detection method and do not need to be treated by cys-gly peptidase (Dug1p). The assay is performed in triplicates and data are means±s.e.m.
To study the kinetic parameters of the ChaC proteins towards glutathione, the cys-gly peptidase (Dug1p)-coupled method was used. For the kinetic experiments, concentration of glutathione ranging from 0.25 to 15 mM were incubated with protein YER163c (1 μg), and ChaC1 (300 ng) was used in 50 μl of reaction mixture, and reactions performed as described above. The Km and Vmax data are means±s.e.m. (n=5 independent experiments)
Total glutathione estimation. ChaC1, YER163c and their mutants ChaC1-E>Q and YER163c-E>Q were expressed in the yeast strain ABC1723, using the yeast expression vector p426GPD. Cells were grown overnight, reinoculated in fresh medium and further grown for 8 h. 10 OD600 of cells expressing each construct were lysed in 800 μl of 5% sulphosalicylic acid using glass bead lysis. Cells were vortexed for 20 min with intervals and centrifuged for 30 min at 4 °C. Supernatant was taken for glutathione estimation. Glutathione estimation was done as described earlier [25]. In brief, 10 μl of supernatant was mixed with 60 μl of DTNB (2 mg/3ml) and 60 μl of glutathione reductase (2.5 unit/ml) in 96-well ELISA plates. Sixty microlitres of NADPH (2 mg/3ml) was added in the above mixtures to start the reaction. Absorbance was taken at every 15 s at 410 nm. Absorbance at 2 min was used to compare the glutathione levels in the different samples.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
A.K. was a recipient of a Research Fellowship from the Council of Scientific and Industrial Research, Government of India. This work was in part supported by a grant-in-aid-project to A.K.B. from the Department of Science and Technology, and Department of Biotechnology, Government of India.
Author contributions: A.K. and A.K.B. conceived the project, planned the experiments, analysed the results and wrote the paper. A.K. performed all the experiments relating to the cloning, assays and enzymology. Amandeep K. performed the haemagglutinin tagging of the proteins and the western blots; S.M., Shantanu S., Sagar S. and S.T. planned the experiments on apoptosis; and S.M. and S.T. executed the experiments on apoptosis.
Footnotes
The authors declare that they have no conflict of interest.
References
- Franco R, Cidlowski JA (2009) Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ 16: 1303–1314 [DOI] [PubMed] [Google Scholar]
- Botta D, Franklin CC, White CC, Krejsa CM, Dabrowski MJ, Pierce RH, Fausto N, Kavanagh TJ (2004) Glutamate-cysteine ligase attenuates TNF-induced mitochondrial injury and apoptosis. Free Radic Biol Med 37: 632–642 [DOI] [PubMed] [Google Scholar]
- Friesen C, Kiess Y, Debatin KM (2004) A critical role of glutathione in determining apoptosis sensitivity and resistance in leukemia cells. Cell Death Differ 11Suppl 1: S73–S85 [DOI] [PubMed] [Google Scholar]
- Circu ML, Aw TY (2008) Glutathione and apoptosis. Free Radic Res 42: 689–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JS, Steinauer KK, Hornung B, Irish JM, Lecane P, Birrell GW, Peehl DM, Knox SJ (2002) Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ 9: 252–263 [DOI] [PubMed] [Google Scholar]
- Franco R, Panayiotidis MI, Cidlowski JA (2007) Glutathione depletion is necessary for apoptosis in lymphoid cells independent of reactive oxygen species formation. J Biol Chem 282: 30452–30465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 13: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11: 381–389 [DOI] [PubMed] [Google Scholar]
- Puthalakath H et al. (2007) ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129: 1337–1349 [DOI] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ (2001) Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol 21: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18: 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins JM et al. (2009) Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J Clin Invest 119: 2925–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair IA (2006) Endogenous glutathione adducts. Curr Drug Metab 7: 853–872 [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT (2005) Metals, toxicity and oxidative stress. Curr Med Chem 12: 1161–1208 [DOI] [PubMed] [Google Scholar]
- Franco R, Cidlowski JA (2006) SLCO/OATP-like transport of glutathione in FasL-induced apoptosis: glutathione efflux is coupled to an organic anion exchange and is necessary for the progression of the execution phase of apoptosis. J Biol Chem 281: 29542–29557 [DOI] [PubMed] [Google Scholar]
- Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel G et al. (2012) Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br J Cancer 106: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey DM, Guffanti AA, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich TA (1993) Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J Biol Chem 268: 11296–11303 [PubMed] [Google Scholar]
- Oakley AJ, Yamada T, Liu D, Coggan M, Clark AG, Board PG (2008) The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle. J Biol Chem 283: 22031–22042 [DOI] [PubMed] [Google Scholar]
- Oakley AJ, Coggan M, Board PG (2010) Identification and characterization of gamma-glutamylamine cyclotransferase, an enzyme responsible for gamma-glutamyl-epsilon-lysine catabolism. J Biol Chem 285: 9642–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn NM, Li Y, Spencer JB (2007) Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem Biol 14: 379–386 [DOI] [PubMed] [Google Scholar]
- Kaur H, Ganguli D, Bachhawat AK. (2012) Glutathione degradation by the alternative pathway (DUG pathway) in Saccharomyces cerevisiae is initiated by (Dug2p-Dug3p)2 complex, a novel glutamine amidotransferase (GATase) enzyme acting on glutathione. J Biol Chem 287: 8920–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bachhawat AK (2010) OXP1/YKL215c encodes an ATP-dependent 5-oxoprolinase in Saccharomyces cerevisiae: functional characterization, domain structure and identification of actin-like ATP-binding motifs in eukaryotic 5-oxoprolinases. FEMS Yeast Res 10: 394–401 [DOI] [PubMed] [Google Scholar]
- Kaur H, Kumar C, Junot C, Toledano MB, Bachhawat AK (2009) Dug1p Is a Cys-Gly peptidase of the gamma-glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys-Gly peptidases. J Biol Chem 284: 14493–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman I, Kode A, Biswas SK (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc 1: 3159–3165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.