Abstract
Overexpression of ERBB2 or ERBB3 is associated with cancer development and poor prognosis. In this study, we show that reactive oxygen species (ROS) induce both ERBB2 and ERBB3 expression in vitro and in vivo. We also identify that miR-199a and miR-125b target ERBB2 and/or ERBB3 in ovarian cancer cells, and demonstrate that ROS inhibit miR-199a and miR-125b expression through increasing the promoter methylation of the miR-199a and miR-125b genes by DNA methyltransferase 1. These findings reveal that ERBB2 and ERBB3 expression is regulated by ROS via miR-199a and miR-125b downregulation and DNA hypermethylation.
Keywords: miR-199a, miR-125b, ERBB2/3, ROS, DNA methylation
Introduction
Many human cancers of epithelial origin are associated with the overexpression of epidermal growth factor receptor family [1, 2]. In particular, ERBB2 and ERBB3 overexpression is linked with cancer development, poor prognosis and drug resistance [3–5]. Except ERBB2/ERBB3 gene amplification, other mechanism(s) of ERBB2 and ERBB3 overexpression remains elusive.
Reactive oxygen species (ROS), including superoxide, hydroxyl radical and hydrogen peroxide, are naturally produced by cells through aerobic metabolism, and mediate a plethora of intracellular signal transduction events [6]. There is a growing interest in ROS biology and cancer. Numerous studies have supported the role of ROS in promoting tumour development [7, 8]. In the context of ovarian cancer, we previously found that ovarian cancer cells spontaneously produce excessive ROS when compared with immortalized ovarian epithelial cells [9]. However, the mechanism by which ROS induce tumour growth remains to be elucidated.
The discovery of microRNAs (miRNAs) helps to reveal new mechanisms of some gene expression. miRNAs are single-stranded RNAs, which are 18–25 nucleotides in length, that regulate gene expression at the post-transcriptional levels through either translation inhibition or the degradation of specific target messenger RNA [10]. The alterations of miRNA profiles are observed in many human cancers with implication in tumour development and growth [11, 12]. Here, we show that ERBB2 and ERBB3 are regulated by ROS in cancer cells and tumour tissues, and reveal a new mechanism of ERBB2 and ERBB3 induction by ROS through miR-199a and miR-125b repression and the DNA hypermethylation via DNA methyltransferase 1 (DNMT1) elevation.
Results and discussion
Endogenous ROS regulate ERBB2/ERBB3 expression
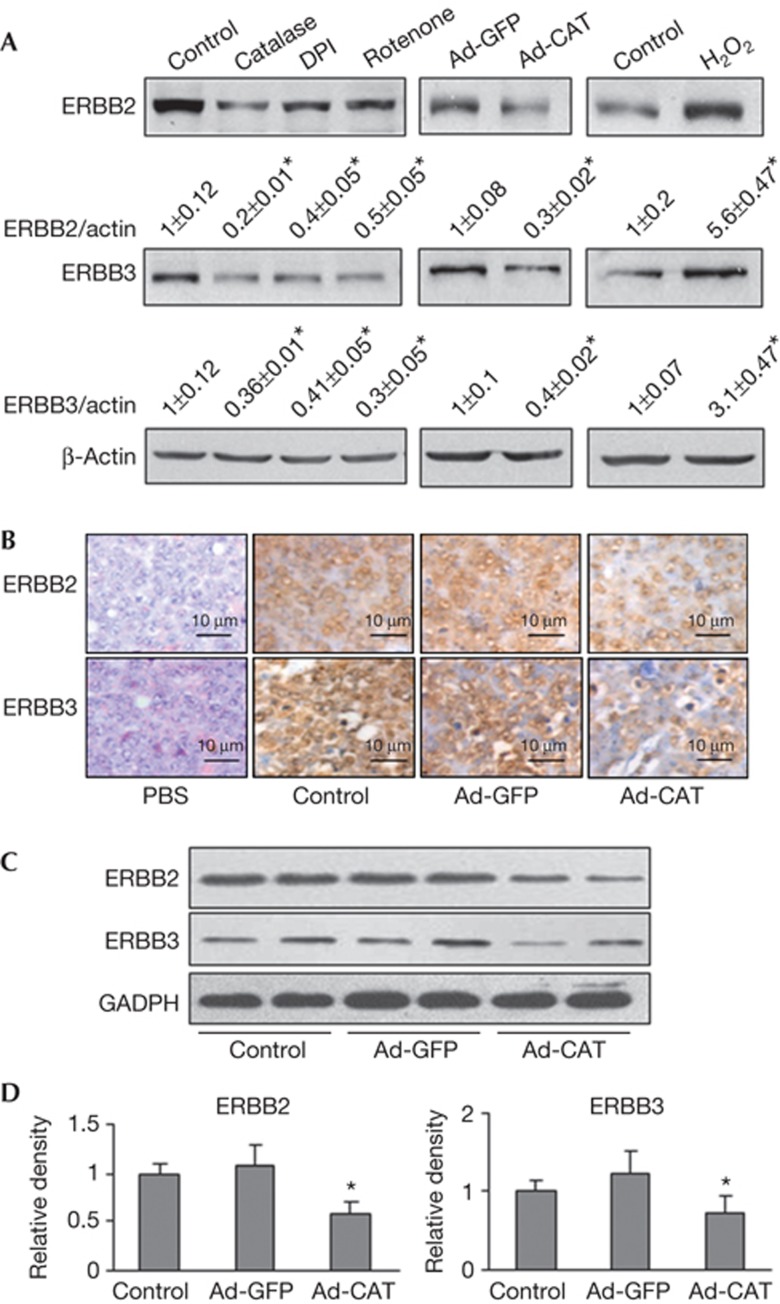
We previously reported that endogenous ROS promote tumour-induced angiogenesis via PI3K pathway [9]. To investigate whether ERBB2 and ERBB3 are regulated by endogenous ROS production, we treated OVCAR-3 cells with different ROS inhibitors: catalase (hydrogen peroxide scavenger), DPI (nicotinamide adenine dinucleotide phosphate oxidase-dependent oxidase inhibitor) and rotenone (the mitochondria complex I inhibitor), respectively. Treatments with the ROS inhibitors resulted in a substantial reduction of ROS in the cells as expected (supplementary Fig S1 online), and decreased the total protein levels of ERBB2 and ERBB3 (Fig 1A). We also infected OVCAR-3 cells with an adenovirus carrying green fluorescent protein (GFP) or catalase and obtained similar results. In addition, hydrogen peroxide treatment further increased both ERBB2 and ERBB3 protein expression levels. We next determined the effect of ROS on ERBB2 and ERBB3 expression in vivo, OVCAR-3 cells were infected with adenovirus carrying GFP or catalase, then implanted into ovaries of nude mice. The ovarian tumours were collected and weighted 30 days after implantation. Catalase treatment significantly inhibited tumour growth (supplementary Fig S2 online). The average tumour weight in the catalase group was 1.20±0.32 g (n=10), and the average weight in GFP control group was 2.03±0.39 g (n=10). The expression levels of ERBB2 and ERBB3 in tumour tissues were evaluated by immunohistochemistry and immunoblotting. The protein levels of ERBB2 and ERBB3 were reduced in tumours generated from the cells expressing catalase than the GFP control (Fig 1B–D). These findings reveal a new role of ROS for inducing ERBB2 and ERBB3 expressions both in vitro and in vivo.
Figure 1.
Reactive oxygen species induce total ERBB2 and ERBB3 expression in vitro and in vivo. (A) OVCAR-3 cells were treated with catalase (3,000 U/ml, 12 h), DPI (1 μM, 12 h), rotenone (2.5 μM, 12 h) or H2O2 (100 μM, 4 h), respectively. The cells were harvested to analyse ERBB2 and ERBB3 protein levels by immunoblotting. Similar experiment was performed when cells were infected with adenovirus carrying GFP or catalase (25 multiplicity of infection) for 60 h. (B) Orthotopic ovarian cancer model in nude mice was established as described in Supplementary Methods. Tumour sections from the nude mice were used for immunohistochemical staining with antibodies against ERBB2 and ERBB3. Representative images from each group are shown. Scale bar, 10 μm. (C) The expression levels of ERBB2 and ERBB3 in ovarian tumour tissues were analysed by immunoblotting. The representative results from each group were shown. (D) The protein levels of ERBB2 and ERBB3 above were quantified by scanning densitometry, normalized to GADPH levels and the control as mean±s.e. (n=6–8). * indicates significant decrease compared with those of the Ad-GFP group (by Student’s t-test, P<0.05). GADPH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein.
miR-199a and miR-125b are inhibited by ROS
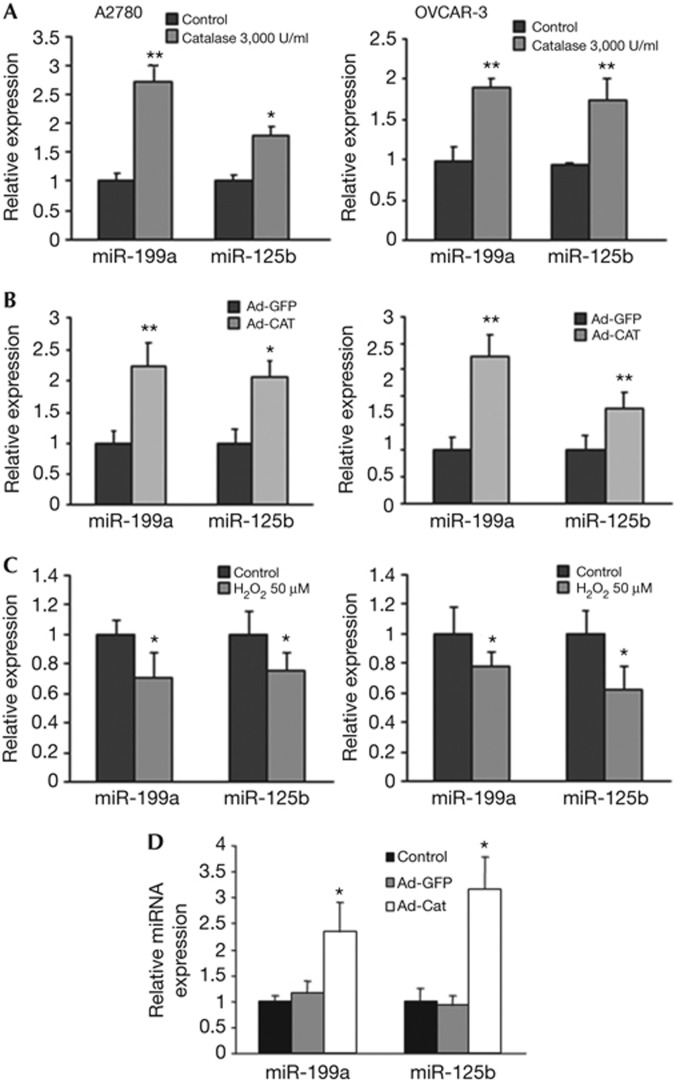
The mechanisms contributing to miRNA dysregulation remain elusive. We previously observed that human cancer cells frequently display high endogenous ROS levels [9]. We first screened miRNAs that might be induced by ROS inhibitors in ovarian cancer cells using microarray, validated those upregulated miRNAs using quantitative PCR (qPCR) method (supplementary Fig S3 online), then we used our newly developed miRNA prediction programme ‘Targetsearch’ to identify ROS-regulated miRNAs that might target ERBB2 and ERBB3, and selected miR-199a and miR-125b for further study. We treated ovarian cancer cells using ROS scavenger catalase, and found that both miR-199a and miR-125b expression levels were significantly induced by catalase treatment in both A2780 and OVCAR-3 cells (Fig 2A). Similar results were obtained by the infection of the cells using adenovirus carrying catalase (Fig 2B). Furthermore, hydrogen peroxide treatment suppressed miR-199a and miR-125b expression (Fig 2C), suggesting that ROS inhibit miR-199a and miR-125b expression. To test whether ROS affect miR-199a and miR-125b expression in vivo, we utilized the orthotopic ovarian cancer model for miRNA analysis, and demonstrated that levels of miR-199a and miR-125b expression were increased by more than 50% in the tumours with catalase overexpression than in the control tumours (Fig 2D). As ROS are widely involved in human development and diseases, oxidative stress-regulated miR-199 and miR-125b expression might have broader implications in normal development and other disease progression that would be interesting to be further investigated in the future.
Figure 2.
Reactive oxygen species inhibit miR-199a and miR-125b expression levels in vitro and in vivo. (A) A2780 and OVCAR-3 cells were treated with catalase at 3,000 U/ml for 12 h. Taqman qRT-PCR was performed to detect the levels of miR-125b and miR-199a in the cells. Results were normalized to U6 expression level and represented as mean±s.e. from three independent replicates. (B) A2780 and OVCAR-3 cells were infected with adenovirus carrying GFP or catalase (25 multiplicity of infection) for 60 h. Taqman qRT was performed as described above. (C) A2780 and OVCAR-3 cells were treated with 100 μM H2O2 for 4 h. Taqman qRT-PCR was performed as described above. (D) Tumour tissues were homogenized for total RNA extraction. The expression levels of miR-199a and miR-125b in tumour tissues from nude mice were determined by Taqman qRT–PCR (n=6–8). All experiments were performed in triplicate and presented as mean±s.e. * indicates significant decrease compared with the control group (by Student’s t-test, P<0.05). GFP, green fluorescent protein; miRNA, microRNA; qRT–PCR, quantitative reverse transcriptase PCR.
MiR-199a and miR-125b target ERBB2 and/or ERBB3
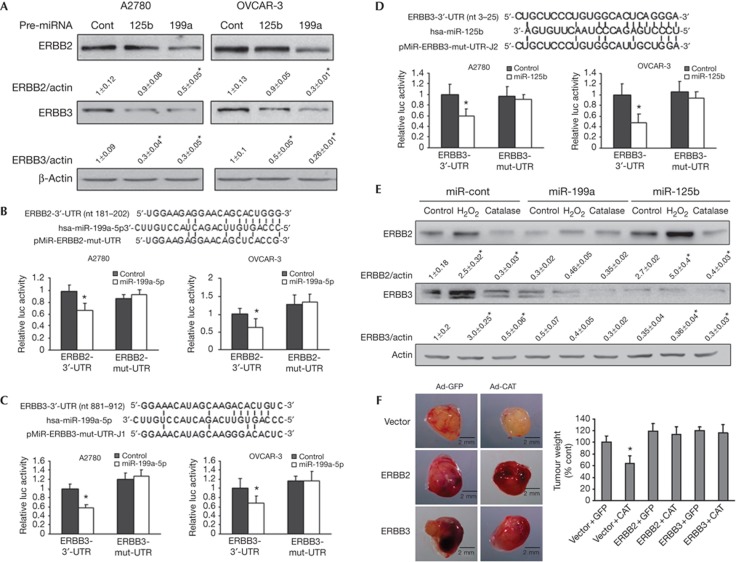
The downregulation of miR-199a and miR-125b was observed by miRNA microarray as previously described [13]. ERBB2 and ERBB3 are putative targets of miR-199a and miR-125b using our newly developed ‘Targetsearch’ programme and RNA22 prediction programme [14]. To experimentally validate the target prediction, we assessed the protein levels of ERBB2 and ERBB3 in OVCAR-3 and A2780 cells transfected with miR-199a or miR-125b precursors. Overexpression of miR-199a (supplementary Fig S4A online) greatly decreased ERBB2 and ERBB3 expression in both cell lines, whereas miR-125b decreased ERBB3 protein level, but not ERBB2 expression (Fig 3A). This indicated that ERBB2 is regulated by miR-199a, whereas ERBB3 is regulated by both miR-199a and miR-125b. To test whether miR-199a directly binds to the 3′-UTR regions of ERBB2 mRNA and ERBB3 mRNA, we constructed the 3′-UTR reporters of ERBB2 and ERBB3 containing the putative miR-199a-binding sites and corresponding mutant constructs downstream of the luciferase reporter gene; and to test whether miR-125b directly binds to the ERBB3 3′-UTR regions, we constructed the 3′-UTR reporters of ERBB3 wild-type and mutant luciferase reporters. Co-transfection of miR-199a or miR-125b precursor with their respective wild-type reporter constructs greatly decreased the luciferase activities in both cell lines, while co-transfection with reporters containing point mutations at putative miRNA-binding sites did not affect the luciferase activities (Fig 3B–D). These results further confirm that ERBB2 is a direct target of miR-199a, and ERBB3 is a direct target of both miR-199a and miR-125b in ovarian cancer cells. MiR-125b was shown to target both ERBB2 and ERBB3 in human breast cancer cell line SKBR3 [15]. However, we did not find that miR-125b inhibited ERBB2 expression in ovarian cancer cells. Observed differences might be owing to the different cell types used in the study or other unknown mechanisms. To determine whether the effect of ROS on ERBB2 and ERBB3 expression is mediated by miR-199a, we established an OVCAR-3 stable cell line overexpressing miR-199a or miR-125b. The expression of miR-199a or miR-125b was much higher in the stable cells compared with the vector control cells (supplementary Fig S4B online). As expected, H2O2 and catalase treatments of vector cells induced and inhibited ERBB2 and ERBB3 expression levels, respectively (Fig 3E); whereas miR-199a stable cells diminished the effects of H2O2 and catalase treatments on ERBB2 and ERBB3 expression. Similarly, overexpression of miR-125b mimicked the effects of catalase, and inhibited H2O2-induced ERBB3 expression. To determine whether ROS scavenger inhibits tumour growth via ERBB2 and ERBB3 in vivo, we performed tumour growth assay using OVCAR-3 stable cells overexpressing ERBB2, ERBB3 or the vector alone. Interestingly, we found that overexpression of ERBB2 led to upregulation of ERBB3, and vice versa (supplementary Fig S4C online). This might be owing to the induction of ERBB2/ERBB3 heterodimer formation, which stabilizes the proteins. Overexpression of ERBB2 or ERBB3 was sufficient to reverse the inhibitory effect of catalase on tumour growth (Fig 3F), indicating that ERBB2 and ERBB3 are functional relevant targets of ROS in vivo.
Figure 3.
miR-199a directly targets ERBB2 and ERBB3, whereas miR-125b targets ERBB3. (A) The cells were transiently transfected with 30 nM pre-miR-199a, pre-miR-125b or pre-miR-miRNA precursor, respectively, and harvested 70 h later. ERBB2 and ERBB3 expression levels were analysed by immunoblotting, and quantified from three replicates. (B–D) Sequence alignment of human miR-199a with 3′-UTR of ERBB2 or ERBB3 and human miR-125b with 3′-UTR of ERBB3. Bottom: mutations in the 3′-UTR of ERBB2 and ERBB3 to create the mutant luciferase reporter constructs. The luciferase activities were presented as relative luciferase activities normalized to those of the cells co-transfected with wild-type 3′-UTR reporter and miRNA precursor control. * indicates significant decrease compared with that of control cells (P<0.05). All tests were performed in triplicate and presented as mean±s.e. (E) OVCAR-3 stable cell lines overexpressing miR-199a, miR-125b or negative control precusor were generated. The cells were treated with catalase (3,000 U/ml, 12 h) or H2O2 (100 μM, 4 h), respectively. ERBB2 and ERBB3 protein expression levels were analysed by immunoblotting. Results from three independent experiments presented as mean±s.e. were shown on the bottom. (F) OVCAR-3 cells stably expressing vector, ERBB2 or ERBB3 were infected with adenovirus carrying GFP or catalase and used for tumour growth assay described in Methods. The images are the representative tumours of each treatment. Right: average tumour weight was normalized to the control group and presented as mean±s.e. (n=8∼10). * indicates significant decrease compared with control group (by one-way analysis of variance, P<0.05).GFP, green fluorescent protein; miRNA, microRNA.
Hypermethylation of miR-199a and miR-125b genes by ROS
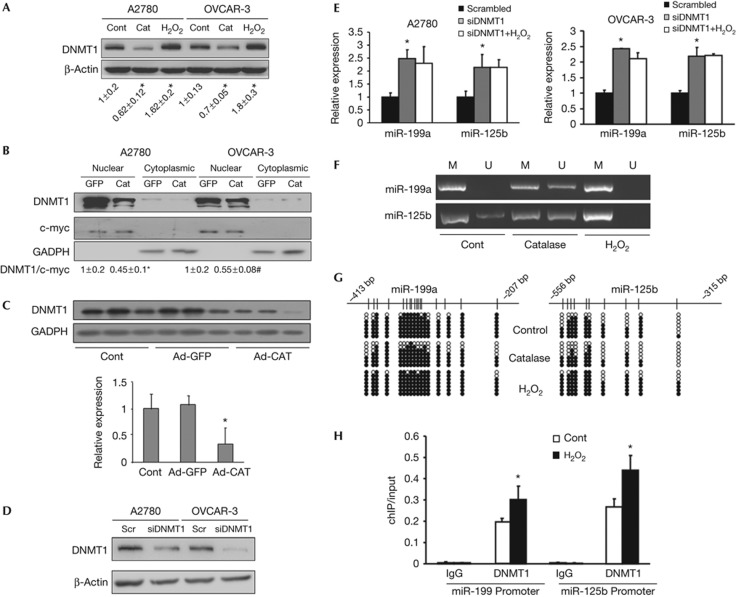
We further investigated how ROS inhibit miR-199 and miR-125b expression. DNA hypermethylation, a principal epigenetic modification occurring in distal CpG-rich regions, has been shown to be associated with aberrant miRNA expression profiles in cancer [16]. DNMT1 is a principal enzyme for DNA methylation, which has been implicated in oxidative stress-induced DNA repair [17]. A recent study demonstrated that ROS recruited DNMT1 to oxidative stress-induced damaged chromatin before the DNA repair process is complete [18]. In addition, several studies linked aberrant DNA hypermethylation to miR-199a and miR-125b suppression in cancer cells and tissues [19, 20]. We therefore propose that endogenous ROS inhibit miR-199a and miR-125b expression by DNMT1-mediated DNA hypermethylation. We found that catalase treatment decreased DNMT1 expression levels in ovarian cancer cells. By contrast, hydrogen peroxide treatment induced DNMT1 expression (Fig 4A). We also showed that DNMT1 protein expression in nuclear fractions was inhibited by catalase (Fig 4B). To determine the relationship between ROS and DNMT1 in vivo, we utilized tumour model to analyse the DNMT1 expression in the tissues. DNMT1 expression levels were significantly lower in the catalase-treated tumours than those of control tumours (Fig 4C), suggesting that DNMT1 protein levels are inhibited by catalase both in vitro and in vivo. To test the direct role of DNMT1 in regulating miR-199a and miR-125b expression, we knocked down DNMT1 in ovarian cancer cells using a short interfering RNA (siRNA) smartpool specifically against DNMT1 (Fig 4D). miR-199a and miR-125b expressions were significantly increased in DNMT1-knockdown cells when compared with the cells transfected with a scrambled siRNA (Fig 4E). Further treatment with H2O2 in DNMT1-knockdown cells did not decreamiR-125b expression. To rule out the possibility of an off-target effect for this pooled siRNA, we repeated the experiments using two unrelated individual siRNAs against DNMT1, and similar results were obtained regarding the miR-199a and miR-125b expression (supplementary Fig S5 online). These results demonstrate that DNMT1 expression regulates miR-199a and miR-125b expression. To investigate whether miR-199a and miR-125b genes are hypermethylated in response to ROS, we used methylation-specific PCR and bisulphite sequencing to analyse the methylation status of the promoter regions of miR-199a and miR-125 in OVCAR-3 cells treated with catalase or H2O2 with represented results shown in Fig 4F–G. We analysed the hypermethylation levels of miR-199a and miR-125b gene promoters using 20 independent clones, and found that catalase treatment decreased hypermethylation levels of CpG islands in the promoter of miR-199a from 83.5 to 70.6% (untreated control, P<0.05) and the miR-125b promoter from 65.5 to 48% (untreated control, P<0.05), whereas H2O2 treatment induced the hypermethylation levels of the miR-199a promoter from 83.5 to 92.4% (the control, P<0.05) and the miR-125b promoter from 65.5 to 78.1% (the control, P<0.05). We next determined whether ROS treatment affects the association of DNMT1 with the promoter regions of miR-199a and miR-125b using Chromatin immunoprecipation (ChIP) assay using DNMT1 antibody, and demonstrated that H2O2 treatment of OVCAR-3 cells significantly increased DNMT1-binding capacity to both miR-199a and miR-125b promoter regions (Fig 4H). Thus, ROS suppress miR-199a and miR-125b expression by inducing the promoter hypermethylation, which is mediated by DNMT1 expression in the cells. However, it remains unclear yet how ROS increase the binding of DNMT1 to the miRNA gene promoters. DNMT1 often functions in a way of DNMT1/transcription factors-including complexes. ROS might help to recruit DNMT1 to the oxidative stress-damaged DNA to silence gene transcription temporarily for the DNA repair and/or miRNA expression. It is also possible that other ROS-responsive transcription factors such as NF-κB which might act as cofactor to promote functional binding of miRNA promoter region in this process. It is likely that ROS have an indirect effect in inducing the binding of DNMT1 to miRNA gene promoters, which would warrant future studies to address this question. Taken together, the current study demonstrates that excessive ROS in cancer cells inhibit expression of miR-199a and miR-125b via DNMT1-mediated DNA methylation, and the repression of miR-199a and miR-125b expression leads to activation of their direct targets ERBB2 and ERBB3 for inducing tumour growth (Fig 5).
Figure 4.
Reactive oxygen species inhibit miR-199a and miR-125 expression through DNMT1-mediated methylation. (A) A2780 and OVCAR-3 cells were treated with 3,000 U/ml catalase for 12 h or treated with 100 μM H2O2 for 4 h. The expression levels of DNMT1 in the cells were analysed by immunoblotting. Relative DNMT1 protein signals were quantified from three independent experiments and presented as mean±s.e. on the bottom. (B) A2780 and OVCAR-3 cells were infected with adenovirus carrying GFP or catalase (25 multiplicity of infection) and cultured for 60 h. DNMT1 protein levels from nuclear and cytoplasmic fractions were extracted and analysed by immunoblotting. Relative DNMT1 protein levels were obtained from three independent experiments. C-myc was used as nuclear marker, whereas GADPH was used as cytosol marker. (C) The expression levels of DNMT1 in ovarian tumour tissues were analysed by immunoblotting. The representative results from each group were shown. The protein levels of DNMT1 were quantified by scanning densitometry, normalized to GADPH levels and the control as mean±s.e. (n=6–8). (D, E) The cells were transfected with 50 nM of a siRNA scramble control or a siRNA smartpool against DNMT1 for 72 h with or without H2O2 treatment (100 μM) for 4 h. The DNMT1 protein levels were detected by immunoblotting, and miR-199a and miR-125b expression levels were analysed by Taqman quantitative reverse transcriptase PCR. (F, G) OVCAR-3 cells were treated with catalase (3,000 U/ml) for 12 h or H2O2 (100 μM) for 4 h. The hypermethylation levels of miR-199a and miR-125b gene promoters were determined by methylation-special PCR analyses, and by bisulphite DNA sequencing showing seven independent clones from each group. Each row represents one cloned PCR product. The representative seven clones from each treatment were shown. The CpG islands are depicted, and each vertical bar illustrates a single CpG site. Black and white cycles represent methylated and unmethylated cytosine, respectively. (H) OVCAR-3 cells were treated with or without H2O2 (100 μM, 4 h). ChIP sample was analysed using antibodies against IgG or DNMT1. The antibody-promoter-binding signals were analysed by quantitative SYBR quantitative PCR, and presented are the mean±s.e. from three independent experiments. * indicate significant difference compared with the control (by Student’s t-test, P<0.05). ChIP, chromatin immunoprecipation; Cont, control; DNMT1, DNA methyltransferase 1; GADPH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; M, methylated state; siRNA, short interfering RNA; U, unmethylated state.
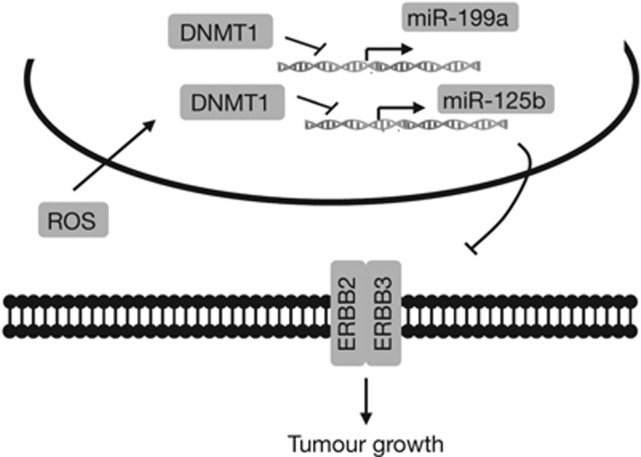
Figure 5.
Schematic diagram. The excessive ROS in cancer cells induce DNMT1 expression, which increases the methylation of miR-199a and miR-125b promoter regions. The DNA methylation leads to miR-199a and miR-125b downregulation, which in turn activates expression of their direct targets ERBB2 and ERBB3 for inducing tumour growth. DNMT1, DNA methyltransferase 1; ROS, reactive oxygen species.
Methods
Reagents, cell culture, transfection and establishment of stable cell lines. Catalase, DPI, rotenone and H2O2 were purchased from Sigma (St Louis, MO, USA). Antibodies against total ERBB2 and ERBB3 (clone 2F12) were from Upstate Biotechnology (Upstate, NY, USA). CM2-DCFH-DA was purchased from Molecular Probes (Eugene, OR, USA). The human ovarian cancer cells OVCAR3 and A2780, and immortalized ovarian epithelial cells IOSE397 were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. siRNAs (SMARTpool) against DNMT1 and scrambled siRNA (SMARTpool) were purchased from Dharmacon (Lafayette, CO, USA). Two unrelated siRNAs against DNMT1 were purchased from Sigma. Transfection of miRNA precursors (Applied Biosystem) or siRNAs were performed using lipofectamine RNAiMax (Invitrogen). To establish stable cells overexpressing miR-199a, OVCAR-3 cells were infected with lentivirus carrying miR-199a, miR-125b or a negative control precursor (Open Biosystems, Huntsville, AL, USA) followed by the selection with puromycin. Stable cell lines of OVCAR-3 cells overexpressing ERBB2 or ERBB3 was generated by transfecting the pReceiver-Lv105 vector expressing ERBB2 or ERBB3 open reading frame into 293FT cell using Trans-lentiviral package kit (Open Biosystems). OVCAR-3 cells were infected by lentivirus carrying ERBB2 or ERBB3, followed by the selection with puromycin to obtain the stable cell lines.
Intracellular H2O2 staining. Cells were cultured in a 6-well plate and incubated at 37 °C overnight to allow attachment, then cultured in serum-free medium overnight. 2',7'-Dichlorodihydrofluorescein diacetate (DCFH-DA) (5 μM) was added and incubated with the cells for 15 min. The cells were then washed with 1 × PBS buffer, and fixed with 10% formaldehyde. Fluorescent images were captured with a fluorescent microcope. The signal quantification was assessed by ImageJ software.
Tumour growth on chicken chrioallantoic membrane. White Leghorn fertilized chicken eggs were incubated at 37 °C under constant humidity. OVCAR-3 cells overexpressing lenti-vector, ERBB2 or ERBB3 were infected with 25 multiplicity of infection (MOI) adenovirus carrying green fluorescence protein or catalase 1 day before implantation. These cells were trypsinized, counted and resuspended in the serum-free medium. The cell suspensions were mixed with Matrigel at 1:1 ratio and implanted onto chorioallantoic membranes (CAM) of 9-day-old chicken embryos. Tumour growth was analysed 10 days after implantation. The average tumour weight was obtained from the CAMs of 8–10 embryos per treatment.
DNA methylation analysis. Genomic DNA was extracted using DNA Mini kit (Qiagen, MD, USA). Total 1 μg genomic DNA was treated with sodium bisulphite using the EpiTect Kit (Qiagen). Bisulphite-treated DNA was then amplified with methylation-specific primers for CpG islands surrounding miR-199a and miR-125b genomic locations. The resultant PCR products were cloned into a pBS vector (Addgene). Twenty positive recombinants were isolated and sequenced by an ABI automated DNA sequencer. The methylation status and levels of individual CpG sites were determined by the comparison of the sequence obtained with the known target sequence. The primers used were described in supplementary Methods online.
ChIP assay. ChIP assay was performed using EpiTect ChIP OneDay Kit (Qiagen) according to the manufacturer’s instruction. DNMT1 antibody (Abcam, Cambridge, MA, USA) was used to pull down the protein–chromatin complexes. Rabbit IgG was used as a negative control. The immunoprecipated DNA was quantified using SYBR Green qPCR (Applied Biosystem). All results were normalized to 1% input value of the same sample.
Statistical analysis. All of the results were obtained from at least three independent experiments. Most results were presented as mean±s.e. from independent experiments, and analysed by Student’s t-test and one-way analysis of variance. All the results were analysed by SPSS for Windows, version 11.5. Differences were considered significant at a value of P⩽0.05.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported in part by the National Key Basic Research Programme of China (2011CB504003), by National Natural Science Foundation of China (81071642 and 30871296) and by National Cancer Institute, NIH (R01CA109460).
Author contributions: J.H., Q.X. and Y.J. planned and performed most of the experiments. X.Q., R.C. and Q.L. performed some of the experiments. J.H., F.A., X.-R.W., S.S.P., Z.L. and B.-H.J. performed the data analysis and manuscript preparation. B.-H.J. coordinated the project.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ono M, Kuwano M (2006) Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin Cancer Res 12: 7242–7251 [DOI] [PubMed] [Google Scholar]
- Steffensen KD, Waldstrom M, Andersen RF, Olsen DA, Jeppesen U, Knudsen HJ, Brandslund I, Jakobsen A (2008) Protein levels and gene expressions of the epidermal growth factor receptors, HER1, HER2, HER3 and HER4 in benign and malignant ovarian tumors. Int J Oncol 33: 195–204 [PubMed] [Google Scholar]
- Sasaki N, Kudoh K, Kita T, Tsuda H, Furuya K, Kikuchi Y (2007) Effect of HER-2/neu overexpression on chemoresistance and prognosis in ovarian carcinoma. J Obstet Gynaecol Res 33: 17–23 [DOI] [PubMed] [Google Scholar]
- Servidei T, Riccardi A, Mozzetti S, Ferlini C, Riccardi R (2008) Chemoresistant tumor cell lines display altered epidermal growth factor receptor and HER3 signaling and enhanced sensitivity to gefitinib. Int J Cancer 123: 2939–2949 [DOI] [PubMed] [Google Scholar]
- Zeineldin R, Muller CY, Stack MS, Hudson LG (2010) Targeting the EGF receptor for ovarian cancer therapy. J Oncol 2010: 414676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M (2006) Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact 160: 1–40 [DOI] [PubMed] [Google Scholar]
- Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI (2008) Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 266: 6–11 [DOI] [PubMed] [Google Scholar]
- Dolado I, Swat A, Ajenjo N, De Vita G, Cuadrado A, Nebreda AR (2007) p38alpha MAP kinase as a sensor of reactive oxygen species in tumorigenesis. Cancer Cell 11: 191–205 [DOI] [PubMed] [Google Scholar]
- Xia C, Meng Q, Liu LZ, Rojanasakul Y, Wang XR, Jiang BH (2007) Reactive oxygen species regulate angiogenesis and tumor growth through vascular endothelial growth factor. Cancer Res 67: 10823–10830 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM (2011) MicroRNAs as therapeutic targets in cancer. Transl Res 157: 216–225 [DOI] [PubMed] [Google Scholar]
- Godshalk SE, Melnik-Martinez KV, Pasquinelli AE, Slack FJ (2010) MicroRNAs and cancer: a meeting summary of the eponymous Keystone Conference. Epigenetics 5: 164–168 [DOI] [PubMed] [Google Scholar]
- Iorio MV et al. (2007) MicroRNA signatures in human ovarian cancer. Cancer Res 67: 8699–8707 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC (2007) Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 282: 1479–1486 [DOI] [PubMed] [Google Scholar]
- Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC (2009) Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 4: 587–592 [DOI] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4: e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM et al. (2011) Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20: 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HH, Davis AJ, Lee TL, Pang AL, Nagrani S, Rennert OM, Chan WY (2011) Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene 30: 3404–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y et al. (2011) miR-125b is methylated and functions as a tumour suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res 71: 3552–3562 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.