Abstract
Phosphatidylinositol (PI) 4-phosphate (PI(4)P) and its metabolizing enzymes serve important functions in cell signalling and membrane traffic. PI 4-kinase type IIα (PI4KIIα) regulates Wnt signalling, endosomal sorting of signalling receptors, and promotes adaptor protein recruitment to endosomes and the trans-Golgi network. Here we identify the E3 ubiquitin ligase Itch as binding partner and regulator of PI4KIIα function. Itch directly associates with and ubiquitinates PI4KIIα, and both proteins colocalize on endosomes containing Wnt-activated frizzled 4 (Fz4) receptor. Depletion of PI4KIIα or Itch regulates Wnt signalling with corresponding changes in Fz4 internalization and degradative sorting. These findings unravel a new molecular link between phosphoinositide-regulated endosomal membrane traffic, ubiquitin and the modulation of Wnt signalling.
Keywords: phosphatidylinositol 4-kinase type IIα, E3 ubiquitin ligase, Itch, endosomes, Wnt signalling
Introduction
Phosphoinositides serve as integrators of intracellular membrane dynamics and cell signalling, thereby crucially regulating cell physiology [1, 2]. Phosphatidylinositol (PI) 4-kinases, including PI 4-kinase type IIα (PI4KIIα), a palmitoylated trans-Golgi network (TGN)/endosomal protein [3], are at the apex of the phosphoinositide cascade [4]. PI4KIIα-mediated synthesis of phosphatidylinositol (PI) 4-phosphate (PI(4)P) [5] is required for the recruitment of clathrin adaptors to endosomes [6] and to the TGN [7]. Loss of PI4KIIα impairs endo-lysosomal degradation of ubiquitinated EGF receptors [8], suggesting that PI4KIIα could control ubiquitin-mediated protein sorting. However, a direct connection between PI(4)P-synthesizing enzymes and the ubiquitin sorting system [9] has not been established. PI4KIIα has also been implicated in canonical Frizzled (Fz)-mediated Wnt signalling [10] via PI(4)P synthesis and association with the signalling adaptor dishevelled (Dvl) [11]. The cellular responsiveness to Wnt signals is further controlled by the ubiquitin-specific protease USP8/UBPY at the level of endo-lysosomal sorting of Fz [12]. These results favour a model whereby endosomal trafficking of activated Fz receptors is regulated by phosphoinositides and ubiquitination.
Here, we identify the TGN/endosomally localized E3 ubiquitin ligase Itch [13] as direct binding partner and enzymatic regulator of PI4KIIα function. Moreover, we show that PI4KIIα and Itch modulate Wnt signalling with corresponding changes in Fz4 internalization and degradative sorting.
Results and Discussion
PI4KIIα undergoes multi-ubiquitination
To better understand the key role of PI4KIIα in TGN/endosomal membrane traffic [4, 6, 7], we immunoprecipitated PI4KIIα from cell lysates. Surprisingly, immunoblot analysis revealed several more PI4KIIα bands at a molecular weight that exceeds that of native PI4KIIα (not shown), consistent with possible ubiquitination of PI4KIIα. Indeed, when haemagglutinin (HA)–PI4KIIα was immunoprecipitated from stably transfected HEK293 cells co-expressing FLAG-ubiquitin (Ub), a ladder of FLAG-Ub-containing bands was detected in the immunoprecipitates (Fig 1A). To characterize the ubiquitination of PI4KIIα further, we used mutant ubiquitin (Ub7R, all Ks exchanged to Rs) unable to generate poly-ubiquitin chains. Ub7R formed Ub conjugates with HA–PI4KIIα indistinguishable from wild-type (WT)-Ub (Fig 1B). Thus, PI4KIIα undergoes multi-ubiquitination.
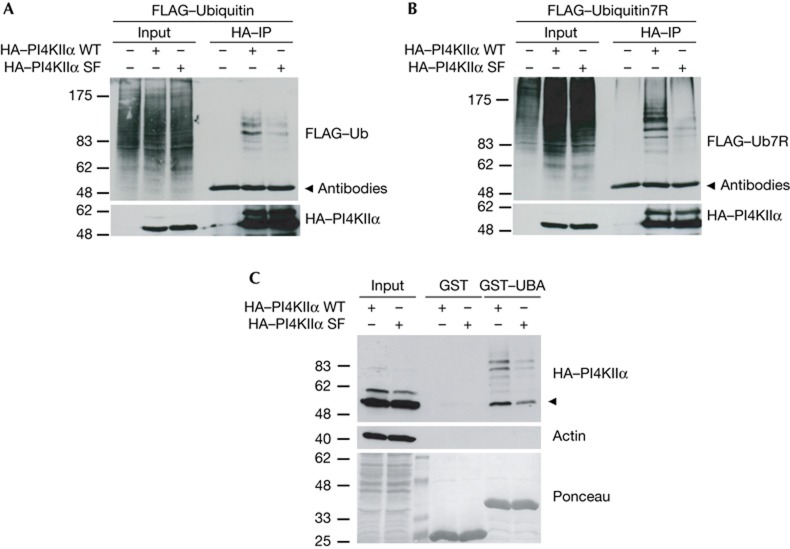
Figure 1.
PI4KIIα undergoes ubiquitination. (A) WT and SF mutant (PPXY to SPXF) HA–PI4KIIα were immunoprecipitated from HEK293 cells and co-transfected with FLAG–ubiquitin. SF is ubiquitinated less efficiently (densitometric analysis: WT 100%, SF 38±9%). (B) As in A but with HEK293 cells expressing FLAG–Ub7R (densitometric analysis: WT 100%, SF 24±8%). (C) Pulldown with GST–UBA3x as a bait from HEK293 cells stably expressing HA–PI4KIIα WT or HA–PI4KIIα SF. Arrowhead indicates unmodified PI4KIIα. Molecular weight markers in kD. GST, glutathione S-transferase; HA, haemagglutinin; IP, immunoprecipitation; PI4KIIα, phosphatidylinositol 4-kinase type IIα; WT, wild-type.
Among the large family of E3 ubiquitin ligases are HECT-type ligases, many of which contain WW domains that recognize PPxY motifs [9, 14, 15]. As PI4KIIα contains a PPxY motif, we analysed whether mutational inactivation of this motif affects PI4KIIα ubiquitination. Mutant PI4KIIα (PI4KIIαSF), in which two conserved residues within the PPxY motif were exchanged (PPxY to SPxF), underwent ubiquitination by either WT-Ub or Ub7R with reduced efficiency (Fig 1A,B). This conclusion was corroborated by the retention of PI4KIIα on a Ub affinity matrix (glutathione S-transferase (GST)–UBA) [16], whereas PI4KIIαSF was retained much less efficiently (Fig 1C). Unexpectedly, the affinity-purified material contained a fraction of unmodified PI4KIIα (Fig 1C, arrow), which could originate from dimerization of non-ubiquitinated PI4KIIα with ubiquitinated PI4KIIα (type II PI kinases form dimers; [17]). These results indicate that PI4KIIα undergoes PPxY motif-regulated multi-ubiquitination, likely involving WW domain-containing HECT-type E3 ubiquitin ligases.
Itch directly associates with and ubiquitinates PI4KIIα
To identify E3 ubiquitin ligases responsible for the ubiquitination of PI4KIIα, we took an MS/MS-based proteomic approach using GST–PI4KIIα as an affinity matrix. In addition to known binding partners for PI4KIIα, including AP-3 [6], we identified several HECT-type E3 ubiquitin ligases, including Itch, NEDD4 and WWP1 (supplementary Fig S1A–C online). Consistent with the high degree of sequence coverage for Itch in MS/MS analyses (supplementary Fig S1C online), we confirmed binding of Itch to GST–PI4KIIα by immunoblotting (Fig 2A) and further by co-immunoprecipitations from native tissue lysates (Fig 2B) or from HA–PI4KIIα expressing HEK293 cells using Itch-specific antibodies (supplementary Fig S1E online). Complex formation involved the PI4KIIα PPxY motif and the WW domain module of Itch (Fig 2C). PI4KIIα was also able to bind to the WW domains of NEDD4.1 or NEDD4.2, suggesting that Itch and NEDD4 might partially overlap with respect to complex formation with PI4KIIα (supplementary Fig S1D online). Analysis of each of the four individual GST-tagged WW domains of Itch revealed that all WW domains except WW4, which lacks one of the conserved W residues required for ligand binding [18], were capable of interacting with native PI4KIIα (supplementary Fig S1F online). Direct association of PI4KIIα–PPxY with the 15N-labelled WW3 domain of Itch was unambiguously verified by NMR spectroscopy. Chemical shift changes induced by titration of the PPxY peptide (Fig 2D) were assigned to amino acids well known to represent the main binding surface on WW domains for proline-rich peptides (Fig 2E), indicative of a canonical PPxY recognition mode [18]. In addition, line broadening beyond detection was observed for a second group of NH peaks; these residues mostly belong to the third β-strand of the WW3 domain suggesting conformational heterogeneity induced by ligand binding (Fig 2E, blue).
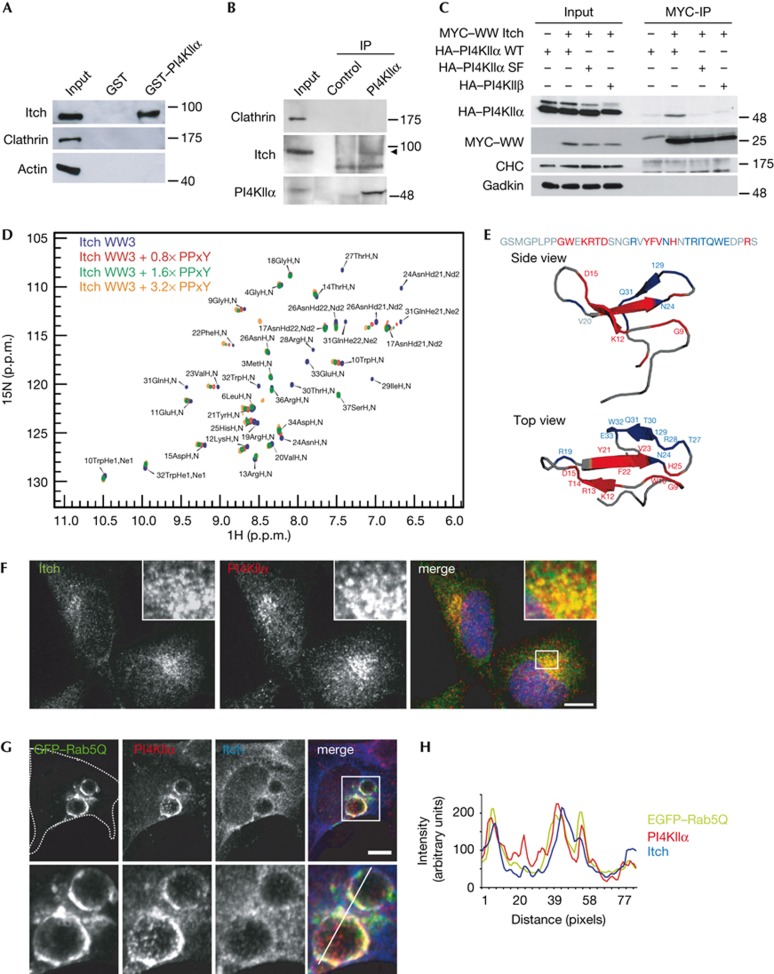
Figure 2.
PI4KIIα directly associates with the WW domains of the E3 ligase Itch. (A) GST–PI4KIIα co-purifies Itch, but not clathrin or actin from rat brain extracts. (B) Endogenous co-immunoprecipitation of PI4KIIα and Itch from uterus membrane extract. Arrow indicates Itch. (C) HEK293 cells stably expressing HA–PI4KIIα WT, HA–PI4KIIα SF or HA–PI4KIIβ were transfected with MYC-Itch WW domains (WW1-4). PI4KIIα WT specifically co-immunoprecipitated with Itch WW domain. Molecular weight markers in kD. (D) Overlay of the 1H, 15N HSQC spectra of 15N-WW3 Itch alone with spectra from samples containing increasing amounts of PI4KIIα PPxY peptide (no peptide, 0.8 × , 1.6 × and 3.2 × molar equivalents). (E) Sequence and cartoon models of Itch-WW3 (PDB ID: 2JO9). Red, NH resonances of residues that show chemical shift changes above average in D. Blue, residues disappearing on ligand addition. (F) Confocal images of endogenous PI4KIIα and Itch in HeLa cells. Pearson: PI4KIIα/Itch: 0.75. Scale bar, 12 μm. (G) Confocal images of endogenous PI4KIIα and Itch in HEK cells overexpressing eGFP–Rab5(Q79L). Cell borders indicated by dotted line. Lower panel, magnification of boxed area. Scale bar, 6 μm. (H) Fluorescence intensities along line shown in G plotted over distance. GST, glutathione S-transferase; HA, haemagglutinin; IP, immunoprecipitation; PI4KIIα, phosphatidylinositol 4-kinase type IIα; WT, wild-type.
To investigate the function of the Itch–PI4KIIα complex, we studied its localization in HeLa cells. PI4KIIα partitioned between the TGN and endosomes [6, 19]. A similar, although less punctuate, distribution was seen for endogenous Itch, which partially colocalizes with PI4KIIα (Fig 2F). PI4KIIα- and Itch-positive structures largely correspond to endosomes as evidenced by their partial colocalization with the endosomal proteins AP-3, LAMP1 and EEA1 (supplementary Fig S2A–E online). Furthermore, PI4KIIα and Itch localize to similar subdomains on enlarged endosomes induced by overexpressing GTP-locked Rab5 (Fig 2G,H). A partial overlap was also seen for the TGN marker TGN46 (supplementary Fig S2F online). Consistent with these observations, facilitated recruitment of Itch to the TGN/endosomal boundary was observed in eGFP–PI4KIIα overexpressing cells (supplementary Fig S4B online). We conclude that Itch and PI4KIIα colocalize at endosomes and partly at the TGN.
Next, we wanted to elucidate if PI4KIIα is a direct substrate of Itch. To this aim, we purified recombinant His6-Itch or a catalytically inactive mutant thereof (C830A) [15]. In vitro ubiquitination reactions containing WT Itch resulted in the formation of several Itch-containing adducts indicative of its auto-ubiquitination that were absent from ItchC830A-containing samples, confirming its catalytic inactivity (not shown). We then analysed the activity of Itch towards GST–PI4KIIα or GST–PI4KIIαSF. Several PI4KIIα-containing high molecular weight bands were observed in immunoblots from samples containing WT Itch, whereas no such bands were seen with the catalytically inactive mutant (C830A) (Fig 3A). In contrast to Itch, recombinant Nedd4.1 failed to ubiquitinate PI4KIIα although it underwent efficient auto-ubiquitination (supplementary Fig S1G online). Itch-binding-defective PI4KIIαSF displayed reduced ubiquitination when compared with its WT counterpart (Fig 3A, B). Conversely, doxycyclin-induced overexression of Myc-Itch in stably transfected HEK293 cells resulted in elevated levels of ubiquitin-conjugated PI4KIIα (Fig 3C), suggesting that Itch also modifies PI4KIIα in cells.
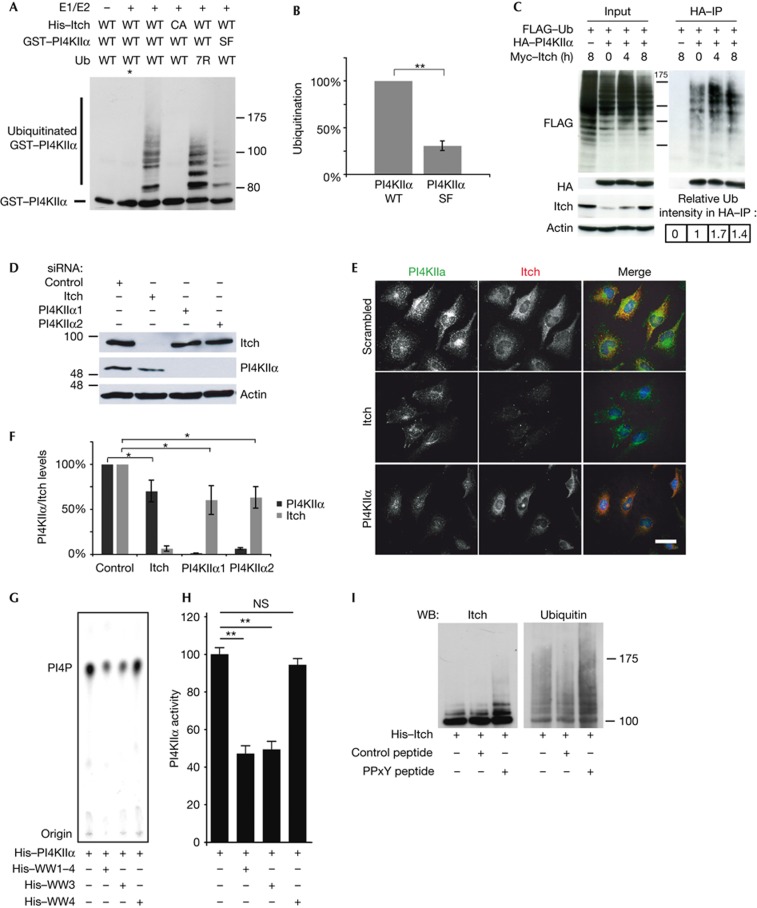
Figure 3.
PI4KIIα and Itch form a functional complex. (A) GST–PI4KIIα was subjected to in vitro ubiquitination assays. WT but not ligase-inactive Itch (CA) ubiquitinates PI4KIIα WT. PI4KIIα SF is ubiquitinated less efficiently. Asterisk denotes reaction at 4 °C. (B) Quantification of Ub signal in lanes 3 and 6 of A (mean±s.d.; n=3, **P<0.01). (C) HA–PI4KIIα and FLAG–ubiquitin were transfected into HEK293 cells, expressing Itch from a doxycyclin-inducible promotor. Enhanced ubiquitination of immunoprecipitated HA–PI4KIIα is observed after 4 h (1.7 times) and 8 h (1.4 times) of Itch induction. (D) siRNA-treated HeLa cells were immunoblotted for Itch, PI4KIIα and actin. (E) Confocal images of siRNA-treated HeLa cells stained with antibodies against PI4KIIα and Itch. Scale bar, 12 μm. (F) Levels of Itch and PI4KIIα analysed by quantitative immunoblotting, on knockdown with Itch and two independent PI4KIIα oligos, as in D. Signals normalized to actin (mean±s.e.m.; n=4, *P<0.05). (G) Analysis of PI4KIIα kinase activity on addition of Itch WW domains, using PI and 32P-γ-ATP as substrates. Samples were analysed by thin layer chromatography and phosphoimaging. (H) Quantification of the data in G, normalized to the amount of PI4P generated by PI4KIIα alone (mean±s.e.m.; n=3, **P<0.01). (I) Immunoblot analysis of Itch autoubiquitination reactions in the presence of PI4KIIα peptide (PPxY) or control peptide (densitometric analysis: Itch alone 100.0%, Itch+control peptide 130.0±0.1%, Itch+PPxY-peptide 201.0±4.6%). GST, glutathione S-transferase; HA, haemagglutinin; IP, immunoprecipitation; PI4KIIα, phosphatidylinositol 4-kinase type IIα; siRNA, short interfering RNA; Ub, ubiquitin; WT, wild-type.
Itch and PI4KIIα reciprocally regulate each other
To obtain functional insights into the interaction between Itch and PI4KIIα, we conducted short interfering RNA (siRNA) knockdowns (Fig 3D). Depletion of Itch caused a notable reduction in the cellular levels of PI4KIIα (Fig 3E,F). Conversely, loss of PI4KIIα reduced the expression levels of Itch compared with control cells (Fig 3E,F). As expression of the individual subunits of multiprotein complexes often is functionally coupled, these data further support the notion that Itch and PI4KIIα form a functional complex in vivo.
Itch and PI4KIIα might also affect each other’s enzymatic activities. We first tested the effect of the Itch WW domain module on PI4KIIα-mediated PI(4)P synthesis. Incubation of 1 μg of recombinant PI4KIIα with PI and radioactive ATP resulted in the time-dependent formation of radiolabelled PI(4)P (supplementary Fig S3A online). Correspondingly lower amounts of PI(4)P were produced if less PI4KIIα was taken for the assay (supplementary Fig S3B online). Addition of WW1-4 or WW3 reduced PI(4)P production by PI4KIIα, whereas WW4, a domain unable to bind to PI4KIIα (supplementary Fig S1F online), had no effect (Fig 3G,H). Itch–WW1-4-mediated inhibition was dose-dependent (supplementary Fig S3D online). Furthermore, a similar inhibitory effect was observed, if full-length recombinant Itch was taken instead of the isolated WW domains (supplementary Fig S3D online). Last, Itch-mediated ubiquitination did not further reduce the PI(4)P-synthesizing activity of PI4KIIα beyond the inhibition induced by catalytically inactive Itch (supplementary Fig S3E online). Collectively, these data identify Itch as a negative regulator of PI4KIIα activity. On the basis of previous data [20], it is conceivable that binding of the PI4KIIα–PPxY motif to Itch alters its ubiquitination activity. To test this, we performed Itch autoubiquitination assays. Addition of the PI4KIIα-derived PPxY peptide activated Itch, whereas a control peptide had no effect (Fig 3I). Thus, Itch and PI4KIIα reciprocally regulate each other’s enzymatic activities, further supporting the hypothesis that both proteins function in the same pathway.
PI4KIIα and Itch modulate Wnt signalling
Previous work has provided evidence for a crucial role of PI4KIIα in activating canonical Wnt signalling at the plasma membrane by local production of PI(4)P and association with Dvl [10, 11]. To corroborate these data and to further investigate the function of PI4KIIα and Itch, we analysed early events in Wnt signalling as well as Wnt target gene expression. Depletion of PI4KIIα inhibited phosphorylation of the Fz co-receptor LRP6, a key component of canonical Wnt signalling (Fig 4A). These results are consistent with earlier findings [10] and confirm a crucial role of PI4KIIα in the formation of Wnt signalling complexes at the cell surface (supplementary Fig S4A online). By contrast, depletion of Itch resulted in increased levels of phospho-LRP6 (Fig 4A), paired with a corresponding increase in the expression of the Wnt target gene axin 2 (Fig 4B), thereby identifying Itch as a putative negative regulator of canonical Wnt signalling.
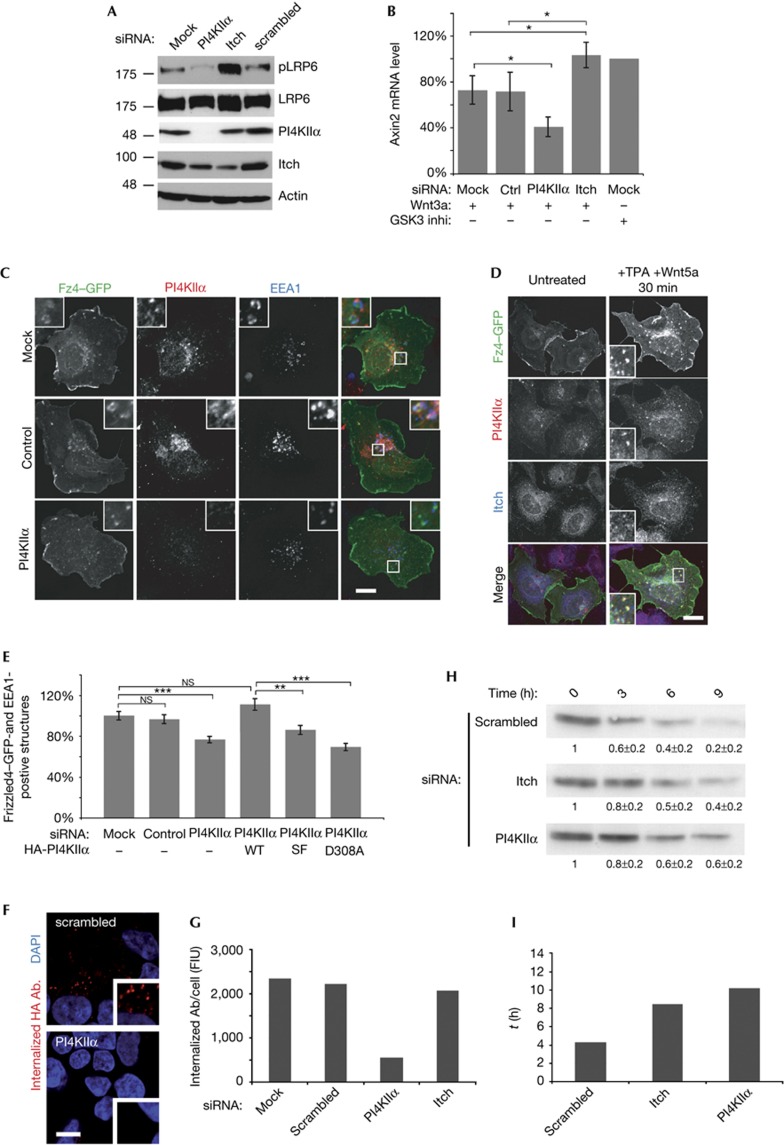
Figure 4.
PI4KIIα/Itch regulate endocytic traffic of Wnt-activated Fz and Wnt target gene expression. (A) Immunoblot of siRNA-treated Cos7 cells labelled for phosphorylated-LRP6 (pLRP6), LRP6, Itch, PI4KIIα and actin (loading control). (B) Axin 2 messenger RNA levels analysed by quantitative reverse transcriptase PCR in Wnt3a-stimulated cells transfected with indicated siRNAs. Axin 2 levels of Wnt3a-treated cells were normalized to mock-transfected cells treated with GSK3 inhibitor. (n=6), *P<0.05. (C) Confocal images of Wnt5a/TPA-stimulated HEK293 cells expressing Fz4–eGFP transfected with the indicated siRNAs or mock-transfected. Cells were stained for EEA1 and PI4KIIα. Scale bar, 12 μm. (D) Confocal images of Wnt5a/TPA-stimulated HEK293 cells expressing eGFP–Fz4, stained for endogenous PI4KIIα and Itch. Scale bar, 12 μm. (E) Quantitative analysis of Fz4–eGFP accumulation in EEA1-positive endosomes as shown in C (mean±s.e.m.; n=3). **P<0.001 and ***P<0.0001. (F) HEK293 cells stably expressing luminally HA-tagged Fz4, were silenced with indicated oligos and allowed to internalize anti-HA antibody for 15 min. Scale bar, 10 μm. (G) Quantification of E (mean from two independent experiments). (H) Degradation of HA–Fz4 in stably transfected HEK293 cells, silenced with the indicated siRNAs. Receptor levels at 0, 3, 6 and 9 h were analysed by immunoblotting for HA (values obtained by densitometry displayed in small inset). (I) τ values for degradation of HA–Fz4 determined from single exponential fits derived from the data exemplified in H (mean from two independent experiments). Both PI4KIIα- and Itch-depleted cells show slowed Fz4 degradation kinetics. DAPI, 4,6-diamidino-2-phenylindole; GFP, green fluorescent protein; HA, haemagglutinin; PI4KIIα, phosphatidylinositol 4-kinase type IIα; siRNA, short interfering RNA; TPA, 12-O-tetradecanoylphorbol-13-acetate.
As activated Fz receptors have been shown to undergo internalization and endosomal sorting [12], it is conceivable that Itch and/or PI4KIIα modulate these pathways. To gain insights into this question, we stimulated cells expressing Fz4–eGFP with Wnt5a to induce its internalization and downstream endosomal sorting during a time course of 40 min. As expected, activated Fz4–eGFP was internalized and sorted to endosomes that also contained EEA1 and PI4KIIα in juxtaposed puncta, possibly representing endosomal subdomains (Fig 4C, upper panel). Itch partially colocalized with PI4KIIα at endosomes positive for internalized Fz4 (Fig 4D) or the Wnt mediator Dvl2 (supplementary Fig S4C online). Depletion of PI4KIIα by siRNA-mediated knockdown reduced Wnt5a-triggered uptake and/or sorting of Fz4–eGFP into EEA1-positive endosomes (Fig 4C,E). This effect was fully rescued by re-expression of siRNA-resistant PI4KIIα (Fig 4E). Enzymatically inactive PI4KIIα (PI4KIIαD308A), although expressed at near identical levels (supplementary Fig S4D online), failed to rescue this phenotype, whereas the WW domain-binding defective, but catalytically active, mutant (PI4KIIαSF) (supplementary Fig S3C online) showed a reduced ability to restore the early endosomal localization of Fz4–eGFP (Fig 4E).
Compromised early endosomal targeting of internalized Fz4 could either reflect defective internalization, defective endosomal sorting or a combination of both. To distinguish between these possibilities, we measured the amount of endocytosed HA–Fz4 15 min post stimulation in antibody uptake experiments. Knockdown of PI4KIIα but not that of Itch inhibited endocytosis of HA–Fz4 (Fig 4F,G). No effect on transferrin uptake was observed under either condition (supplementary Fig S4F online). These results indicate that PI4KIIα is required for Fz internalization, whereas Itch is dispensable. Next, we carried out pulse-chase experiments to follow the degradative sorting of HA–Fz4. In control cells, HA–Fz4 was degraded with a tau value of about 4 h. Depletion of either PI4KIIα or Itch substantially delayed degradation of HA–Fz4 (Fig 4H,I). These data suggest that loss of Itch impairs degradative sorting of internalized Fz receptors, consistent with aggravated Wnt signalling and elevated Wnt target gene expression in Itch-knockdown cells (Fig 4A,B). Our results identify Itch as a negative regulator of canonical Wnt signalling and indicate a tentative model (supplementary Fig S4A online) whereby complex formation between Itch and PI4KIIα at endosomes facilitates degradative sorting of internalized Fz receptors.
We provide several lines of evidence for a functional interaction between PI4KIIα and the E3 ubiquitin ligase Itch. Furthermore, we show that Itch presumably via complex formation with PI4KIIα regulates degradative endocytic sorting of active Fz receptors. These results extend recent data about a crucial role for PI4KIIα [10, 11] and the ubiquitin machinery [12] in regulating Wnt/Fz signalling by providing a direct molecular link between both systems. Wnt-induced activation of Fz/LRP6 has been shown to trigger PI(4)P and subsequent PI(4,5)P2 synthesis [10] via Dvl-mediated stimulation of PI4KIIα [11] at the plasma membrane. Our data favour a model according to which PI4KIIα-mediated local production of PI(4,5)P2 facilitates canonical Wnt signalling [10] and PI(4,5)P2-mediated Fz receptor internalization into endosomes [21]. Association of Itch with PI4KIIα on Fz-containing endosomes might have a dual regulatory role. Itch-mediated inhibition of PI4KIIα-mediated PI(4)P synthesis on endosomes limits the Wnt signalling response, whereas ubiquitination of PI4KIIα might regulate further Ub-dependent degradative sorting of internalized Fz receptors [12]. Consistent with this possibility, ubiquitinated PI4KIIα preferentially interacted with endosomal sorting proteins, such as the VHS domain protein Tom1 and myoferlin. Conversely, ubiquitination of PI4KIIα inhibited its association with endocytic proteins, including SNX9, SNX18, endophilin and SGIP1 (supplementary Table S1 online). Hence, Itch-mediated ubiquitination of PI4KIIα might act as a switch that redirects the enzyme from the plasma membrane to a degradative endosomal pathway. In agreement with this hypothesis, absence of Itch leads to hyperactivation of Wnt signalling consistent with the increased half-life of Fz receptors under these conditions. Opposite effects on Wnt signalling and Fz degradation have been reported on loss of the de-ubiquitinating enzyme UBPy/USP8 [12]. Irrespective of the precise molecular function of PI4KIIα ubiquitination by Itch and perhaps other HECT domain E3 ligases, our findings unravel a new molecular link between phosphoinositide-regulated endosomal membrane traffic, ubiquitin and the modulation of Wnt signalling. It is conceivable if not likely that PI4KIIα–Itch have a similar regulatory role during endocytic sorting of other signalling receptors [8, 22, 23]. Future studies will need to address these issues.
Methods
Previously published methods were used for recombinant protein expression, cell culture, microscopy, affinity chromatography, immunoprecipitation and lipid kinase assays [24]. Supplementary methods: full description of materials and detailed protocols for the above, NMR spectroscopy, antibodies, plasmids and statistics.
Immunoprecipitation of PI4KIIα. For immunoprecipitating endogenous PI4KIIα from native tissue, eight uteri of adult mice were homogenized in 10 ml homogenization buffer (Buffer A: 20 mM Hepes pH 7.4, 100 mM NaCl or KCl, 2 mM MgCl2) supplemented with 1 mM PMSF and protease inhibitor cocktail. The crude homogenate was centrifuged for 20 min at 2,500g to remove unbroken material, and subsequently for 45 min at 80,000g. The resulting pellet was thoroughly resuspended in 5 ml of buffer A containing 1% Triton-X100. The lysate was cleared by centrifugation at 80,000g for 30 min. One millilitre of cleared lysate containing 1 mg of protein was then incubated with antibodies immobilized on protein A/G PLUS-agarose. Beads were washed extensively and eluted with SDS–polyacrylamide gel electrophoresis (PAGE) sample buffer.
Ubiquitination assays. For in vitro ubiquitination assays, 5 μg His6–Itch or His6–Itch (D830A) was incubated with 2.5 μg GST–PI4KIIα or GST–PI4KIIαSF in 100 μl reaction buffer containing 100 mM KCl, 20 mM Hepes pH 7.4, 2 mM MgCl2, 1 mM dithiothreitol, 4 mM ATP, 2 μg/ml E1 (UBE1, Boston Biochem), 2 μg/ml E2 (UbcH7, Boston Biochem) and 50 μg/ml mammalian ubiquitin (WT or 7R) (Boston Biochem). Samples were incubated for 30 min at 37 °C and the reactions were stopped by addition of SDS–PAGE sample buffer. Samples were analysed by SDS–PAGE and staining with Coomassie Blue or by immunoblotting.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Annie Angers and Klaus-Peter Knobeloch for plasmids. Supported by grants from the DFG (SFB740/C8; HA2686/4-1).
Author contributions: Experiments were carried out by J.M., M.K. and M.W. (cell biology, biochemistry, protein purification for NMR), F.G. and C.F. (NMR), and E.K. (MS/MS). J.M. and V.H. wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA (2008) Function and dysfunction of the PI system in membrane trafficking. EMBO J 27: 2457–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko B, Mao YS, Wlodarski P, Jung G, Binns DD, Sun HQ, Yin HL, Albanesi JP (2009) Palmitoylation controls the catalytic activity and subcellular distribution of phosphatidylinositol 4-kinase II{alpha}. J Biol Chem 284: 9994–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Balla T (2006) Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol 16: 351–361 [DOI] [PubMed] [Google Scholar]
- Szentpetery Z, Varnai P, Balla T (2010) Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA 107: 8225–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Craige B, Wainer BH, Guo J, De Camilli P, Faundez V (2005) Phosphatidylinositol-4-kinase type II alpha is a component of adaptor protein-3-derived vesicles. Mol Biol Cell 16: 3692–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL (2007) PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell 18: 2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minogue S, Waugh MG, De Matteis MA, Stephens DJ, Berditchevski F, Hsuan JJ (2006) Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci 119: 571–581 [DOI] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I (2011) The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 12: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W et al. (2008) Wnt3a-mediated formation of phosphatidylinositol 4,5-bisphosphate regulates LRP6 phosphorylation. Science 321: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Li L, Pan W, Wu D (2009) Regulation of phosphatidylinositol kinases and metabolism by Wnt3a and Dvl. J Biol Chem 284: 22544–22548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S (2010) Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J 29: 2114–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers A, Ramjaun AR, McPherson PS (2004) The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J Biol Chem 279: 11471–11479 [DOI] [PubMed] [Google Scholar]
- Otte L, Wiedemann U, Schlegel B, Pires JR, Beyermann M, Schmieder P, Krause G, Volkmer-Engert R, Schneider-Mergener J, Oschkinat H (2003) WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci 12: 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10: 398–409 [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 10: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH (1998) Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell 94: 829–839 [DOI] [PubMed] [Google Scholar]
- Morales B, Ramirez-Espain X, Shaw AZ, Martin-Malpartida P, Yraola F, Sánchez-Tilló E, Farrera C, Celada A, Royo M, Macias MJ (2007) NMR structural studies of the ItchWW3 domain reveal that phosphorylation at T30 inhibits the interaction with PPxY-containing ligands. Structure 15: 473–483 [DOI] [PubMed] [Google Scholar]
- Craige B, Salazar G, Faundez V (2008) Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell 19: 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund T, Pelham HR (2009) Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep 10: 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ (2003) Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science 301: 1391–1394 [DOI] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL (2003) The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell 5: 709–722 [DOI] [PubMed] [Google Scholar]
- Omerovic J et al. (2007) The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J 21: 2849–2862 [DOI] [PubMed] [Google Scholar]
- Krauss M, Kukhtina V, Pechstein A, Haucke V (2006) Stimulation of phosphatidylinositol kinase type I-mediated phosphatidylinositol (4,5)-bisphosphate synthesis by AP-2mu-cargo complexes. Proc Natl Acad Sci USA 103: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.