Biological research is in a crisis: the advent of high-throughput data generation has resulted in a deluge of empirical information from biological systems at every level of organization. In addition, scientists are increasingly realizing that biological systems, even the simplest ones, are extraordinarily complex. Generally, biology suffers from a lack of theoretical frameworks that could integrate both complexity and the amount of data so as to orient future research. In an attempt to address this problem, Nobel laureate Sydney Brenner argued that Turing machines have much in common with cells and living organisms; that cells and living organisms are, in fact, the best examples of Turing's and von Neumann's machines [1]. This is not a new concept; Brenner's comment is just the latest one among many others who have tried to explain the behaviour and abilities of living systems by comparing these to machines or computers.
However, we think that this analogy between cells and machines is misguided. It has not, and will not, contribute to our understanding of biological systems. In our opinion, such a mechanistic, bottom-up doctrine does not explain the true living nature of organisms as it ignores the top-down, context-dependent and agent-based aspects of living nature [2, 3, 4, 5, 6].
…biology suffers from a lack of theoretical frameworks that could integrate both complexity and the amount of data so as to orient future research
Similarly to Martin Nowak's explanation model for the evolution of human language 10 years earlier [5], or Manfred Eigen's attempts to explain genetic code by cybernetic information theory based on the Turing and von Neumann automaton model, Brenner confuses the mathematical theory of information, as it was developed by Alan Turing and John von Neumann, with information processing by living organisms. The former is a mathematical procedure, which depends on set algorithms to perform any tasks, and which only processes quantifiable sets of signs according to algorithm-based rules. By contrast, living organisms generate new sequences and new information without involving any algorithms. They are characterized by an inherent competence to use content-loaded signs, relevant for their survival, for signalling and communication [5]. These content-loaded signs are used at several levels of organization and complexity, starting with single molecules up to complex ecosystems. Moreover, to decode the context-dependent content of these signs, biological agents must be capable of their correct interpretation, that is, so-called context involvement. At the level of a single cell, the complexity of the content-loaded signs and their interpreter networks is already immense. Living systems are composed of interacting networks of autonomous agents, all capable of sensing signs, interpreting their content according to their context and acting on their own behalf [2,3,4].
…Brenner confuses the mathematical theory of information […] with information processing by living organisms
The crucial difference between a Turing and von Neumann machine and biological cell-based organisms is that the formal language needed to operate machines is not compatible with the natural languages of genetic codes and signalling networks. Whereas the first depends on formalizable algorithms, the latter cannot be formalized [6]. Natural language and communication are context dependent. This means that the same syntactic structure of content-loaded signs can transport different and even contradictory meanings for the language-using receiver, dependent on the context in which the language sequence is used [7]. By contrast, formalizable languages use a strict syntax to determine meaning independently of the contextual circumstances. ‘The shooting of the hunters’ can transport different meanings, which is obvious for every language-competent child; algorithm-based machines that use a formalized language are not able to pick up these differences.
In this respect, Brenner steps into a ‘mechanistic mouse trap’ when he matches artificial machines with natural organisms that he terms ‘natural machines’. How, then, should a natural machine look? Machines, no matter how complex and sophisticated, are inevitably the result of machine-building competences of human engineers, but an organism is not the mere result of a blueprint represented by its genes. First, context-dependent epigenetic markers control the activity of genes. These markers are not inscribed in the genome but are the result of internal processes, environmental factors and developmental history. Second, agency on its own behalf and interpretation of meaning—context-decoding—are obviously, and essentially, already present at the level of single proteins or RNA molecules [8,9,10]. Living organisms competently use species-specific signals to transport meanings to coordinate common cellular behaviour [5,6,7,11,12,13,14]. Organisms can even generate de novo sequences to adapt appropriately [11]. A machine can only simulate biocommunication, but it cannot communicate in such non-formalizable terms. Whereas children that use a natural language competently can speak in everyday communication about mathematics, and can change between mathematical assumptions and non-mathematical terms, this is completely impossible for machines.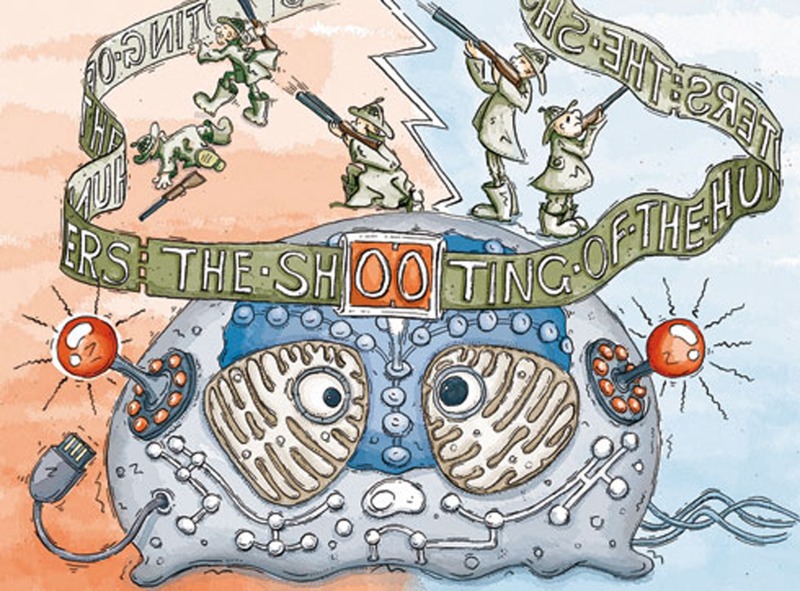
…living organisms generate new sequences and new information without involving any algorithms
The origin of genetic variations that are the substrate for selection and evolutionary change are probably the result of “errors” in the “copying machine”; therefore such “mutations would provide inheritable changes in the progeny” [1]. Even this concept does not accommodate the abundance of empirical data demonstrating that changes in the genetic code, which are of evolutionary relevance, are the result of fine-tuned processes by a large network of mobile genetic elements and persistent viruses that alter DNA sequences and that are the driving force behind the evolution of biological complexity [15,16,17]. As within this text, errors cannot generate new content-loaded sequences, but living organisms that are competent in generating correct sequences are generators, senders, receivers and interpreters of natural languages or codes. No natural language speaks itself as no natural code codes itself; it always requires living agents acting on their own behalf [2,3,4] that are competent in generating and interpreting these natural codes. In this respect, Brenner fails with his attempt to restore the Turing and von Neumann automaton model for biology. This approach cannot explain competent living agent-based signalling and communications between cells, tissues, organs and organisms [2,3,4,5,6,7]. To explain these competences of living agents and organisms in a non-reductive way, we have to include pragmatic communication theory that provides relevant explanations of signalling and communication in organisms. In addition to the three biological questions that Sydney Brenner highlighted—‘how does it work’, ‘how is it built’ and ‘how did it get that way’ [1]—we also need to ask ‘what does it mean?’ and ‘in which context?’.
Finally, we also do not agree with Brenner's criticism of Erwin Schrödinger for his statement that chromosomes act as an “architect's plan and builder's craft in one” [1]. The claim that chromosomes just represent a code script is wrong as chromosomes contain DNA sequences as well as proteins and RNA molecules. In fact, the amounts of proteins and RNAs are much higher than in DNA. Moreover, proteins and RNAs determine a three-dimensional architecture of the chromatin complex of interphase chromosomes, which then controls DNA expression. In this respect, the structural code reigns over the script code. In organisms, the upward (bottom-up) and downward (top-down) causations are inherently intertwined between numerous feedback interactions [18]. Therefore, claims that the ‘book of life’ is written in the genetic programme of the genome [1,19] are not embracing the whole complexity of living organisms.
No natural language speaks itself as no natural code codes itself; it always requires living agents […] that are competent in generating and interpreting these natural codes
In conclusion, the concept of genes as a symbolic representation of an organism—a code script—is not the whole story. Discoveries about viruses [20,22,23,24], most of which do not use DNA but rather RNA as a code script, also highlight the non-centrality of DNA-based code script for living agents. Agency, context, meaning, interpretation and behaviour are features of biological organization that can be independent of the DNA-based code script. In living organisms, these cognitive features feed back, in a top-to-bottom manner, into the DNA sequences [18,25].

Günther Witzany

František Baluška
Footnotes
The authors declare that they have no conflict of interest.
References
- Brenner S (2012) Life's code script. Nature 482: 461. [DOI] [PubMed] [Google Scholar]
- Kauffman SA (2000) Investigations. Oxford, UK: Oxford University Press [Google Scholar]
- Kauffman SA, Clayton P (2006) On emergence, agency, and organization. Biol Philos 21: 501–521 [Google Scholar]
- Kauffman SA (2007) Question 1: origin of life and the living state. Orig Life Evol Biosph 37: 315–322 [DOI] [PubMed] [Google Scholar]
- Witzany G (2011) Can mathematics explain the evolution of human language? Commun Integr Biol 4: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzany G (2000) Life: The Communicative Structure. A New Philosophy of Biology. Hamburg, Germany: Libri Books on Demand [Google Scholar]
- Baluška F, Witzany G (2012) Why biocommunication of plants? In Biocommunication of Plants (eds Witzany G, Baluška F), pp. v–vii. Berlin, Germany: Springer [Google Scholar]
- Málaga-Trillo E, Sempou E (2009) PrPs: proteins with a purpose. Lessons from the zebrafish. Prion 3: 129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fabret C, Namy O (2011) Are prions part of the dark matter of the cell? Prion 5: 129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ (2011) The good viruses: viral mutualistic symbioses. Nat Rev Microbiol 9: 99–108 [DOI] [PubMed] [Google Scholar]
- Shapiro JA (2011) Evolution: A View from the 21st Century. Upper Saddle River, New Jersey, USA: FT Press Science [Google Scholar]
- Baluška F (2011) Evolution in revolution. Paradigm shift in our understanding of life and biological evolution. Commun Integr Biol 4: 521–523 [Google Scholar]
- Trewavas A, Baluška F (2011) The ubiquity of consciousness. EMBO Rep 12: 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas A (2005) Plant intelligence. Naturwissenschaften 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Witzany G (2009) Non-coding RNAs. Persistent viral agents as modular tools for cellular needs. Ann NY Acad Sci 1178: 244–267 [DOI] [PubMed] [Google Scholar]
- Baluška F (2009) Cell-cell channels, viruses and evolution: via infection, parasitism and symbiosis toward higher levels of biological complexity. Ann NY Acad Sci 1178: 106–119 [DOI] [PubMed] [Google Scholar]
- Villarreal LP, Witzany G (2010) Viruses are essential agents within the roots and stem of the tree of life. J Theor Biol 262: 698–710 [DOI] [PubMed] [Google Scholar]
- Noble D (2012) A theory of biological relativity: no privileged level of causation. Interface Focus 2: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (2010) Sequences and consequences. Philos Trans R Soc Lond B Biol Sci 365: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM (2006) Viruses take center stage in cellular evolution. Genome Biol 7: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal LP (2005) Viruses and the Evolution of Life. Washington DC, USA: ASM [Google Scholar]
- Hegde NR, Maddur MS, Kaveri SV, Bayry J (2009) Reasons to include viruses in the tree of life. Nat Rev Microbiol 7: 615. [DOI] [PubMed] [Google Scholar]
- Ruiz-Saenz J, Rodas JD (2010) Viruses, virophages, and their living nature. Acta Virol 54: 85–90 [DOI] [PubMed] [Google Scholar]
- Forterre P (2010) Giant viruses: conflicts in revisiting the virus concept. Intervirology 53: 362–378 [DOI] [PubMed] [Google Scholar]
- Ellis GFR (2012) Top-down causation and emergence: some comments on mechanisms. Interface Focus 2: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]


