Abstract
A novel extensin gene has been identified in soybean (Glycine max L.) that encodes a hydroxyproline-rich glycoprotein (SbHRGP3) with two different domains. In this study expression of SbHRGP3 was investigated during soybean root development. SbHRGP was expressed in roots of mature plants, as well as seedlings, and showed a distinct pattern of expression during root development. The expression of SbHRGP3 increased gradually during root development of seedlings and reached a maximum while the secondary roots were maturing. The maximum expression level was contributed mainly by the secondary roots rather than by the primary root. Furthermore, expression of SbHRGP3 was preferentially detected in the regions undergoing maturation of the primary and secondary roots. These results imply that the expression of SbHRGP3 is regulated in an organ- and development-specific manner and that in soybean SbHRGP3 expression may be required for root maturation, presumably for the cessation of root elongation. Wounding and sucrose in combination enhanced expression of SbHRGP3 in roots, whereas both wounding and sucrose were required for the expression of SbHRGP3 in leaves.
The plant cell wall is complex in structure and consists of various macromolecules, such as carbohydrates, proteins, lignin, and cutin (Varner and Lin, 1989). There is cumulative information about the deposition of specific cell wall components and their gene expression in response to external or internal signals (Keller and Lamb, 1989; Ye and Varner, 1991; Wycoff et al., 1995). Structural arrangement of the cell wall components and their interactions have also been investigated (Varner and Lin, 1989). It is well documented that cellulose is the most abundant among the cell wall components and that the cellulose framework of the cell wall is interpenetrated by a cross-linked matrix of noncellulosic molecules, such as hemicellulose and pectin. Another network in the plant cell wall is made of structural proteins, which include HRGPs, GRPs, PRPs, solanaceous lectins, and arabinogalactan proteins (Showalter, 1993).
Extensins are a family of HRGPs and constitute the major protein components in cell walls of dicots (Showalter, 1993). A distinctive characteristic of dicot extensins is their repetitive Ser-Pro4 pentapeptides (Chen and Varner, 1985; Showalter and Varner, 1989). The pentapeptide blocks are conserved in almost all dicot extensins (Smith et al., 1986; Showalter and Varner, 1989) and have been reported in a gymnosperm extensin (Fong et al., 1992). A phylogenetic analysis revealed three major groups of extensins in dicots (Ahn et al., 1996). Extensins are glycoproteins composed of a polypeptide backbone and attached short carbohydrates. The majority of Pro residues in extensin polypeptides are hydroxylated, and most Hyp and Ser residues are glycosylated by posttranslational modifications (Kieliszewski and Lamport, 1994). Short arabinoside chains are attached to the Hyp residues, and single galactosyl residues are attached to some Ser residues. These carbohydrates are thought to be important in maintaining the structure of HRGPs as linear, rod-like molecules (Stafstrom and Staehelin, 1988). When secreted to the cell wall, extensins become insoluble, presumably due to the formation of covalent cross-linkages such as IDT bridges (Biggs and Fry, 1990; Iiyama et al., 1994; Qi et al., 1995).
It is known that the expression of extensin genes is not constitutive in the whole body of healthy plants, but they are regulated in a tissue-specific (Ye and Varner, 1991; Ye et al., 1991) and/or development-specific manner (Keller and Lamb, 1989). Expression of extensin genes is also induced and/or enhanced by various conditions and treatments (Showalter, 1993). This expression is induced by many cellular and environmental factors, such as mechanical wounding, fungal infection, viral infection, elicitors, and developmental signals. These indicate that conditions and tissue localization of extensin gene expression apparently vary from plant to plant and among cell and tissue types, presumably in accordance with the different functions in different cell types and tissues (for review, see Showalter, 1993).
We previously reported the isolation of a novel extensin gene (SbHRGP3) encoding an HRGP from soybean (Glycine max), which consists of two different domains in its polypeptide (Ahn et al., 1996). Histochemical analyses of transgenic tobacco (Nicotiana tabacum L.) seedlings carrying the SbHRGP3-β-glucuronidase chimeric gene (SbHRGP3-GUS) showed that SbHRGP3-GUS expression was activated by the maturation of the primary root and then inactivated; however, reactivation occurred just before the secondary root initiation specifically at the epidermis of the zone from which the secondary root was to be initiated. Its expression was also observed in the mature part of the secondary root. Furthermore, the expression of SbHRGP3-GUS was not induced by wounding alone in the transgenic plants; Suc was also required for its wound-inducible expression.
Because the expression of SbHRGP3 was regulated developmentally and spatially in transgenic tobacco seedlings, we further investigated the expression of SbHRGP3 in soybean to elucidate its actual role during soybean root development. Here we report that the expression of SbHRGP3 is organ specific and correlated with maturation of roots in soybean. We also show that SbHRGP3 is preferentially expressed in the region undergoing maturation of roots in soybean seedlings. Wounding and Suc in combination enhanced the expression of SbHRGP3 in soybean roots.
MATERIALS AND METHODS
Soybean (Glycine max cv Paldal) was used for all experiments. For RNA extraction from roots of soybean seedlings, seeds were germinated on wet filter paper and seedlings were grown in a modified 0.5× Hoagland solution (Hoagland and Arnon, 1950) in a 16-h light/8-h dark cycle at 28°C.
For stage-specific expression of SbHRGP3, total RNA was prepared from roots of 1-, 2-, 3-, 5-, 7-, 9-, and 13-d-old seedlings. The primary and secondary roots were harvested from seedlings and sectioned into slices for spatial expression of SbHRGP3.
Induction of SbHRGP3 Expression
To examine the effect of wounding and Suc on expression of SbHRGP3 in soybean roots, roots from 9-d-old seedlings were cut into 3- to 5-mm slices with a razor blade and floated on the modified 0.5× Hoagland solution with or without 3% Suc for 24 h. Unwounded roots were also floated on the solution with or without 3% Suc for 24 h. The same strategies were applied to leaves of soybean plants grown in field conditions (Ahn et al., 1996).
Isolation of Total RNA
Total RNA was isolated from various tissues of soybean as described by Chomczynski and Sacchi (1987). Excessive polysaccharides were removed by precipitation with sodium acetate according to the method of Mansson et al. (1985).
RNA Gel-Blot Analysis
An equivalent amount (10 μg) of total RNA was run on a 1% agarose gel containing formaldehyde (Sambrook et al., 1989) and transferred onto a Hybond N+ membrane (Amersham) as recommended by the manufacturer. The equivalence of RNA loading into each lane in the gel was verified by staining RNA with EtBr. Transferred RNA was fixed onto the membrane by UV cross-linking (Spectronics, Rochester, NY). The hybridization probe was a fragment containing the coding region of SbHRGP3, which was amplified by PCR, using the EXTP5 primer (5′-GGCAGTACCAAAACCCAAGATGGGGTCTC-3′ from positions −6 to +23) and EXTP6 primer (5′-TGAAGAAGCTTACTACTCGGGAAGCTAAG-3′ from positions +1325 to +1353; Ahn et al., 1996). The membrane was hybridized overnight with the random-primer-labeled probe in a solution of 50% formamide, 5× SSPE (1× SSPE is 0.15 m NaCl, 10 mm sodium phosphate, and 1 mm EDTA, pH 7.4), 5× Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% PVP, and 0.02% BSA), 0.1 mg/mL sheared salmon sperm DNA, and 0.5% SDS at 42°C. The hybridized membrane was washed in 2× SSPE and 0.1% SDS at room temperature, then in 1× SSPE and 0.1% SDS twice at 65°C, and finally in 0.1× SSPE and 0.1% SDS at 65°C for 5 min. For quantitation, the membrane was analyzed with a bioimaging analyzer (BAS-1500, Fuji, Tokyo, Japan).
Examination of Anatomical Features of Roots
The primary roots of soybean seedlings at various times after germination were serially cross-sectioned by hand using a double-edged razor blade. Differences in anatomical features, including maturation of xylem vessel members in the regions at the various distances from the root tip, were examined under a microscope (Optiphot-2, Nikon). The secondary roots of various lengths were also serially cross-sectioned and examined similarly.
RESULTS
Differential Regulation of SbHRGP3 during Soybean Development
In previous studies, SbHRGP3 was identified to be expressed in roots and hypocotyls of soybean seedlings (Hong et al., 1994; Ahn et al., 1996). Furthermore, SbHRGP3 was expressed at the specific stages of root development in transgenic tobacco seedlings (Ahn et al., 1996).
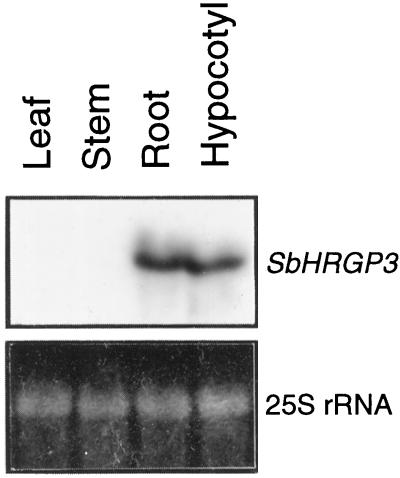
To examine the expression pattern of SbHRGP3 in mature soybean plants, we carried out an RNA gel-blot analysis with total RNAs isolated from leaves, stems, and roots of mature plants. Total RNA isolated from hypocotyls of 13-d-old seedlings of soybean was used as a positive control, because SbHRGP3 was first identified in hypocotyls of soybean seedlings (Hong et al., 1994). The expression pattern of SbHRGP3 in the organs of mature soybean plants is shown in Figure 1. Whereas SbHRGP3 mRNA was detected in roots, no expression of SbHRGP3 was observed in either leaves or stems. The expression level of SbHRGP3 was similar in the roots and hypocotyls. The size of the hybridized mRNA was 1.6 kb, as expected from the cDNA and genomic sequences (Hong et al., 1994; Ahn et al., 1996). These indicate that SbHRGP3 is expressed differentially in different organs during soybean development.
Figure 1.
Organ-specific expression of SbHRGP3 in soybean. Total RNA was extracted from leaves, stems, and roots of mature soybean plants grown in the field and subjected to gel-blot analysis. The RNA gel blot was hybridized with SbHRGP3-specific probe, as described in Methods. Total RNA prepared from hypocotyls of 13-d-old seedlings was used as a positive control. The bottom panel shows the EtBr-stained 25S rRNA in the gel, indicating that equal amounts of RNA were loaded in each lane.
Expression Pattern of SbHRGP3 during Root Development
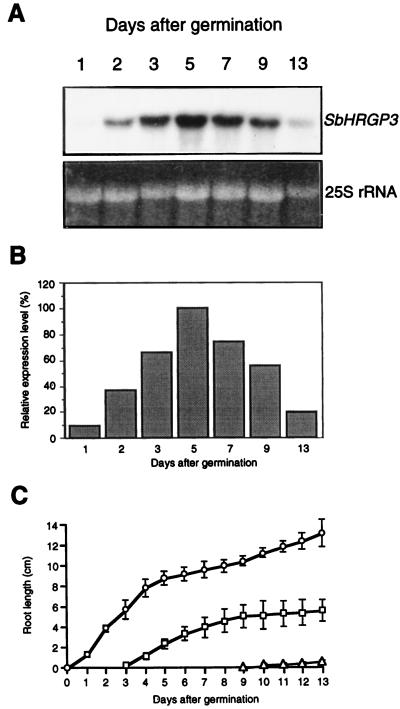
Because continuous expression of SbHRGP3 was observed only in roots during soybean development, we evaluated expression levels of SbHRGP3 in roots at different developmental stages. Total RNAs were extracted from roots of 1-, 2-, 3-, 5-, 7-, 9-, and 13-d-old seedlings and analyzed on an RNA gel blot. As shown in Figure 2, A and B, expression of SbHRGP3 was very low at the beginning of seed germination. Its expression in the roots subsequently increased until 5 DAG and then decreased gradually. This suggests that the expression of SbHRGP3 may be modulated in a development-specific manner during root development in soybean.
Figure 2.
Expression of SbHRGP3 in roots of soybean seedlings during development. A, Total RNA was extracted from roots of 1-, 2-, 3-, 5-, 7-, 9-, and 13-d-old seedlings and its expression levels were analyzed on gel blots. The bottom panel shows the EtBr-stained 25S rRNA in the gel. B, Relative expression levels of SbHRGP3 in roots of the seedlings in A were compared. C, The growth patterns of the primary, secondary, and tertiary roots in soybean seedlings were analyzed during seedling development. ○, Primary root; □, secondary root; and ▵, tertiary root.
Changes in the length of the primary, secondary, and tertiary roots of soybean during root development are shown in Figure 2C. After soybean seeds were germinated, the primary root grew rapidly until 4 DAG, and then its growth gradually decelerated. From 9 DAG, the primary root of soybean seedlings resumed its growth slowly. The secondary roots were developed from the primary root 3 DAG and extended until 8 DAG. The tertiary roots emerged 9 DAG. The aboveground parts began to extend 7 DAG; a pair of opposite, compound leaves developed first, followed by alternate leaves composed of small trifoliates.
We compared the expression pattern of SbHRGP3 with the growth pattern of soybean roots. As the primary root grew rapidly, the expression level of SbHRGP3 in the primary root increased simultaneously at least until 3 DAG (Fig. 2, A and B). The expression level of SbHRGP3 5 DAG reached the maximum during the root development, which was contributed by both the primary and secondary roots at the developmental stage. Deceleration of the secondary root growth from 7 DAG was followed by a decrease in the expression level of SbHRGP3. This indicates that the expression of SbHRGP3 in the roots is tightly linked with root development in soybean.
Predominant Expression of SbHRGP3 in the Secondary Roots
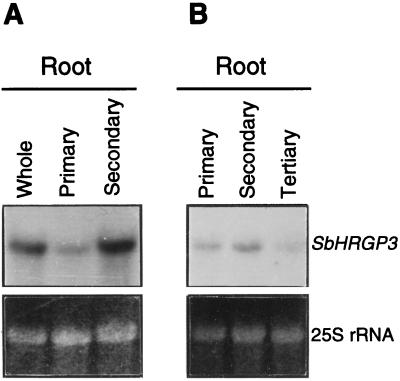
Because the maximum level of SbHRGP3 expression in the 5-d-old seedlings was contributed by both the primary and secondary roots (Fig. 2, A and B), we compared the relative expression levels of SbHRGP3 in the primary and secondary roots of the 5-d-old seedlings. Total RNAs of the primary roots and the secondary roots harvested separately from the seedlings were analyzed on RNA gel blots. As shown in Figure 3A, although the expression of SbHRGP3 was observed in both the primary and secondary roots, SbHRGP3 was mainly expressed in the secondary roots of the 5-d-old seedlings. The expression level in the secondary roots was about 4-fold higher than that in the primary root. This indicates that the expression level of SbHRGP3 in the primary root was highest in 3-d-old seedlings, whereas SbHRGP3 expression in the secondary roots reached the maximum in 5-d-old seedlings (Fig. 2, A and B). These results suggest that the expression of SbHRGP3 may be regulated in concert with the development of the primary and secondary roots.
Figure 3.
Expression of SbHRGP3 in roots of seedlings 5 DAG (A) and 20 DAG (B). The primary, secondary, and tertiary roots were dissected separately from the soybean seedlings. Total RNA was extracted from the roots and analyzed on gel blots. Total RNA extracted from the whole roots was used as a control in A. The bottom panels show the EtBr-stained 25S rRNA in the gels.
We also compared levels of SbHRGP3 mRNA in the primary, secondary, and tertiary roots of the 20-d-old plants. As shown in Figure 3B, the level of SbHRGP3 mRNA was still highest in the secondary roots, although the overall expression level in the 20-d-old plants was lower than that in the 5-d-old seedlings.
Preferential Expression of SbHRGP3 in the Region Undergoing Maturation in Soybean Roots
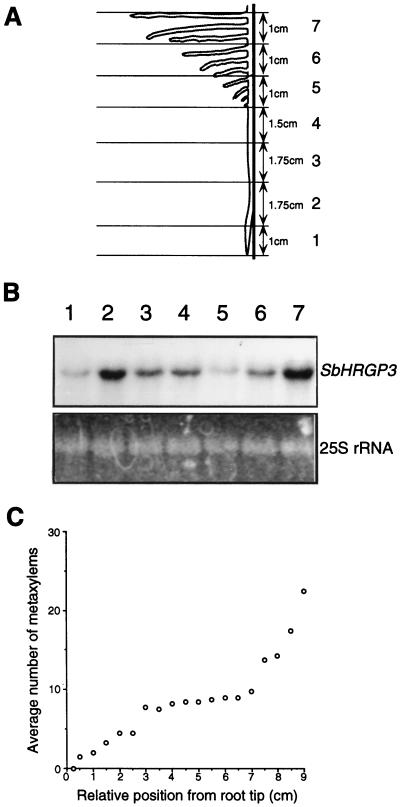
Because expression of SbHRGP3 is regulated in a development-specific manner during root development in soybean, we carried out an RNA gel-blot analysis to examine the spatial distribution of SbHRGP3 mRNA along the primary root axis in the 6-d-old seedlings that reached the decelerating phase (Fig. 2C). The primary root was serially dissected along the axis (Fig. 4A). Sections 5 to 7 also contained the secondary roots initiated from the primary root. Total RNAs isolated from each section were subjected to the RNA gel-blot analysis as shown in Figure 4B. Although the expression level along the primary root axis varied depending on the sections, it gradually increased and then decreased basipetally (Fig. 4B, lanes 1–4). The decrease in the SbHRGP3 expression level occurred ahead of the secondary root initiation.
Figure 4.
Spatial distribution of SbHRGP3 mRNA along the primary root axis of 6-d-old seedlings. A, The primary root was serially dissected along the root axis as shown. B, Total RNA was extracted from each section in A and analyzed on a gel blot. The bottom panel in B shows the EtBr-stained 25S rRNA in the gel. Sections 5 to 7 also included the secondary roots initiated from the primary root. C, The number of metaxylems in each xylem arch of the regions at the various distances from the root tip.
It was interesting that section 5, from which the secondary roots began to emerge (Fig. 4A), showed the lowest level of expression among the sections. However, as sections moved toward the root-hypocotyl transition zone, the expression levels gradually increased again (Fig. 4B, lanes 5–7). The increase in the expression level of SbHRGP3 from section 5 to section 7 was contributed mainly by the secondary roots (Fig. 4A). This suggests that the distribution of SbHRGP3 mRNA may vary spatially along the primary root axis, depending on the developmental stages of the sections.
To investigate anatomical features of the root region showing the maximum level of SbHRGP3 expression in the primary root, we cross-sectioned the primary root along the axis and analyzed the number of metaxylems in each xylem arch as shown in Figure 4C. The soybean root typically contained a tetrarch structure in each position; however, the number of metaxylems in each arch of the different regions varied. The region of 3 cm from the root tip increased gradually in the number of metaxylems basipetally in each xylem arch. The region 3 to 7 cm from the root tip maintained nearly the same number of metaxylems (10), whereas the region from 7 cm to the root-hypocotyl transition zone increased again in the number of metaxylems, because of the structural transition of the tetrarch pattern into the typical ring-like form found in stems. The first metaxylem appeared in the region 0.5 to 0.75 cm from the root tip in the 6-d-old seedlings, which includes the regions of cell division and elongation (Rost and Baum, 1988). Therefore, the region 0.75 to 3 cm from the root tip is considered to be undergoing maturation, whereas the region from 3 to 7 cm is the mature region.
As shown in Figure 4B, SbHRGP3 was expressed at the highest level in the region 1 to 2.75 cm from the root tip. This indicates that in soybean the SbHRGP3 expression begins in the region of elongation, reaches a maximum level in the region undergoing maturation, and then ceases in the mature region of the primary root.
Expression Pattern of SbHRGP3 in the Secondary Root of Soybean
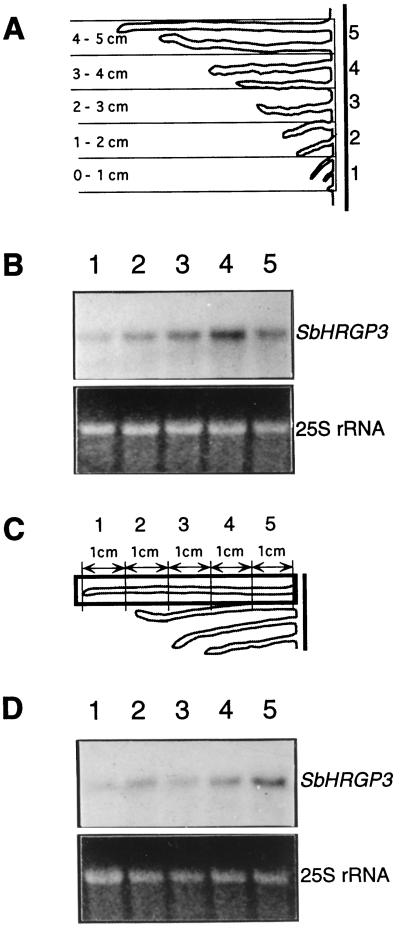
Because expression of SbHRGP3 in the 5-d-old seedlings was mainly observed in the secondary roots (Fig. 3), we also investigated the stage-specific expression of SbHRGP3 during the secondary root development. The secondary roots were harvested according to their lengths from the 5-d-old seedlings (Fig. 5A), and total RNAs isolated from the secondary roots were subjected to RNA gel-blot analysis (Fig. 5B). As shown in Figure 5B, expression levels of SbHRGP3 increased as the secondary roots grew, reaching a maximum in the secondary roots (3–4 cm) and then decreasing as the secondary root growth decelerated. This indicates that expression of SbHRGP3 is regulated temporally during secondary root development, as observed in the primary root (Fig. 2).
Figure 5.
Temporal and spatial expression levels of SbHRGP3 in secondary roots during seedling development. A, The secondary roots of soybean were dissected from the primary root and grouped according to their lengths. B, Total RNA was prepared from each group and analyzed on an RNA gel blot. C, The secondary 5-cm-long roots were dissected. D, Total RNA was prepared from each section and analyzed on a gel blot. The bottom panels in B and D show the EtBr-stained 25S rRNA in the gels.
We also carried out RNA gel-blot analysis to elucidate the spatial distribution of SbHRGP3 mRNA in the secondary roots. The 5-cm long secondary roots that reached the decelerating phase of growth (Fig. 2C) were harvested from the seedlings and were serially dissected as depicted in Figure 5C. Total RNAs from the sections were subjected to RNA gel-blot analysis. As shown in Figure 5D, SbHRGP3 mRNA was detected in the entire secondary roots; however, levels of SbHRGP3 mRNA gradually increased toward the primary-secondary root transition zone.
In the secondary root, unlike in the primary root, two protoxylem members first differentiate centripetally into the metaxylems, which is similar to the diarch structure. The other two protoxylem members subsequently develop into metaxylems, thereby forming the typical tetrarch structure, as observed in the primary root. Investigation of the anatomical features of the secondary root indicated that the whole 5-cm-long secondary root was still undergoing maturation (data not shown). This indicates that the secondary root differentiates slowly and that its developmental process is different from that of the primary root.
As shown in Figures 2 and 3, the expression level of SbHRGP3 in the primary root reached a maximum 3 DAG, whereas the expression level in the secondary root reached a maximum 5 DAG. Furthermore, the sections exhibiting the maximum level of SbHRGP3 expression in the primary (Fig. 4A, section 2) and secondary roots (Fig. 5C, section 5) were undergoing maturation. This suggests that the expression of SbHRGP3 may be closely related to maturation and, presumably, cessation of cell elongation in roots of soybean.
Wounding and Suc in Combination Enhanced the Expression of SbHRGP3 in Soybean Roots
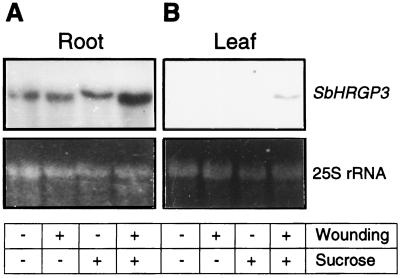
We previously demonstrated that wounding and Suc in combination induce the expression of SbHRGP3 in leaves of transgenic tobacco plants (Ahn et al., 1996). To examine effects of wounding and Suc on the SbHRGP3 expression in soybean roots, we wounded roots of 9-d-old seedlings and floated them on 3% Suc solution for 24 h. Total RNA was extracted from the roots and subjected to RNA gel-blot analysis. As shown in Figure 6A, SbHRGP3 was inherently expressed in soybean roots, and neither wounding nor Suc alone affected the expression. However, wounding and Suc in combination enhanced the expression of SbHRGP3 2-fold. The enhancement in the expression by both wounding and Suc was not limited to the 9-d-old seedlings; it was also observed in 5-d-old seedlings (data not shown). In contrast to the roots, both wounding and Suc were required for the induction of SbHRGP3 expression in soybean leaves (Fig. 6B).
Figure 6.
Enhancement of the inherent expression of SbHRGP3 by the combination of wounding and Suc in roots of 9-d-old soybean seedlings. A, Roots of 9-d-old seedlings were wounded and floated on the solution with 3% Suc for 24 h, and then total RNA was extracted and analyzed on a gel blot. Total RNAs were also prepared from wounded roots floated on the solution without Suc, unwounded roots on the solution with 3% Suc, and unwounded roots on the solution without Suc for 24 h. B, Levels of SbHRGP3 mRNA in leaves of mature soybean plants grown in the field were analyzed on an RNA gel blot similarly to the method used for roots. The bottom panels show the EtBr-stained 25S rRNA in the gels.
DISCUSSION
We examined the expression pattern of a novel extensin gene, SbHRGP3, which encodes an HRGP with two different domains during root development in soybean. SbHRGP3 was differentially regulated in different organs at different developmental stages, showing root-specific expression in mature soybean plants and hypocotyl and root expression in seedlings (Hong et al., 1994; Ahn et al., 1996). Its expression was also modulated in a development-specific manner in roots during seedling development. Furthermore, SbHRGP3 was expressed mainly in the regions undergoing maturation in the primary and secondary roots, as was also observed in transgenic tobacco plants (Ahn et al., 1996). Assuming that the levels of SbHRGP3 mRNA actually correlate with those of SbHRGP3 protein, these results suggest that SbHRGP3 may be involved in the cessation of cell elongation in soybean roots.
Expression of SbHRGP3 Is Temporally Regulated during Root Development in Soybean
Recently, the root system has been focused on as a useful system for understanding organ development because roots are relatively simple organs, their growth pattern is uniform, and they have a small number of differentiated cell types (Aeschbacher et al., 1994). In particular, the development of new roots from existing roots provides a novel opportunity to investigate cellular development and differentiation. Anatomical examinations have shown that there are considerable differences between the primary root and the secondary root (Dolan et al., 1993), suggesting that the organization of the secondary root could be controlled by a different set of factors than the primary root. Furthermore, the process of secondary root development is divided into a number of specific stages (Laskowski et al., 1995).
In this study we investigated the pattern and location of SbHRGP3 expression during root development in soybean. Because SbHRGP3 expression was observed in roots of mature soybean plants (Fig. 1), we evaluated the expression pattern of SbHRGP3 in roots during seedling development in more detail. As shown in Figure 2, SbHRGP3 expression increased until 5 DAG and then decreased. When the expression level of SbHRGP3 reached a maximum in the root system of seedlings, this occurred mainly in the secondary roots (Fig. 3). Furthermore, SbHRGP3 was mainly expressed in the maturing regions of both the primary and secondary roots just before their growth decelerated (Figs. 4 and 5). These results indicate that the expression of SbHRGP3 is related to the growth of the root system as a whole.
Although it is known that the organization and development of secondary roots are different from those of the primary root (Dolan et al., 1993; Laskowski et al., 1995), much is still unknown about the genetic program of secondary root development. This is partly because only a few genes related to the organization and development of the secondary roots have been isolated and characterized so far. Proteins encoded by these genes include an HRGP from tobacco (HRGPnt3) (Keller and Lamb, 1989; Vera et al., 1994) and a novel protein from Arabidopsis thaliana (Lateral Root Primordia 1; Smith and Fedoroff, 1995). HRGPnt3 was transiently expressed in a subset of the pericycle and endodermis during initiation of the secondary roots in tobacco, and its expression continued in the cells at the tip of the emerging secondary roots.
Expression of the gene for Lateral Root Primordia 1 was activated during the early stage of root primordium development and was turned off prior to the emergence of the secondary roots from the primary root. SbHRGP3 also seems to play an important role in secondary root development. SbHRGP3 expression was specific to the epidermal cells of the zone from which the secondary root was to be initiated in the transgenic tobacco plants (Ahn et al., 1996). The region of the primary root showing a high level of SbHRGP3 expression (Fig. 4A, section 4) was ahead of the region from which the secondary roots were emerging (Fig. 4A, section 5). This confirms that SbHRGP3 has a special structural function not only in maturation but also in hardening of the epidermal cells in the primary root to reduce the severe damage caused by emerging secondary roots. Therefore, HRGPnt3 and SbHRGP3 seem to share some common features in that they are regulated developmentally during secondary root development. It is likely that HRGPnt3 plays a role in the initiation of the secondary roots, whereas SbHRGP3 is involved in the cell wall reformation required for initiation of the secondary roots.
Histochemical assays with mature transgenic tobacco plants containing SbHRGP3-GUS support the role of SbHRGP3 in root maturation, as was previously suggested in transgenic tobacco seedlings (Ahn et al., 1996). The expression of SbHRGP3-GUS was dispersed in the epidermal cells of maturing regions in roots of mature, transgenic plants (data not shown).
SbHRGP3 Is Involved in Maturation of Roots in Soybean
HRGPs have been known to play an important structural role in the plant cell wall. Associated with this structural role, the glycoproteins probably play another active role in cell growth and development, because levels of HRGP expression vary depending on tissue types and developmental stages (Showalter, 1993). It was previously proposed that high levels of HRGPs are correlated with the cessation of cell elongation (i.e. extension; Cleland and Karlsnes, 1967; Sadava and Chrispeels, 1973; Monro et al., 1974). As extension ceases, the cell walls become more rigid and less extensible, probably because of increased cross-linking of cell wall polysaccharides, increased amounts of the cell wall proteins, and/or increased peroxidase activity in the region of maturation of the root (Lyndon, 1990). In addition, Carpita and Gibeaut (1993) recently suggested a possible role of extensins in locking primary walls into a definite shape at the end of cell elongation.
The growing root has been traditionally divided into three regions: the regions of cell division, elongation, and maturation basipetally along the primary root axis (Esau, 1977; Raven et al., 1992). However, these regions cannot be clearly delimited from each other but, rather, overlap to some extent. Most cell division takes place in the region of cell division, which includes the apical meristem and the nearby portion of root. Behind this region is the region of elongation, which is usually only a few millimeters in length. Most of the increase in root length results from cell elongation in this region. The region of elongation is followed by the region of maturation, in which most of cells of the primary tissues mature. Protoxylem elements also begin to mature basipetally in the region of maturation. Other anatomical features, including the cell number of the cortex, epidermis, and pericycle, as well as their cell size and number of cell layers, showed little variation along the entire primary root axis. Therefore, protoxylem maturation is considered to be a marker to delimit the region of maturation from other regions in the root (Fahn, 1990).
We examined the number of mature metaxylem vessel members during soybean root development, especially in the 6-d-old seedlings (Fig. 4C), and found that the region with the increasing number of metaxylem vessel members 0.75 to 3 cm from the root tip is undergoing maturation, whereas the region from 3 to 7 cm, which maintains a constant number of metaxylem vessel members, has completed maturation. Because the expression of SbHRGP3 reached a maximum in the region undergoing maturation (Figs. 4 and 5), SbHRGP3 may play an important role in root maturation in soybean.
SbHRGP3 has two domains, each having different motifs (Ahn et al., 1996). In addition to the Ser-Pro4 pentapeptide blocks, the repeat unit in domain 1 contains the Tyr-Tyr-Tyr-His block, whereas the repeat unit in domain 2 contains the Val-Tyr-Lys-Tyr-Lys block. The two blocks have been suggested to be involved in intra- and intermolecular IDT cross-linking (Kieliszewski and Lamport, 1994).
The repeat unit of domain 2 also contains unusual repetitive sequences (Tyr-Lys-Tyr-Pro) that may also participate in the IDT bridge formation. The presence of these putative functional sites suggests that SbHRGP3 could be classified as an inter- and intramolecular cross-linking extensin (Kieliszewski and Lamport, 1994). The repeat unit in domain 1 of SbHRGP3 has the Tyr-rich block with His instead of conventional Lys residues and contains Ser-Pro4-Lys-His-Ser instead of the palindromic Ser-Pro4-Ser-Pro-Ser-Pro4 sequence. The repeat unit has been reported for only SbHRGP3 and a bean extensin (Wycoff et al., 1995). The His residues of SbHRGP3 have been suggested to serve as ionic interaction sites for protein-carbohydrate or protein-protein interactions in cell wall formation (Ahn et al., 1996). All of these features suggest that SbHRGP3 may actively interact with other cell wall components and/or with itself in soybean roots, thereby forming a rigid network in the cell wall.
Root elongation is considered to occur as a result of expansion of the epidermal cells in the region of elongation (Hasenstein and Evans, 1988). Once cell elongation has been completed in roots, the primary wall is locked into shape. The locking mechanism may involve extensin as one component (Carpita and Gibeaut, 1993). Incorporation of extensin into the cell wall prepares the cell to stop elongating. Along with the structural properties of SbHRGP3 and its expression in the epidermal cells of roots (Ahn et al., 1996), high levels of SbHRGP3 expression in the region undergoing maturation suggest that SbHRGP3 deposition in the cell wall may be required for the maturation of epidermal cells in soybean roots. It also suggests that SbHRGP3 in the cell walls of epidermal cells may cross-link to itself and/or to the other cell wall components, thereby leading not only to the cessation of cell elongation but also to the prevention of further elongation.
Expression of PRPs, another major family of structural proteins, has also been described during organ development in soybean (Ye et al., 1991; Showalter, 1993). A recent study showed that secretion of PRP2 was correlated with lignification of the secondary walls of protoxylem elements in soybean hypocotyls (Ryser et al., 1997). PRP2 and GRP1.8 were shown to be involved in the formation of a network interconnecting the ring- or spiral-shaped secondary wall thickenings of protoxylem elements and xylem parenchyma cells. PRP2 and GRP1.8 had different functions in the terminal phase of protoxylem differentiation in soybean hypocotyls. Another PRP, SbPRP1, was also found to be localized in several cell types of soybean hypocotyls (Wyatt et al., 1992). SbPRP1 expression increased within epidermal cells in the elongating and mature regions of the hypocotyl. Furthermore, its promoter drove GUS expression in the apical and elongating regions of both primary and lateral roots, most strongly at the epidermis in transgenic tobacco (Suzuki et al., 1993). SbPRP1 expression was localized to the actively growing region of the root, and this expression was temporally regulated during very early stages of seedling growth. These results suggest that different cell wall proteins may play different roles in various cell types during root development.
Inherent Expression of SbHRGP3 in Roots Was Enhanced by Wounding and Suc
Wounding and Suc in combination enhanced the inherent expression of SbHRGP3 in soybean roots, whereas both wounding and Suc were required for the induction of the expression of SbHRGP3 in soybean leaves (Fig. 6). Many extensin genes were reported to be wound inducible, with rare exceptions (Showalter, 1993). SbHRGP3 was the first extensin gene with a wound-inducible expression that required Suc. Although it was previously proposed that Suc is an important factor in wound response (Johnson and Ryan, 1990; Kim et al., 1991), it only enhanced the wound inducibility of other plant genes.
Signal transduction pathways for sugar are well characterized in prokaryotes (Vyas et al., 1988; Åqvist and Mowbray, 1995); however, relatively little is known about the molecular and biochemical mechanisms underlying sugar responses in multicellular eukaryotes, especially in sugar-producing higher plants. Because higher plants synthesize and accumulate sugars through photosynthesis, sugars were thought to be energy sources for growth and development rather than signal molecules. However, it has become apparent that sugars are physiological signals repressing or activating plant genes involved in many essential processes (Cheng et al., 1992; Jang and Sheen, 1994; Mita et al., 1995; Ni et al., 1996). Recently, Jang et al. (1997) reported that hexokinases might act as a sugar sensor in the signal transduction pathway mediating Glc-regulated gene expression in various organisms, including yeast, mammals, and plants. This suggests that the role of hexokinase in sugar sensing and signaling is evolutionarily conserved in all eukaryotes.
We have shown that SbHRGP3 is regulated in an organ- and development-specific manner during root development in soybean. Wounding and Suc affect the expression of SbHRGP3 in various tissue types, suggesting that distinct signal transduction pathways are probably involved in the regulation of SbHRGP3 in different tissues. Further study will be needed to elucidate the regulation mechanisms of SbHRGP3 in response to different signals in different tissues.
Abbreviations:
- DAG
days after germination
- EtBr
ethidium bromide
- GRP
Gly-rich protein
- HRGP
Hyp-rich glycoprotein
- IDT
isodityrosine
- PRP
Pro-rich protein
- SbHRGP3
soybean HRGP3
Footnotes
This work was supported in part by the Korea Science and Engineering Foundation through the Research Center for Cell Differentiation (J.S.L.) and the Research Center for Agricultural Bio-Materials (Y.D.C.) at Seoul National University.
LITERATURE CITED
- Aeschbacher RA, Schiefelbein JW, Benfey PN. The genetic and molecular basis of root development. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:25–45. [Google Scholar]
- Ahn JH, Choi Y, Kwon MY, Kim S-G, Choi YD, Lee JS. A novel extensin gene encoding a hydroxyproline-rich glycoprotein requires sucrose for its wound-inducible expression in transgenic plants. Plant Cell. 1996;8:1477–1490. doi: 10.1105/tpc.8.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åqvist J, Mowbray SL. Sugar recognition by a glucose/galactose receptor. J Biol Chem. 1995;270:9978–9981. [PubMed] [Google Scholar]
- Biggs KJ, Fry SC. Solubilization of covalently bound extensin from Capsicum cell walls. Plant Physiol. 1990;92:197–204. doi: 10.1104/pp.92.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Varner JE. An extracellular matrix protein in plants: characterization of a genomic clone for carrot extensin. EMBO J. 1985;4:2145–2151. doi: 10.1002/j.1460-2075.1985.tb03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-L, Acedo GN, Cristinsin M, Conkling M. Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA. 1992;89:1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidine thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cleland R, Karlsnes AM. A possible role of hydroxyproline-containing proteins in the cessation of cell elongation. Plant Physiol. 1967;42:669–671. doi: 10.1104/pp.42.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organization of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants, Ed 2. New York: John Wiley & Sons; 1977. [Google Scholar]
- Fahn A. Plant Anatomy, Ed 4. Oxford, UK: Butterworth-Heinemann; 1990. [Google Scholar]
- Fong C, Kieliszewski MJ, de Zacks R, Leykam JF, Lamport DTA. A gymnosperm extensin contains the serine-tetrahydroxyproline motif. Plant Physiol. 1992;99:548–552. doi: 10.1104/pp.99.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenstein KH, Evans ML. Effects of cations on hormone transport in primary roots of Zea mays. Plant Physiol. 1988;86:890–894. doi: 10.1104/pp.86.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DAI (1950) The water culture method of growing plants without soil. Calif Agric Ext Serv Circ 347
- Hong JC, Cheong YH, Nagao RT, Bahk JD, Cho MJ, Key JL. Isolation and characterization of three soybean extensin cDNAs. Plant Physiol. 1994;104:793–796. doi: 10.1104/pp.104.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiyama K, Lam TB-T, Stone BA. Covalent cross-links in the cell wall. Plant Physiol. 1994;104:315–320. doi: 10.1104/pp.104.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, León P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Sheen J. Sugar sensing in higher plants. Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Ryan CA. Wound-inducible potato inhibitor II genes: enhancement of expression by sucrose. Plant Mol Biol. 1990;14:527–536. doi: 10.1007/BF00027498. [DOI] [PubMed] [Google Scholar]
- Keller B, Lamb CJ. Specific expression of a novel cell wall hydroxyproline-rich glycoprotein gene in lateral root initiation. Genes Dev. 1989;3:1639–1646. doi: 10.1101/gad.3.10.1639. [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. Extensin: repetitive motifs, functional sites, posttranslational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- Kim S-R, Costa MA, An G. Sugar response element enhances wound response of potato proteinase inhibitor II promoter in transgenic tobacco. Plant Mol Biol. 1991;17:973–983. doi: 10.1007/BF00037137. [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex I. Formation of lateral root meristem is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Lyndon RF (1990) Plant Development: The Cellular Basis. Unwin Hyman, London
- Mansson P-E, Hsu D, Stalker D. Characterization of fruit specific cDNAs from tomato. Mol Gen Genet. 1985;200:356–361. [Google Scholar]
- Mita S, Suzuki-Fujii K, Nakamura K. Sugar-inducible expression of a gene for β-amylase in Arabidopsis thaliana. Plant Physiol. 1995;107:895–904. doi: 10.1104/pp.107.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro JA, Bailey RW, Penny D. Cell wall hydroxyproline-polysaccharide association in Lupinus hypocotyls. Phytochemistry. 1974;13:375–382. [Google Scholar]
- Ni M, Cui D, Gelvin SB. Sequence-specific interactions of wound-inducible nuclear factors with mannopine synthase 2′ promoter wound-responsive elements. Plant Mol Biol. 1996;30:77–96. doi: 10.1007/BF00017804. [DOI] [PubMed] [Google Scholar]
- Qi X, Behrens BX, West PR, Mort AJ. Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension cultures. Evidence for a covalent cross-link between extensin and pectin. Plant Physiol. 1995;108:1691–1701. doi: 10.1104/pp.108.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven PH, Evert RF, Eichhorn SE (1992) Biology of Plants, Ed 5. Worth Publishers, New York
- Rost TL, Baum S. On the correlation of primary root length, meristem size and protoxylem tracheary element position in pea seedlings. Am J Bot. 1988;75:414–424. [Google Scholar]
- Ryser U, Schorderet M, Zhao GF, Studer D, Ruel K, Hauf G, Keller B. Structural cell-wall proteins in protoxylem development: evidence for a repair process mediated by a glycine-rich protein. Plant J. 1997;12:97–111. doi: 10.1046/j.1365-313x.1997.12010097.x. [DOI] [PubMed] [Google Scholar]
- Sadava D, Chrispeels MJ. Hydroxyproline-rich cell wall protein (extensin): role in the cessation of elongation in excised pea epicotyls. Dev Biol. 1973;30:49–55. doi: 10.1016/0012-1606(73)90047-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Varner JE (1989) Plant hydroxyproline-rich glycoproteins. In A Marcus, ed, The Biochemistry of Plants: A Comprehensive Treatise, Vol 15. Academic Press, New York, pp 485–520
- Smith DL, Fedoroff NV. LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell. 1995;7:735–745. doi: 10.1105/tpc.7.6.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Muldoon EP, Willard JJ, Lamport DTA. Tomato extensin precursors P1 and P2 are highly periodic structures. Phytochemistry. 1986;25:1021–1030. [Google Scholar]
- Stafstrom JP, Staehelin LA. Antibody localization of extensin in cell walls of carrot storage roots. Planta. 1988;174:321–332. doi: 10.1007/BF00959517. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Fowler TJ, Tierney ML. Deletion analysis and localization of SbPRP1, a soybean cell wall protein gene, in roots of transgenic tobacco and cowpea. Plant Mol Biol. 1993;21:109–119. doi: 10.1007/BF00039622. [DOI] [PubMed] [Google Scholar]
- Varner JE, Lin LS. Plant cell wall architecture. Cell. 1989;56:231–239. doi: 10.1016/0092-8674(89)90896-9. [DOI] [PubMed] [Google Scholar]
- Vera P, Lamb C, Doerner PW. Cell-cycle regulation of hydroxyproline-rich glycoprotein HRGPnt3 gene expression during the initiation of lateral root meristems. Plant J. 1994;6:717–727. [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- Wyatt RE, Nagao RT, Key JL. Patterns of soybean proline-rich protein gene expression. Plant Cell. 1992;4:99–110. doi: 10.1105/tpc.4.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wycoff KL, Powell PA, Gonzales RA, Corbin DR, Lamb C, Dixon RA. Stress activation of a bean hydroxyproline-rich glycoprotein promoter is superimposed on a pattern of tissue-specific developmental expression. Plant Physiol. 1995;109:41–52. doi: 10.1104/pp.109.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Song Y-R, Marcus A, Varner JE. Comparative localization of three classes of cell wall proteins. Plant J. 1991;1:175–183. doi: 10.1111/j.1365-313x.1991.00175.x. [DOI] [PubMed] [Google Scholar]
- Ye Z-H, Varner JE. Tissue-specific expression of cell wall proteins in developing soybean tissues. Plant Cell. 1991;3:23–37. doi: 10.1105/tpc.3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]