Abstract
Opening of the MPT (mitochondrial permeability transition) pore is a critical event in mitochondrial-mediated cell death. However, with the exception of CypD (cyclophilin D), the exact molecular composition of the MPT pore remains uncertain. C1qbp (complement 1q-binding protein) has recently been hypothesized to be an essential component of the MPT pore complex. To investigate whether C1qbp indeed plays a critical role in MPT and cell death, we conducted both gain-of-function and loss-of-function experiments in MEFs (mouse embryonic fibroblasts). We first confirmed that C1qbp is a soluble protein that localizes to the mitochondrial matrix in mouse cells and tissues. Similarly, overexpression of C1qbp in MEFs using an adenovirus resulted in its exclusive localization to mitochondria. To our surprise, increased C1qbp protein levels actually suppressed H2O2-induced MPT and cell death. Antithetically, knockdown of endogenous C1qbp with siRNA (small interfering RNA) sensitized the MEFs to H2O2-induced MPT and cell death. Moreover, we found that C1qbp could directly bind to CypD. Therefore C1qbp appears to act as an endogenous inhibitor of the MPT pore, most likely through binding to CypD, and thus protects cells against oxidative stress.
Keywords: C1qbp, cell death, cyclophilin D, mitochondria, mitochondrial permeability transition (MPT), oxidative stress
INTRODUCTION
The MPT (mitochondrial permeability transition) pore is a large, non-specific channel that spans the inner mitochondrial membrane and is known to mediate the lethal permeability changes that initiate mitochondrial-driven cell death [1–3]. MPT pore opening leads to loss of mitochondrial electrochemical potential (Δψm), cessation of ATP synthesis, increased ROS (reactive oxygen species) production, mitochondrial swelling and rupture, and, eventually, cell death [1–3]. Thus, the MPT pore has been shown to be a causative factor in the pathogenesis of many diseases states, including ischaemia/reperfusion injury [4,5], muscular dystrophy [6,7], neurodegenerative diseases [8,9] and drug-induced organ toxicities [10,11].
However, the exact components that make up the MPT pore remain unknown. It was originally proposed that the MPT pore consisted of VDAC (voltage-dependent anion channel) in the outer membrane, ANT (adenine nucleotide translocase) in the inner membrane and CypD (cyclophilin D) in the matrix [1–3]. Unfortunately, studies in mice genetically deficient for each of these components have questioned the validity of VDAC and ANT as MPT constituents, leaving only CypD as a key component of the MPT pore [4,12–16]. Moreover, CypD is only a regulator of the MPT pore and, being a small soluble matrix protein, cannot act as the pore-forming channel itself. Thus there is a dire need to identify the remaining members of the MPT pore complex, especially the pore-forming protein itself.
Recently, C1qbp (complement 1q-binding protein; also known as gC1qr, p32, HAPB1 or SF2p32) has been proposed to be a novel component of the MPT pore [17]. C1qbp was originally thought to act as plasmalemmal receptor that enabled the binding of C1q to the cell surface [18]. However, since then C1qbp has been reported to exist in multiple subcellular compartments, including mitochondria [18–22]. Previous studies have suggested that binding to mitochondrial C1qbp is required for the pro-death actions of Bcl2 family protein Hrk [21] and the tumour suppressor protein p14ARF (alternative reading frame) [22]. Overexpression of C1qbp was also reported to increase mitochondrial ROS production and lead to the loss of Δψm, cytochrome c release and cell death in rat fibroblasts [23]. Moreover, the crystal stucture for C1qbp has been solved and indicates that C1qbp can form a pore-like homotrimer [24].
Together, these data suggest that C1qbp plays a role in mitochondrial-driven cell death and, therefore, could indeed be part of the MPT machinery. Nevertheless, this concept has yet to be tested. Consequently, the purpose of the present study was to test the role of C1qbp in MPT and cell death in cultured fibroblasts using both gain-of-function and loss-of-function approaches. However, to our surprise, we found that C1qbp in fact acts as an inhibitor, rather than a promoter, of MPT and subsequent cell death. Moreover, this inhibitory effect appears to be due to the ability of C1qbp to directly interact with CypD.
EXPERIMENTAL
Animals
All experiments involving the harvesting of mouse tissues and embryos were approved by the University of Missouri-Columbia Animal Care and Use Committee and conformed to the NIH guidelines for the use and care of animals.
Reagents
Calcein-AM (calcein-acetoxymethyl ester), Mitotracker-CMXRos, PI (propidium iodide) and Lipofectamine RNAiMAX were from Invitrogen; DMEM (Dulbecco’s modified Eagle’s medium), FBS (fetal bovine serum) and HBSS (Hanks buffered saline solution) were from Hyclone; recombinant His-tagged CypD, C1qbp and GST (glutathione transferase)-tagged proteins were from Prospec; recombinant GST–CypD and GST–C1qbp were from Abnova; the HisPur Co2+ resin purification kit was from Thermo Pierce; Protein A/G beads were from Santa Cruz Biotechnology; glutathione–Sepharose beads were from GE Lifesciences; and all other chemicals/reagents were from Sigma–Aldrich.
Cell culture
Primary cultured MEFs (mouse embryonic fibroblasts) were obtained from E (embryonic day) 13.5–15.5 C57/B6 mouse embryos by trypsin digestion as described previously [4,15]. The MEFs were then maintained in DMEM supplemented with 10 % FBS.
Subcellular fractionation
MEFs, mouse hearts and mouse livers were subfractionated into mitochondrial, light membrane and cytosolic fractions by differential centrifugation as described previously [4,15]. In some experiments the purified mitochondria were subjected to limited digestion with proteinase K (0.1–1.6 μg/ml) for 15 min on ice. The digestion was then stopped by the addition of 1 mM PMSF and the mitochondria were then subjected to Western blotting. For the analysis of mitochondrial membrane compared with soluble proteins, mouse liver mitochondria were incubated in 0.1 M Na2CO3, pH 11.0, for 30 min. The insoluble membrane proteins were then pelleted by centrifugation at 100 000 g for 30 min. Both the soluble and membrane fractions were then subjected to Western blotting.
Adenoviruses and siRNAs (small interfering RNAs)
Replication-deficient adenoviruses for β-galactosidase and mouse C1qbp (with a C-terminal Myc tag) were generated using the AdEasy adenoviral system (Stratagene). MEFs were typically infected with adenovirus at a MOI (multiplicity of infection) of 30–120 plaque-forming units for 2 h at 37 °C. The cells were then cultured for another 48 h in virus-free medium before analysis. An siRNA against mouse C1qbp (5′-ggagggauacaaacuauacacucaa-3′), as well as a non-specific control siRNA, were obtained from Invitrogen. MEFs were transfected with 100 nM siRNA using Lipofectamine RNAiMAX and then cultured for 48 h prior to the experiments.
Fluorescence microscopy
MPT pore opening was assessed by calcein/CoCl2 staining [4,15]. Briefly, after treatment, MEFs were incubated with 1 μM calcein-AM plus 1 mM CoCl2 in HBSS for 30 min at 37 °C. Dye-loaded cells were then washed twice with HBSS and fluorescence images were collected using an inverted fluorescent microscope (Olympus IX51) connected to a digital camera. Cell death was determined by PI exclusion as described previously [4,15]. For immunocytochemistry, cells were fixed in 4 %paraformaldehyde. After permeabilization the slides were incubated overnight with anti-Myc (Santa Cruz Biotechnology), anti-C1qbp (Sigma) or anti-ATP synthase (Mitosciences). The cells were then incubated with the appropriate fluorophore-conjugated secondary antibody (Alexa, Invitrogen) and visualized. For some co-localization studies, the cells were first incubated with 100 nM Mitotracker-CMXRos for 30 min at 37 °C prior to fixing.
Immunoprecipitations and GST-based pulldowns
Cardiac mitochondrial lysate (1 mg) was incubated with 1 μg of anti-C1qbp antibody (Sigma) or normal rabbit IgG plus Protein A/G beads overnight at 4 °C. After washing 3 times with homogenization buffer, the immunocomplexes were then subjected to Western blotting. The GST-based pulldowns were conducted as for the immunoprecitations, except using recombinant GST, GST–CypD or GST–C1qbp plus glutathione–Sepharose beads. For the direct interaction studies, His-tagged recombinant CypD was incubated with recombinant C1qbp overnight in homogenization buffer, and the resultant complexes were purified using Co2+ affinity columns.
Western blotting
Proteins were resolved by SDS/PAGE using 10–15 %acrylamide, transferred on to PVDF membranes, and blotted using the following commercially available antibodies: anti-C1qbp, anti-lactate dehydrogenase and anti-monoamine oxidase B from Abcam; anti-ANT, anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and anti-mitofusin-1 from Santa Cruz Biotechnology; anti-CypD, anti-VDAC and anti-ATP Synthase from Mitosciences; anti- Na+/K+-ATPase and anti-Myc-tag from Cell Signaling; and anti-HSP60 and anti-apoptosis-inducing factor from BD Biosciences. The polyclonal anti-mitochondrial phosphate carrier antibody was custom made for us by Yenzyme. Membranes were then incubated with the appropriate alkaline phosphatase-linked secondary antibody (Santa Cruz Biotechnology) and visualized by enhanced chemifluorescence (Amersham Biosciences).
Statistical analyses
Statistical significance was calculated by the Student’s t test. A P value <0.05 was considered statistically significant.
RESULTS
C1qbp localizes to the mitochondrial matrix in mouse cells and tissues
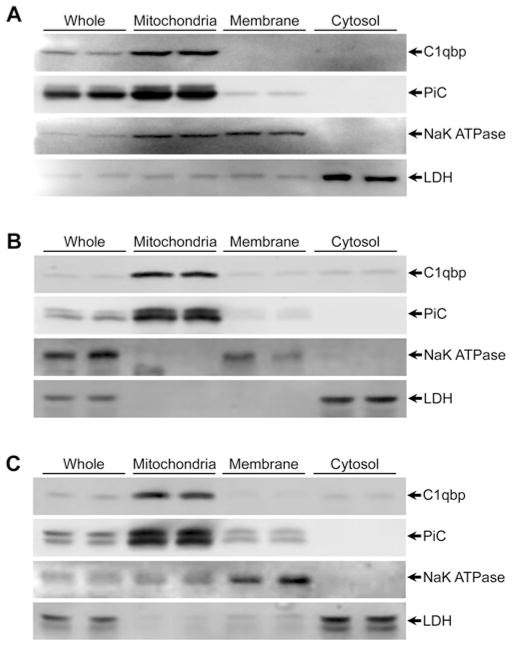
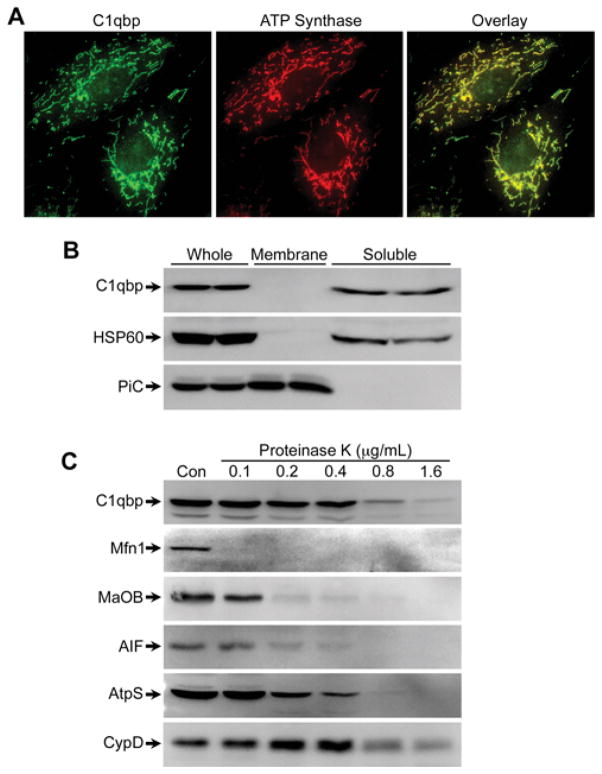
Given the conflicting reports that C1qbp exists in multiple subcellular compartments, we first conducted experiments to confirm that C1qbp indeed localizes specifically to the mitochondrion. We therefore subfractionated MEFs into mitochondrial, membrane and cytosolic compartments. Although this subcellular preparation of the cultured cells still resulted in a significant amount of Na+/K+-ATPase in the mitochondrial fraction, C1qbp expression mirrored only that of the mitochondrial phosphate carrier (Figure 1A). Similar results were observed when we probed for C1qbp in subcellular fractions from mouse heart and liver (Figure 1B and 1C), with both organs showing exclusive localization of C1qbp to the mitochondria. Consistent with these findings, immunocytochemistry demonstrated the mitochondrial localization of endogenous C1qbp in MEFs (Figure 2A). Alkali precipitation of liver mitochondrial membranes confirmed that C1qbp is a soluble protein, rather than an integral membrane protein (Figure 2B). To assess in which soluble compartment of the mitochondrion C1qbp resides, we incubated liver mitochondria in increasing amounts of proteinase K. Outer membrane proteins such as mitofusin-1 and monoamine oxidase B are fully digested at relatively low protease concentrations (Figure 2C), with internal proteins such as apoptosis-inducing factor (inter-membrane space), ATP synthase (inner membrane), and CypD (matrix) being increasingly resistant. The digestion profile of C1qbp matched that of CypD, being only fully digested at the highest proteinase K concentration (Figure 2C), indicating that C1qbp is localized in the mitochondrial matrix.
Figure 1. C1qbp localizes to the mitochondria in mouse cells and tissues.
Subcellular mitochondrial, light membrane, and cytosolic fractions were prepared by differential centrifugation from MEFs (A), mouse hearts (B) and mouse livers (C). The fractions were then Western blotted for C1qbp, the mitochondrial phosphate carrier (PiC), plasmalemmal Na+/K+-ATPase (NaK ATPase) and cytosolic lactate dehydrogenase (LDH). Results shown are representative of four independent experiments performed in duplicate.
Figure 2. C1qbp is a soluble matrix protein.
(A) Immunocytochemistry for endogenous C1qbp in MEFs. An antibody against ATP synthase was used to identify mitochondria. (B) Mitochondrial membranes were alkali-precipitated from mouse liver mitochondria, and the resultant membrane and soluble fractions immunoblotted for C1qbp, mitochondrial phosphate carrier (PiC) and heat-shock protein 60 (HSP60). (C) Mouse liver mitochondria were incubated with increasing concentrations of proteinase K and then immunoblotted for C1qbp, mitofusin-1 (Mfn1), monoamine oxidase B (MaOB), apoptosis-inducing factor (AIF), ATP synthase (AtpS) and CypD. Results shown are representative of four independent experiments performed in duplicate.
Overexpression of C1qbp inhibits MPT and cell death in cultured MEFs
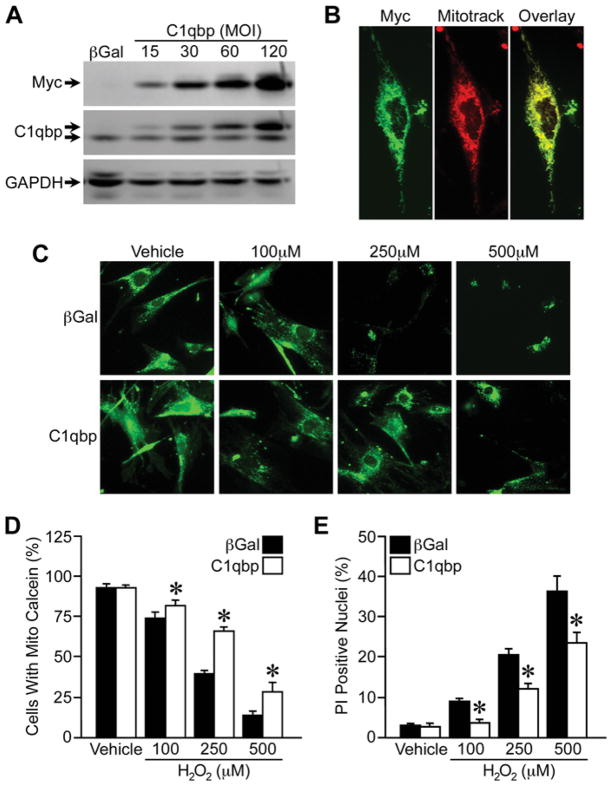
C1qbp has been reported to be a mediator of oxidative stress-induced cell death [23]. Consequently, we next tested whether overexpression of C1qbp would induce or exacerbate MPT and cell death in cultured MEFs. We constructed an adenovirus encoding for mouse C1qbp with a C-terminal Myc tag, which resulted in a gene dose-dependent increase in C1qbp protein in infected MEFs (Figure 3A). An adenovirus encoding βGal (β-galactosidase) was used as a control. Immunocytochemical staining for the Myc tag confirmed that the overexpressed C1qbp was still correctly targeted to the mitochondria (Figure 3B). To assess the effect of C1qbp on MPT, we used the calcein/CoCl2 method to fluorescently label mitochondria in cultured MEFs (Figure 3C). Incubation of MEFs with increasing concentrations of H2O2 elicited a dose-dependent decrease in mitochondrial calcein staining, indicative of MPT. Overexpression of C1qbp did not affect calcein fluorescence at baseline. However, to our surprise, increased mitochondrial levels of C1qbp greatly suppressed the oxidative stress-induced MPT (Figure 3C). Consistent with this finding, we observed considerably less cell mortality in response to H2O2 in the C1qbp-overexpressing MEFs when compared with βGal-infected cells (Figure 3D).
Figure 3. Overexpression of C1qbp attenuates H2O2-induced MPT and cell death.
MEFs were infected with an adenovirus encoding Myc-tagged C1qbp or βGal (control), and then analysed 48 h later. (A) Western blotting for Myc and C1qbp in MEFs infected with either βGal adenovirus or increasing MOI of C1qbp adenovirus. The upper band in the C1qbp blot is the exogenous Myc-tagged C1qbp, whereas the lower band is the endogenous protein. GAPDH was used to demonstrate equivalent loading. (B) Immunocytochemistry for Myc-tagged C1qbp in infected MEFs. Mitotracker CMXRos was used to label the mitochondria. (C) MPT determined by calcein/CoCl2fluorescence in βGal and C1qbp adenovirus-infected MEFs exposed to increasing concentrations of H2 O2 for 6 h. (D) Quantification of the calcein fluorescence data. (E) Cell death measured by PI staining in βGal and C1qbp adenovirus-infected MEFs exposed to increasing concentrations of H2 O2 for 6 h. The results shown are representative of three or four independent experiments performed in duplicate. Error bars indicate S.E.M., *P < 0.05 compared with βGal.
Depletion of C1qbp exacerbates MPT and cell death in cultured MEFs
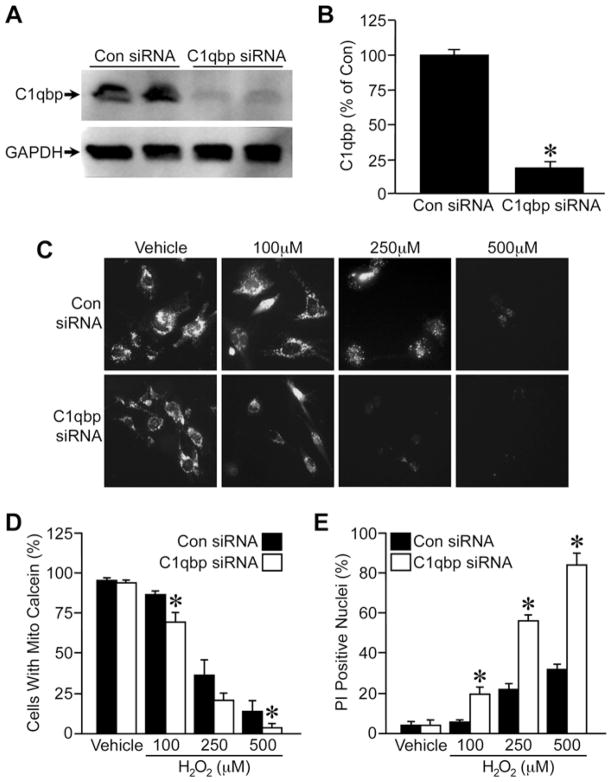
These data suggested that C1qbp could suppress oxidative stress-induced MPT and cell death. To examine this concept further, we next conducted antithetical siRNA knockdown studies. Incubation of MEFs with a C1qbp-specific siRNA resulted in an ~ 80 % reduction in C1qbp protein levels after 48 h (Figures 4A and 4B). MEFs depleted of C1qbp tended to exhibit a greater loss of calcein fluorescence at lower concentrations of H2O2 than control siRNA-transfected cells (Figure 4C), and were more sensitive to H2O2 cytotoxicity (Figure 4D).
Figure 4. Depletion of C1qbp sensitizes cells to H2O2-induced MPT and death.
MEFs were transfected with control or C1qbp-specific siRNAs, and then analysed 48 h later. (A) Western blotting for C1qbp in control and C1qbp siRNA-transfected MEFs. GAPDH was used to demonstrate equivalent loading. (B) Quantification of C1qbp expression in the transfected MEFs. (C) MPT determined by calcein/CoCl2 fluorescence in control and C1qbp siRNA-transfected MEFs exposed to increasing concentrations of H2 O2 for 6 h. (D) Quantification of the calcein fluorescence data. (E) Cell death measured by PI staining in control and C1qbp siRNA-transfected MEFs exposed to increasing concentrations of H2 O2 for 6 h. The results shown are representative of three or four independent experiments performed in duplicate. Error bars indicate S.E.M., * P < 0.05 compared with control siRNA (Con siRNA).
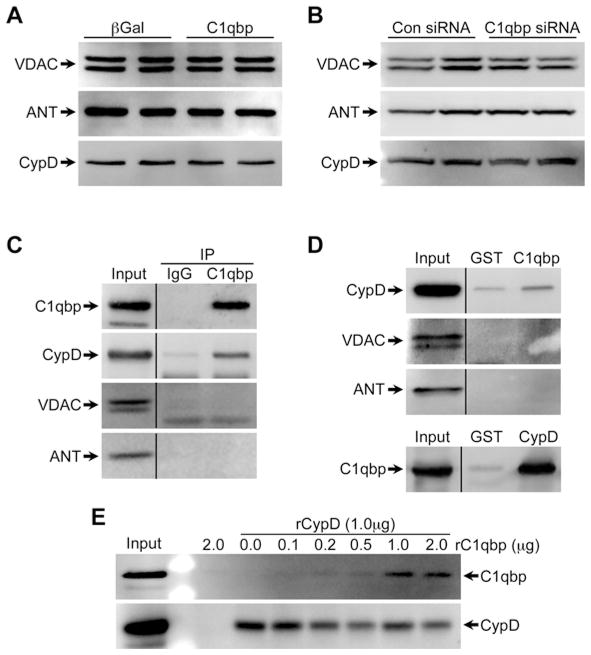
C1qbp directly interacts with CypD
To better understand the mechanism by which C1qbp inhibits MPT, we examined the levels of various putative MPT pore components in the C1qbp-modified cells. However, neither overexpression nor depletion of C1qbp affected the cellular levels of ANT, VDAC or CypD (Figures 5A and 5B), suggesting that some other post-translational mechanism was at work. We therefore immunoprecipitated C1qbp from MEF lysates, and then blotted the resultant complexes for ANT, VDAC and CypD. Interestingly, C1qbp co-precipitated with CypD but not with ANT or VDAC (Figure 5C). This interaction was confirmed using GST-based pulldowns of either C1qbp or CypD (Figure 5D). In order to assess whether such an interaction was direct or indirect, we co-incubated His-tagged CypD with increasing amounts of recombinant C1qbp and purified the complexes on a Co2+-agarose column. We observed a dose-dependent increase in the amount of C1qbp that could interact with CypD (Figure 5E). Interestingly, the maximum interaction was observed at 1μg of C1qbp, with no further increase seen at 2 μg.
Figure 5. C1qbp directly interacts with CypD.
(A) Western blots for VDAC, ANT and CypD in βGal and C1qbp adenovirus-infected MEFs. (B) Western blots for VDAC, ANT and CypD in control (Con) and C1qbp siRNA-transfected MEFs. (C) C1qbp was immunoprecipitated (IP) from mouse cardiac mitochondria and the complexes were immunoblotted for C1qbp, CypD, VDAC and ANT. Immunoprecipitation with non-specific IgG was used as a control. (D) Mouse cardiac mitochondrial lysates were incubated with GST, GST–C1qbp or GST–CypD fusion proteins and the complexes immunoblotted for C1qbp, CypD, VDAC and ANT. (E) Increasing amounts of recombinant C1qbp (rC1qbp) were incubated with recombinant His-tagged CypD (rCypD). The complexes were purified on a Co2+-agarose column and then immunoblotted for C1qbp and CypD. All results shown are representative of three or four independent experiments performed in duplicate.
DISCUSSION
The results of the present study provide novel and compelling evidence that C1qbp acts as an endogenous inhibitor of the MPT pore. We found that C1qbp overexpression could inhibit oxidative stress-induced MPT and cytotoxicity, whereas depletion of C1qbp had the opposite effect and instead sensitized the cells to MPT and cell death. Thus these findings contradict the recent postulation that C1qbp may be a necessary component of the MPT pore [17]. Moreover, they are in conflict with previous reports that C1qbp itself can induce mitochondrial ROS production and that it is required for ARF and Hrk-induced mitochondrial-dependent cell death [21–23].
The reasons for the conflicting results between the present and previous studies are not clear. In the study suggesting a role of C1qbp in ARF-mediated apoptosis, the vast majority of the studies revolved around the overexpression of the various proteins in cancer cell lines [22], and the endogenous functions of C1qbp in normal cells were not tested. Chowdhury et al. [23] reported that overexpression of C1qbp in rat fibroblasts induced Bax expression, cytochrome c release, ROS production and apoptosis at ~ 60 h post-transfection. Although most of our studies involving C1qbp overexpression were carried out 48 h post-infection, we conducted pilot studies examining the cells at 72 h post-infection and failed to find any significant changes in Bax expression or cell death (results not shown). Moreover, the theory that C1qbp could be the MPT pore is based upon the assumption that C1qbp, as a homotrimer, is inserted into the inner membrane [17,21]. However, there is no evidence to suggest that C1qbp can act as a membrane protein. Indeed, similar to other reports [25,26], in the present study we found that C1qbp is a soluble protein that co-purifies with the mitochondrial matrix rather than the membrane fraction. Whether C1qbp can insert into the membrane under different conditions remains to be tested.
Instead, our present results are more in line with several reports that C1qbp is up-regulated in several different types of cancer cell lines and tumours [27–30], which suggests that C1qbp is in fact cytoprotective. Indeed, induction of the ‘hormetic’, i.e. cell survival response, was associated with up-regulation of C1qbp in hepatoma cells [31]. Moreover, the prognosis of breast cancer patients has been shown to be inversely proportional to the amount of C1qbp expression in their tumours [30]. Further support for this line of reasoning comes from the fact that knockdown of C1qbp in MDA-MB-231 tumour cells sensitizes them to glucose deprivation-induced cell death [27]; similar to our present results showing that C1qbp knockdown sensitizes MEFs to MPT and cell death.
With regard to the mechanism(s) by which C1qbp affects MPT inhibition, we found that genetic manipulation of C1qbp did not affect the expression levels of any of the putative components of the MPT pore. However, we did observe that C1qbp was able to directly interact with CypD, a critical regulator of the MPT pore. Interestingly, this interaction was maximal when C1qbp and CypD were at a 1:1 ratio, and further increasing the amount of C1qbp did not increase the amount of CypD binding. To the best of our knowledge, these are the first data demonstrating an interaction between C1qbp and CypD, and suggest that C1qbp’s ability to inhibit the MPT pore lies in its capacity to bind to CypD. Whether the resulting MPT inhibition is simply due to C1qbp sequestering CypD away from other pore components, or if a more complex mechanism is at play is the subject of ongoing investigation in our laboratory. In addition, we cannot rule out at this juncture that other secondary effects of C1qbp on mitochondrial function could indirectly influence MPT sensitivity. For example, C1qbp has been reported to affect complex-I activity [23,27]. This in turn could conceivably alter ROS and ATP production, both of which can influence MPT pore opening.
In conclusion, we have demonstrated for the first time that C1qbp can inhibit the MPT pore ostensibly through an interaction with the MPT pore component CypD. These data suggest that C1qbp represents an important clinical target. Activation of C1qbp could be used to protect organs against MPT-driven cell death, whereas inhibition of C1qbp and the resultant cell death could be a viable intervention for the treatment of cancers.
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant numbers HL092327 and HL094404 (to C.B.)].
Abbreviations used
- ANT
adenine nucleotide translocase
- ARF
alternative reading frame
- βGal
β-galactosidase
- C1qbp
complement 1q-binding protein
- calcein-AM
calcein-acetoxymethyl ester
- CypD
cyclophilin D
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GST
glutathione transferase
- HBSS
Hanks buffered saline solution
- MEF
mouse embryonic fibroblast
- MOI
multiplicity of infection
- MPT
mitochondrial permeability transition
- PI
propidium iodide
- ROS
reactive oxygen species
- siRNA
small interfering RNA
- VDAC
voltage-dependent anion channel
Footnotes
AUTHOR CONTRIBUTION
Christopher Baines conceived and designed the projects. Allison McGee and Christopher Baines performed the experiments and analysed the data. Christopher Baines wrote the manuscript.
References
- 1.Halestrap AP. What is the mitochondrial permeability transition pore? J Mol Cell Cardiol. 2009;46:821–831. doi: 10.1016/j.yjmcc.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 2.Baines CP. The molecular composition of the mitochondrial permeability transition pore. J Mol Cell Cardiol. 2009;46:850–857. doi: 10.1016/j.yjmcc.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zorov DB, Juhaszova M, Yaniv Y, Nuss HB, Wang S, Sollott SJ. Regulation and pharmacology of the mitochondrial permeability transition pore. Cardiovasc Res. 2009;83:213–225. doi: 10.1093/cvr/cvp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 5.Clarke SJ, McStay GP, Halestrap AP. Sanglifehrin A acts as a potent inhibitor of the mitochondrial permeability transition and reperfusion injury of the heart by binding to cyclophilin-D at a different site from cyclosporin A. J Biol Chem. 2002;277:34793–903479. doi: 10.1074/jbc.M202191200. [DOI] [PubMed] [Google Scholar]
- 6.Millay DP, Sargent MA, Osinska H, Baines CP, Barton ER, Vuagniaux G, Sweeney HL, Robbins J, Molkentin JD. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat Med. 2008;14:442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiepolo T, Angelin A, Palma E, Sabatelli P, Merlini L, Nicolosi L, Finetti F, Braghetta P, Vuagniaux G, Dumont JM, et al. The cyclophilin inhibitor Debio 025 normalizes mitochondrial function, muscle apoptosis and ultrastructural defects in Col6a1−/− myopathic mice. Br J Pharmacol. 2009;157:1045–1052. doi: 10.1111/j.1476-5381.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du H, Guo L, Fang F, Chen D, Sosunov AA, McKhann GM, Yan Y, Wang C, Zhang H, Molkentin JD, et al. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer’s disease. Nat Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forte M, Gold BG, Marracci G, Chaudhary P, Basso E, Johnsen D, Yu X, Fowlkes J, Rahder M, Stem K, et al. Cyclophilin D inactivation protects axons in experimental autoimmune encephalomyelitis, an animal model of multiple sclerosis. Proc Natl Acad Sci USA. 2007;104:7558–7563. doi: 10.1073/pnas.0702228104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masubuchi Y, Suda C, Horie T. Involvement of mitochondrial permeability transition in acetaminophen-induced liver injury in mice. J Hepatol. 2005;42:110–116. doi: 10.1016/j.jhep.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama H, Chen X, Baines CP, Klevitsky R, Zhang X, Zhang H, Jaleel N, Chua BH, Hewett TE, Robbins J, et al. Ca2+- and mitochondrial-dependent cardiomyocyte necrosis as a primary mediator of heart failure. J Clin Invest. 2007;117:2431–2444. doi: 10.1172/JCI31060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 13.Schinzel AC, Takeuchi O, Huang Z, Fisher JK, Zhou Z, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc Natl Acad Sci USA. 2005;102:12005–12010. doi: 10.1073/pnas.0505294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basso E, Fante L, Fowlkes J, Petronilli V, Forte MA, Bernardi P. Properties of the permeability transition pore in mitochondria devoid of Cyclophilin D. J Biol Chem. 2005;280:18558–18561. doi: 10.1074/jbc.C500089200. [DOI] [PubMed] [Google Scholar]
- 15.Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol. 2007;9:550–555. doi: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, Jones DP, MacGregor GR, Wallace DC. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starkov AA. The molecular identity of the mitochondrial Ca2+ sequestration system. FEBS J. 2010;277:3652–3663. doi: 10.1111/j.1742-4658.2010.07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghebrehiwet B, Lim BL, Kumar R, Feng X, Peerschke EI. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunol Rev. 2001;180:65–77. doi: 10.1034/j.1600-065x.2001.1800106.x. [DOI] [PubMed] [Google Scholar]
- 19.Storz P, Hausser A, Link G, Dedio J, Ghebrehiwet B, Pfizenmaier K, Johannes FJ. Protein kinase Cμ is regulated by the multifunctional chaperon protein p32. J Biol Chem. 2000;275:24601–24607. doi: 10.1074/jbc.M002964200. [DOI] [PubMed] [Google Scholar]
- 20.Sengupta A, Banerjee B, Tyagi RK, Datta K. Golgi localization and dynamics of hyaluronan binding protein 1 (HABP1/p32/C1QBP) during the cell cycle. Cell Res. 2005;15:183–186. doi: 10.1038/sj.cr.7290284. [DOI] [PubMed] [Google Scholar]
- 21.Sunayama J, Ando Y, Itoh N, Tomiyama A, Sakurada K, Sugiyama A, Kang D, Tashiro F, Gotoh Y, Kuchino Y, Kitanaka C. Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ. 2004;11:771–781. doi: 10.1038/sj.cdd.4401418. [DOI] [PubMed] [Google Scholar]
- 22.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury AR, Ghosh I, Datta K. Excessive reactive oxygen species induces apoptosis in fibroblasts: role of mitochondrially accumulated hyaluronic acid binding protein 1 (HABP1/p32/gC1qR) Exp Cell Res. 2008;314:651–667. doi: 10.1016/j.yexcr.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 24.Jiang J, Zhang Y, Krainer AR, Xu RM. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J Biol Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 26.Seytter T, Lottspeich F, Neupert W, Schwarz E. Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae is related to the human complement receptor gC1q-R. Yeast. 1998;14:303–310. doi: 10.1002/(SICI)1097-0061(19980315)14:4<303::AID-YEA217>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol. 2010;30:1303–1318. doi: 10.1128/MCB.01101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinstein DB, Stortchevoi A, Boosalis M, Ashfaq R, Ghebrehiwet B, Peerschke EI, Calvo F, Guillaume T. Receptor for the globular heads of C1q (gC1q-R, p33, hyaluronan-binding protein) is preferentially expressed by adenocarcinoma cells. Int J Cancer. 2004;110:741–750. doi: 10.1002/ijc.20105. [DOI] [PubMed] [Google Scholar]
- 29.Fogal V, Zhang L, Krajewski S, Ruoslahti E. Mitochondrial/cell-surface protein p32/gC1qR as a molecular target in tumor cells and tumor stroma. Cancer Res. 2008;68:7210–7218. doi: 10.1158/0008-5472.CAN-07-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YB, Jiang CT, Zhang GQ, Wang JS, Pang D. Increased expression of hyaluronic acid binding protein 1 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol. 2009;100:382–386. doi: 10.1002/jso.21329. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh SY, Hsu CY, He JR, Liu CL, Lo SJ, Chen YC, Huang HY. Identifying apoptosis-evasion proteins/pathways in human hepatoma cells via induction of cellular hormesis by UV irradiation. J Proteome Res. 2009;8:3977–3986. doi: 10.1021/pr900289g. [DOI] [PubMed] [Google Scholar]