Abstract
Complement factor H (FH) attenuates C3b molecules tethered via their thioester domains to self-surfaces and thereby protects host tissues. FH is a cofactor for initial C3b proteolysis that ultimately yields a surface-attached fragment (C3d), corresponding to the thioester domain. We used NMR and X-ray crystallography to study the C3d:FH19–20 complex in atomic detail. NMR further identified glycosaminoglycan-binding residues in FH module 20 of the C3d:FH19–20 complex. Mutagenesis justified the merging of the C3d:FH19–20 structure with an existing C3b:FH1–4 crystal structure. The merged structure was concatenated with the available FH6–8 crystal structure and new SAXS-derived FH1–4, FH8–15 and FH15–19 envelopes. The combined data suggests a bent-back FH molecule, binding via its termini to two sites on one C3b molecule and simultaneously to adjacent polyanionic host-surface markers.
The ~30 proteins of the complement system cooperate in a vital contribution to innate immunity 1. The complement system is important for clearance of immune complexes and cellular debris, and for augmenting cell-based immunity, but is most famous for rapid targeting and elimination of pathogens. Host tissues may also sustain complement-mediated damage; several diseases, including age-related macular degeneration, atypical hemolytic uremic syndrome (aHUS) and dense deposit disease, link to mutations and single nucleotide polymorphisms (SNPs) in complement genes (reviewed in 2).
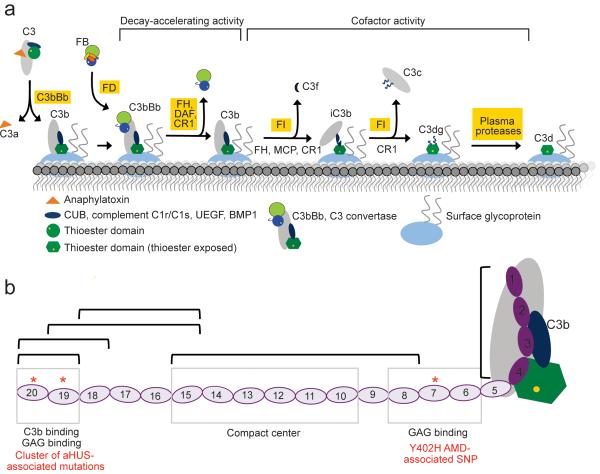
Key to the rapid responsiveness of complement is spontaneous, ubiquitous, low-level C3b production. In this “alternative pathway” (AP) of complement activation, cleavage of C3 to C3b is accompanied by a major rearrangement of its domains 3 (Fig. 1). Notably, the thioester-containing domain (TED) migrates, rotates, and exposes its activated thioester thus allowing rapid attachment of C3b to nucleophiles on nearby particles, basement membranes or cell surfaces. C3b is both an important opsonin and a component of three out of four convertase complexes that drive the complement amplification cascade. For example, the bimolecular C3bBb complex, a C3 convertase, cleaves C3 to form more C3b (Fig. 1), while the trimolecular (C3b)2Bb complex is a C5 convertase generating C5b thereby instigating the cytolytic terminal pathway of complement. Rigorous regulation is necessary to minimize complement-mediated damage to host.
Figure 1.
Introduction to C3b/C3d and complement factor H (FH). (a) The central event in complement activation is C3 cleavage to C3b by C3 convertase accompanied by attachment to surfaces mediated by the TED. In the presence of additional complement regulatory molecules C3b may be further degraded sequentially to iC3b, C3c, C3dg and C3d. C3d corresponds to the TED and remains surface-bound on the cell surface. (b) Complement FH has 20 CCPs and possesses multiple binding sites for different ligands. Disease-linked mutations have been reported throughout FH. Several recombinant fragments of FH were utilized in the current studies and are indicated by black brackets. aHUS - atypical hemolytic uremic syndrome; AMD - age-related macular degeneration.
Complement factor H (FH) limits promulgation of C3b 4-6, and this activity is essential to ensure the AP of complement activation operates in a manner that is both proportionate and targeted. This 155-kDa soluble glycoprotein consists exclusively of 20 complement control modules (CCPs) 7 (Fig. 1), each containing approximately 60 amino acid residues. FH acts in the fluid phase and on self-surfaces and displays several related activities. It competes with complement factor B for binding to C3b thus blocking C3Bb formation and also accelerates the irreversible decay of any C3bBb that does form. Finally FH is a cofactor for factor I-mediated C3b cleavage to iC3b that cannot bind factor B. The iC3b fragment retains the intact TED and thus remains surface attached (Fig. 1). The TED survives a series of further degradation steps that may occur resulting, sequentially, in surface-bound fragments, C3dg (~39 kDa)(prepro-C3 numbering 955–1303) and C3d (~34 kDa)(prepro-C3 numbering 996–1303); the latter corresponds almost precisely to the original TED and is a ligand for complement receptor type 2 (CR2), and thereby an adjuvant for humoral immunity.
FH engages most effectively with C3b or C3bBb when these are attached to self-surfaces carrying specific polyanionic markers such as glycosaminoglycans (GAGs) and sialic acid 8,9. The consequent enrichment of FH on such surfaces compared to its paucity on, for example, bacterial surfaces, helps innate immunity discriminate between host and pathogens. Two FH regions bind to C3b (Fig. 1): the N-terminal four CCPs (FH1–4), which harbor the factor I cofactor and Bb decay acceleration activities; and, more strongly, the two C-terminal CCPs (FH19–20) 10-13. The FH1–4:C3b interaction is well characterized 14 (Fig. 1) while the FH19–20:C3b interaction is not. This is despite the presence in the FH C-terminus of a mutation cluster linked to the kidney disease aHUS 15-17 together with a polyanion-binding site required for self-surface recognition by FH 18,19. These two C3b-binding sites of FH confer avidity 12 but the role of the intervening 14 CCPs remains controversial although CCP7 contains a second polyanion-binding site 12,16,20,21 and, intriguingly, harbors the SNP most strongly linked to the risk of developing age-related macular degeneration 22-25.
How these multiple binding sites for C3b and polyanions, located towards either end of the 20-CCP length of FH, cooperate to engage with C3b preferentially on a GAG-rich self-surface remains a mystery despite much effort. This dearth of knowledge impedes understanding of the role of FH mutants and sequence variants in disease. Here we show that a novel atomic-resolution crystal structure of the C3d:FH19–20 complex emulates the interaction between the FH C-terminus and C3b. We have combined this structure with further experimental data to build an insightful model of the complex formed by FH with C3b tethered to a self-surface.
RESULTS
The crystal structure of C3:FH19–20 solved at 2.1-Å resolution
Useable co-crystals of C3d and FH19–20 were obtained at pH 7.0 and pH 9.0. In both cases the asymmetric unit contains three molecules each of FH19–20 and C3d. Although crystal packing was not identical, similar intermolecular interactions are observed in both crystal structures (Supplementary Fig. 1). The higher-pH structure has better resolution (2.1 Å versus 3.5 Å; Table 1) and is discussed further.
Table 1.
Data collection and refinement statistics
| C3d:CFH19-20 crystal | |
|---|---|
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 74.67, 82.99, 85.60 |
| α, β, γ (°) | 112.75, 110.13, 99.96 |
| Resolution (Å) | 41.59-6.64 (2.21-2.10) |
| Rmerge | 9.9 (46.5) |
| I / σI | 14.9 (5.1) |
| Completeness (%) | 95.9 (96.4) |
| Redundancy | 4.6 (4.7) |
| Refinement | |
| Resolution (Å) | 41.59-2.10 |
| No. reflections | 93199 |
| Rwork / Rfree | 17.06/20.59 |
| No. atoms | |
| Protein | 9990 |
| Ligand/ion | 49 |
| Water | 947 |
| Average B-factors | |
| Protein | 35.30 |
| Ligand/ion | 44.23 |
| Water | 39.13 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.018 |
Values in parentheses are for highest-resolution shell.
A total of eight sets of heterotypic contacts were observed between FH19–20 molecules and C3d molecules in the crystal structure (Supplementary Fig. 1, Supplementary Tables 1-2), with the two most extensive sets being nearly identical. This duplicated interface – unlike others within the crystal structure – could correspond to the physiologically critical interface formed between the FH C-terminus and C3b: FH19–20 does not experience any steric clashes when this interface is extrapolated onto the corresponding TED domain of C3b (as judged from the multiple crystal structures of C3b 3,14,26, see Supplementary Fig. 2); nor would it sterically hinder the functionally critical covalent linkage of C3d to biological surfaces, centered around Gln19 (Gln1013; prepro-C3 numbering indicated in parentheses hereon). Strikingly, this interface also correlates well with the distribution of a number of disease-associated variants of FH and C3 (discussed further below). The alternative complexes that might be inferred from other, less extensive, candidate interfaces in the crystal structure (Supplementary Tables 1-2) could not occur in the complex of FH with surface-bound C3b since they would involve significant steric clashes between FH19–20 and C3b, or they would mask the residues involved in surface tethering (Supplementary Fig. 2). Moreover, the distributions of disease-associated mutations within FH and C3 are inconsistent with the alternative interfaces. Further experimental validation of the physiologically relevant C3d:FH19–20 complex is described in detail below.
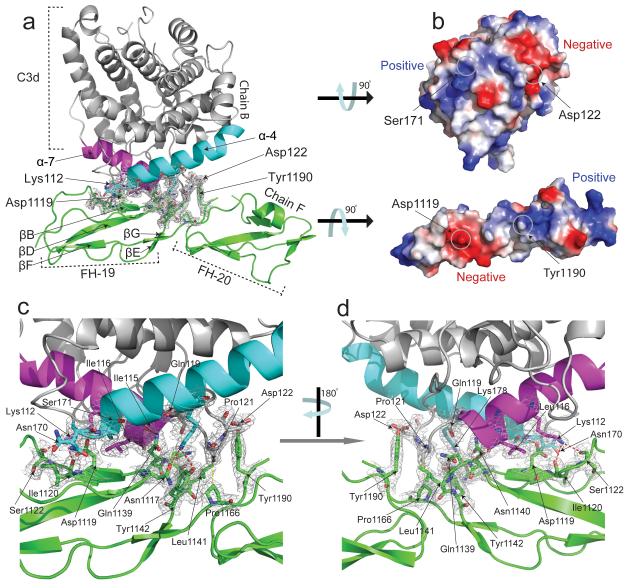
Our structure confirms that discontinuous stretches of the FH19–20 polypeptide form a contiguous binding surface for C3d (Fig. 2a,b; Supplementary Fig. 1) burying ~750 Å2 of surface area in total. The C-terminal half of C3d α-helix 4 (α4) contacts a region centered on β-strand B (βB) of FH CCP19 that also includes the hypervariable region following βB, the C-terminal part of βD, and the early part of the βD-βE loop. The α4-α5 loop of C3d protrudes, occupying the cleft between the two FH CCPs. Its side-chains contact βC in CCP20 as well as the intermodular linker. Also of note is substantial burial in the intermolecular interface of Tyr1190 (in CCP20) and van der Waals interactions between inter-modular linker residue Pro1166 and C3d-residue Prol121 (Pro1114 in C3). The N-terminal half of C3d α7 also participates: Lys178 (Lys1171) forms H-bonds with CCP19 residues Asn1117 and also with Gln1139 and Tyr1142 of the CCP19 βD-βE loop. Thus a network of side chain-backbone and side chain-side chain H-bonds and van der Waals’ contacts (Fig. 2c-d, Supplementary Fig. 1) stabilize the intermolecular interface that involves both modules 19 and 20 of FH.
Figure 2.
Structure of the C3d:FH19–20 complex showing interacting domains and overall molecular architecture. All interacting amino acid residues are shown as sticks; H bonds are represented by dashed lines. The resolution of the electron density (2Fo-Fc map) map is 2.1 Å and is contoured at 1σ. (a) Modules 19 (residues 1108–1166) and 20 (residues 1167–1231) of FH are shown in green. Amino acids belonging to two interacting helices (α7 (cyan, residues 170–189) and α4 (purple, residues 106–118) of C3d (shown in grey) form the majority of interactions with FH. (b) Electrostatic surface representations of C3d and FH19–20 (calculated using the APBS plug-in for Pymol 51) rotated 90° counter-clockwise and clockwise, respectively, from the orientations shown in (a). ((c) and (d)) Enlarged views of the C3d and FH19–20 interface.
NMR identifies C3b-interacting segments in both modules 19 and 20
NMR spectroscopy was used to independently address the C3d:FH19–20 complex. Following addition of (non-isotopically labeled) C3d a marked reduction of NH signal intensities occurred in the [1H,15N]-HSQC spectrum of 15N-FH19–20 (Fig 3;Supplementary Fig. 3). This reflects formation of the C3d:FH19–20 complex that would be expected to have slower molecular tumbling and shorter T2 relaxation times compared to FH19–20. No such changes were observed in a control in which 15N-FH19 replaced 15N-FH19–20 (not shown). Signal losses and concomitant line broadening were, however, not uniform during titration of 15N-FH19–20. This indicates that, for selected residues, the overall relaxation rate contains a contribution from chemical exchange on the intermediate timescale. Such behavior is well documented 27,28; it occurs for amides that exhibit larger chemical shift differences between bound and free forms. Corresponding residues very likely contribute directly to binding, or participate in conformational changes upon binding 29,30.
Figure 3.
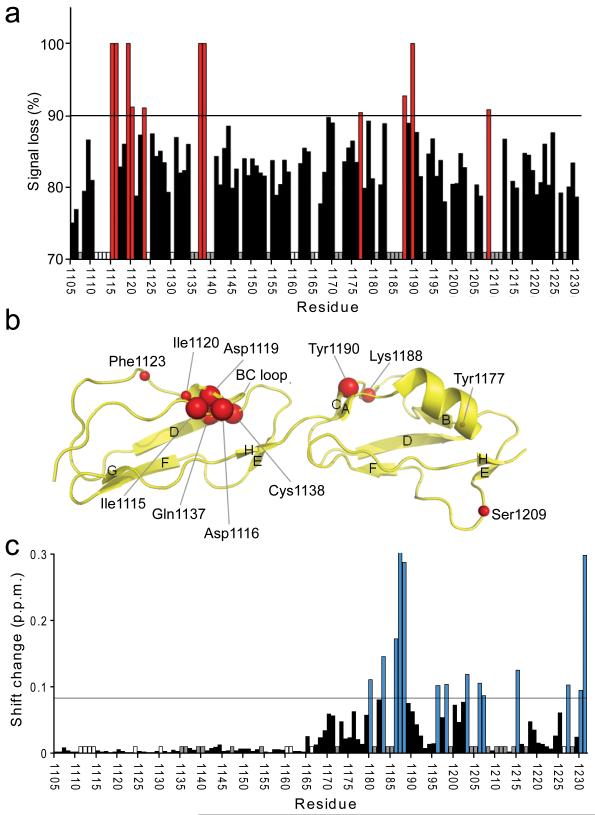
Heteronuclear NMR spectroscopy used to map the C3d and dp8 binding sites on FH19–20. (a) The percentage broadening observed for unequivocally assigned backbone amides upon addition of C3d is indicated: Red bars signify > 90% broadening; white bars indicate proline residues; and grey bars indicate residues whose assignments were missing or whose intensities were weak in free FH19–20. (b) Amides which experience a line-broadening > 90% are schematically shown as red spheres on the structure of FH19–20 (2G7I). The sizes of the spheres have been adjusted to correlate with the degree of line-broadening that each amide cross-peak experiences: Larger spheres represent complete disappearance of the signal; smaller spheres represent signals broadened by > 90% but which are still detectable. (c) Combined amide chemical shift changes upon addition of 8.5-fold excess of dp8. Blue bars indicate shift changes larger than the threshold (double the average shift change; corresponding residues mapped onto the structure of FH19–20 in Fig. 5d), white and grey bar have the same meaning as in (a).
Eleven cross-peaks originally present in the 15N-FH19–20 spectrum were broadened upon addition of C3d to the extent that they lost more than 90% of their signal intensities (Fig. 3, Supplementary Fig. 3). Mapping the positions of these most strongly affected amides on the FH19–20 structure (Fig. 3) provides strong evidence for the C3d-binding surface identified in the crystal-structure: Within CCP19, β-strand B (βB) (including Asp1119) and the first few residues of the βB-βC hypervariable loop plus the end of βD are implicated; within CCP20, βC appears to be involved. Only Tyr1177 and Ser1209 (whose signal loss was just above the 90% threshold) lie outside of this interaction region.
Mutagenesis studies of the C3d:FH19–20 binding interface
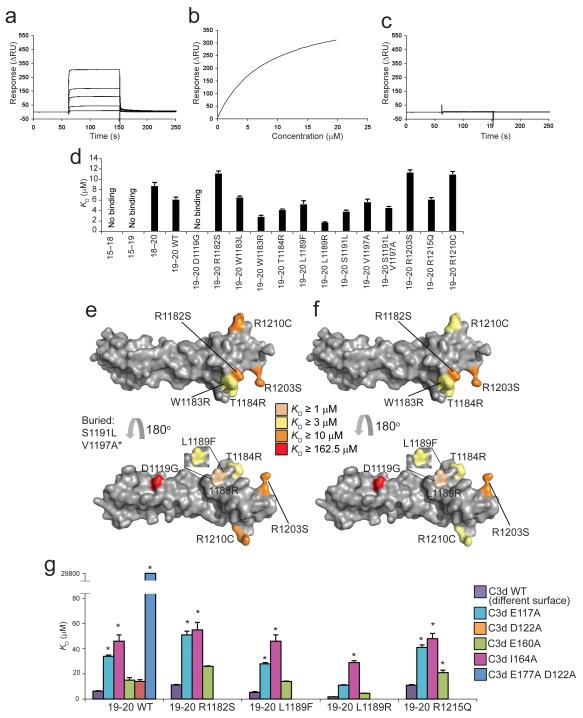
We combined new and existing mutagenesis data to corroborate our structural findings. Using surface plasmon resonance, we tested previously generated functionally critical mutations within the structure of FH19–20 31 (Fig. 4a-d). The FH mutation that was observed to have the most deleterious effect on the binding of FH to C3d and C3b, and that also abrogates activity in a hemolysis-protection assay, is D1119G within CCP19 (Fig. 4c-e, Supplementary Fig. 4 and Supplementary Fig. 5); this is consistent with its key position in the interface identified from NMR and X-ray crystallography. A second key interface residue according to our X-ray crystallographic structure is Gln1139 of CCP19; in a previously reported mutagenesis study the Q1139A mutant of FH19–20 exhibited greatly reduced capacity to inhibit wild-type FH binding to C3d 32. A number of other functionally significant mutations occupy several faces of CCP20 but do not appear to delineate a discrete binding surface (Fig. 4d-f).
Figure 4.
SPR studies of FH19–20 mutants binding to C3d. (a) Sensorgram showing the binding of wild-type FH19–20 to amine-coupled plasma-derived C3d. (b) KD fitting of (a). (c) Sensorgram showing the very weak binding of FH19–20 D1119G to amine-coupled plasma-derived C3d. (d) Summary of affinity constants of FH fragments and mutants for recombinant C3d. (e) Surface representation of FH19–20 (2G7I) highlighting residues corresponding to mutations that change C3b binding 31. (f) Surface representation of FH19–20 (2G7I) highlighting residues corresponding to mutations that change C3d binding. Ser1191 and Val1197 are buried residues. *Val1197 has been analyzed in the context of the double mutant S1191L V1197A. Leu1189 has been analyzed in the context of two distinct mutants: L1189R and L1189F. (g) Summary of the KD values for complexes of mutants of FH19–20 and C3d mutants E117A (E1110A), D122A (D1115A) and E160A (E1153A), I164A (I1157A, with wild-type FH19–20 only) and E117A D122A (E1110A D1115A, with wild-type FH19–20 only). The KD of the latter was ~30 mM. * indicates that the corresponding KD value was extrapolated. Error bars (standard error of the mean) for the KD measurements are indicated.
Affinities were also measured between selected FH19–20 mutants and five C3d mutants (Fig. 4g, Supplementary Fig. 4): E117A (E1110A), D122A (D1115A), and E117A D122A (E1110A D1115A) occupy the NMR and crystallography-derived interface, while E160A (E1153A) and I164A (I1157A) occur within the ‘concave’ surface of this molecule away from the putative FH19–20 binding site. Strikingly, our E117A D122A (E1110A D1115A) double mutation resulted in a decrease in affinity by a factor of ~1,600 (Supplementary Table 3; Supplementary Fig. 4). With regard to single mutants, in general (and with the exception of the aforementioned D1119G), loss of a positive charge in FH19–20 or loss of a negative charge in C3d decreased affinity, even away from the binding interface. However, it is noteworthy that the C3d single-site E117A (E1110A) and D122A (D1115A) mutations attenuate FH19–20 binding by a factor of two or three when compared to E160A and the non-charge substitution I164A, and by a factor of four to six when compared to wild-type C3d, respectively (Supplementary Table 3).
Thus mutagenesis confirms that Glu117 (Glu1110) and Asp122 (Asp1115) in C3d and Asp1119 in CCP19 and Gln1139 in CCP19 are critical interface residues. Other charged residues explored in this study presumably participate in electrostatic steering effects. Such long-range charge effects have previously been reported for the interactions that occur between C3d and CR2, and between C3d and the staphylococcal immune evasion molecule Efb-C 33,34.
FH19–20 binds to both C3d and C3b in the same way
We investigated whether structural information concerning the C3d:FH19–20 complex can be extrapolated to the physiologically important C3b:FH complex. We measured affinities for C3d (and, in some cases, for C3b) of FH15–19, FH18–20, FH19–20 and a panel of FH19–20 mutants (Supplementary Fig. 4 and Supplementary Table 3). Affinities of FH18-20 and FH19–20 for C3d are similar (Fig. 4a,b,d and Supplementary Table 3). Moreover, both bind equally well to C3b and C3d, while FH15–19 binds to neither C3d nor C3b (Fig. 4d and Supplementary Fig. 4). Thus we further confirmed that both the C3b:FH and C3d:FH interfaces involve CCP19 and CCP20 (in good agreement with 35), but not CCP18. Importantly, each of these FH19–20 mutants binds similarly well, or similarly poorly, to both C3d (Fig. 4d and Supplementary Fig. 4) and C3b (based on KD values reported by 31). Notably, FH19–20 D1119G binds to neither of these C3 fragments (Fig. 4c and 31). A comparison of the mutagenesis data, summarized in Figures 4e and 4f, thus illustrates that the C3d:FH19–20 interface must emulate the interface that is found between the C-terminus of FH and C3b. A logical extension of this finding is that the site in C3d that interacts with FH19–20 must correspond to an accessible face of the TED within the C3b. As outlined above, this requirement is met by the C3d:FH19–20 interface identified by both NMR and crystallography.
Extrapolation to a C3b:FH1–4:FH19–20 complex
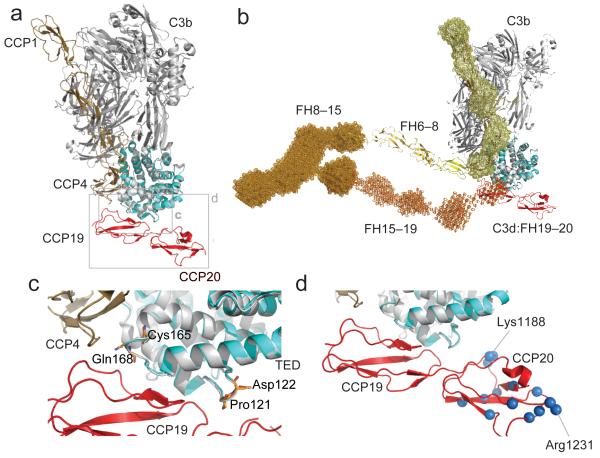
The very good superposition of the C3d polypeptide within the C3d:FH19–20 complex onto the relevant residues of the TED domain within the crystal structure of the C3b:FH1–4 complex (PDB accession code: 2WII 14) is shown in Figure 5. The resulting merged model of a ternary complex of C3b with two segments of FH suggests that CCP19 of FH19–20 lies immediately adjacent to CCP4 of FH1–4 but that the two fragments occupy distinct regions of C3b consistent with simultaneous binding. The merged model also suggests that, in the complex, the majority of CCP20 is exposed, and positioned to interact with polyanionic surface markers.
Figure 5.
Potential model of FH engagement with surface bound C3b (a) Superposition (C3d on TED) of the C3d:FH19–20 complex and C3b:FH1–4 complex demonstrating the close proximity of FH modules 4 and 19. (b) The FH19–20:C3d structure (red and cyan) is shown superimposed (C3d on TED) onto the FH1–4:C3b structure (tan and gray). This FH model has been constructed by juxtaposition or superposition of the structures of FH5 (yellow) and FH6–8 (yellow-orange) and the SAXS-shape envelopes of FH8-15 (bright orange) and FH15-19 (orange). Structures are shown in cartoon representation; SAXS shape envelopes are shown in mesh. Note, the FH1–4 SAXS shape envelope (pale yellow) is superimposed onto the C3b:FH1–4 structure. (c) Close up view of the α4-α5 loop and the α6-α7 loop of C3d. Highlighted in orange are the residues Pro121 (Pro1092), Asp122 (Asp1093), Cys165 (Cys1136) and Gln168 (Gln1139) for which disease-associated mutations have been reported. (d) Close-up view of the FH19–20 interaction with TED highlighting residues also like to interact with self-surface markers. Amides experiencing considerable chemical shift perturbations upon exposure to dp8 are highlighted as blue spheres (corresponding to their nitrogen atoms). Residues Lys1188 and Arg1231, which exhibit the greatest perturbations upon addition of dp8, are indicated.
CCP20 within C3d-bound FH19–20 recognizes surface-associated GAG molecules
Experiments were performed to ascertain whether CCP20 within the C3d:FH19–20 complex can still engage with model GAGs (that are surrogates for self-surface markers), and to identify which FH residues are critical to GAG-binding.
In initial benchmarking work, an octasaccharide fraction of heparin, dp8, was titrated into 15N-labeled FH19–20 (i.e. no C3d present) (Supplementary Fig. 3). The largest chemical-shift changes (brought about by an 8.5-fold dp8:FH19–20 ratio) (Fig. 3c) occur in residues that belonging to CCP20. The distribution of affected residues (Fig. 5), is consistent with two (or more) dp8 molecules binding with similar affinities to electropositive patches around Lys1186–Lys1188, and Arg1231; this agrees with a previous NMR study on FH19–20 performed with a tetrasaccharide heparin fragment 16.
Titration with up to a 15-fold excess of dp8 into the C3d:15N-FH19–20 complex caused chemical shift changes but, importantly, did not alter spectral intensities or line-widths (Supplementary Fig. 3). In separate experiments, a 20-fold excess (over FH19–20) of a pentasaccharide GAG (Fondaparinux) did not inhibit FH19–20 binding to amine-coupled C3d according to SPR (Supplementary Fig. 4); furthermore molar excesses of up to 200-fold (over C3d) of dp8 failed to inhibit C3d binding to FH19–20 in an ELISA-based assay (Supplementary Fig. 6). All these experiments support the existence of a ternary (or higher order) complex formed between C3d, FH19–20 and GAGs.
It is notable that the subset of FH19–20 NH resonances experiencing substantial dp8-induced chemical shift changes is essentially the same whether FH19–20 is in complex with C3d or not (compare Supplementary Fig. 3b and Supplementary Fig. 3c). The amide resonances of some CCP20 residues (exemplified by Lys1188) previously implicated in C3d binding remain very weak when GAGs are added, consistent with C3d proximity; but they still exhibit GAG-associated chemical-shift perturbations similar to those observed in the absence of C3d, which is consistent with GAG-binding and ternary complex formation. While a mixture of binary and tertiary complexes may be present in the NMR tube we can nevertheless conclude that at least one dp8 molecule binds to the C3d:FH19–20 complex at a region of CCP20 distinct from the C3d-binding site (Fig. 5) and proximal to the surface to which C3d/TED is tethered. Our data are in excellent agreement with previous studies in which K1186A, K1188A, K1188Q, L1189R and K1230A FH19–20 mutants, amongst others, were shown to have altered heparin affinity 31,32. Thus the GAG-binding face originally delineated by FH19–20 chemical shift perturbations in the absence of C3d 16, is now shown to also exist in the C3d:FH19–20 complex.
Factor H bends back to allow both termini to interact with C3b
In the modeled ternary complex of C3b, FH1–4 and FH19–20, the C-terminus of CCP4 and the N-terminus of CCP19 lie close together on the same side of C3b, but importantly, do not clash (Fig. 5a-c). This is consistent with a 1:1 complex of C3b and FH in which intervening FH CCP modules form a compact bent-back structure (Fig. 5b), or with a 1:2 FH:C3b complex in which a more extended single FH molecule bridges two C3b molecules. To complement the available incomplete experimental evidence on FH architecture 36-38, we collected SAXS curves for FH1–4, FH8–15 and FH15–19, and performed ab initio shape calculations (Fig. 5b, Supplementary Fig. 7). The resulting ab initio model of FH1–4 is in excellent agreement with the corresponding crystal structure 14, while the FH8–15 and FH15–19 shape envelopes (for which no structures are available) describe a relatively compact bent-back structure, and an extended rod-like structure, respectively. The compact shape of FH8–15 model (as previously described also for both FH10–15 and FH11–14 37) involves significant intermodular contacts that could stabilize bent-back structures for both free and C3b-bound full-length FH. By manually concatenating our SAXS-derived envelopes for FH8–15 and FH15–19 with the X-ray derived structures of FH6–8 36 and C3d:FH19–20 it was possible to model FH modules 6–20 in complex with C3b (Fig. 5b). This model was readily adjusted so that CCP6 and the C-terminus of CCP4 (in the C3b:FH1–4 complex overlaid via TED on C3d) are separated by a space large enough to be spanned by the known structure of CCP5 39 thus creating a model of a 1:1 C3b:FH complex.
DISCUSSION
Here we illuminate how FH binds to its principal target. This crucial event in regulation of C3b promulgation on self-surfaces is central to protection of host tissues from complement-mediated damage. Building on previous indications that FH19–20 binds to both C3b and the C3b-derived fragment, C3d 32, we showed that the C3d:FH19–20 interaction is a surrogate for the less experimentally accessible FH:C3b interaction. Our new observations, when combined with published data, show how the FH molecule bends back on itself such that its N terminus performs cofactor and decay accelerating activities while the C terminus recognizes a composite consisting of the TED of C3b and nearby polyanionic carbohydrates. This allows distinction of a self-surface from most bacterial ones.
Mindful of controversy surrounding the physiological validity of a crystal structure of C3d complexed with CR2 CCP1-2 40, we employed mutagenesis and NMR to select the physiologically relevant intermolecular interface from several possibilities within our crystal structure of C3d:FH19–20. Moreover this interface of C3d is consistent with other observations. (i) It is accessible in TED (i.e. within the context of C3b) tethered to a surface; moreover the orientation of FH19–20 means that CCP18 is projected clear of C3b, consistent with our finding that it binds neither C3d nor C3b (Supplementary Fig. 4 and Supplementary Table 3). (ii) It does not overlap the FH1–4 interaction site on C3b 14, consistent with avidity between sites 12. (iii) It is directly adjacent to (but non-overlapping with) the Efb-C-binding site 41 (Supplementary Fig. 8); this is important because Efb-C enhances binding of C3b to FH19–20 42. (iv) The C3d and FH residues involved are conserved (Supplementary Fig. 9).
We confirmed that previously identified GAG-binding residues of CCP20 are still accessible within this complex. This is compelling structural evidence for the hypothesis that FH19–20 interacts with a composite site consisting of the TED and proximal polyanions. Previous attempts to dock C3d onto the surface of FH19–20 now prove to have been unsuccessful 16,43. In both of these models of C3d:FH19–20, only CCP20 makes contact with C3d; moreover the interfaces on C3d differ from that identified in our co-crystal structure.
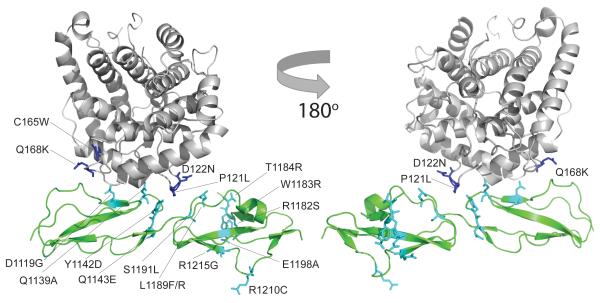
Our new data reveal locations, relative to key interaction surfaces, of disease-linked mutations and sequence variations (Fig. 6). Our structure explains the key role in C3b binding we observed for the disease-linked mutation D1119G, despite suggestions from others that this mutation is functionally null 32. Our complex also shows that the following aHUS-linked FH mutations 44 affect the C3d:FH19–20 interface and thereby perturb the fine balance of affinities needed for proper FH operation: Y1142D and Y1142C, since Tyr1142 H-bonds to C3d’s Lys178 (Lys1171); Q1143E due to its proximity to Tyr1142; L1189F, L1189R and S1191L, since Tyr1190 H-bonds to Asp122 (Asp1115) of C3d and is partially buried at the interface. Other aHUS-linked FH mutations are in the GAG-binding regions of CCP20 and/or could disrupt putative electrostatic steering events (E1198A, R1203A, R1210C, R1215G, R1182S, W1183R and T1184R). Another subset of aHUS-linked FH mutants likely promotes structural perturbations (W1157R, V1134G, G1194D, V1197A, F1199S and P1226S). Regarding the C3b side of the interaction, the following aHUS-linked mutations likely disrupt the binding interface: P121L (P1114L) 45 and D122N (D1115N) 46 are in the C3d α4-α5 loop occupying the FH19–20 intermodular cleft; C165W (C1158W) and Q168K (Q1161K) 46 are in the C3d α6–α7 loop that, intriguingly, coincides with closest approach of the N-terminal and C-terminal C3b-binding sites of FH (Fig. 5c).
Figure 6.
The location of mis-sense mutations associated with the development of (aHUS) (and Q1139A) mapped onto the structure of the C3d:FH19–20 complex. The side-chains of C3b mis-sense mutations are shown as blue sticks; side-chains of FH19–20 mis-sense mutations are shown as cyan sticks.
In a previously solved structure of the FH1–4:C3b complex 14, the four N-terminal CCPs of FH form an elongated contact with C3b running largely parallel with the long axis of C3b. If C3b in this complex were surface-attached via its thioester, CCP1 would be furthest, and CCP4 closest, to that surface. In other AP regulators, a similar three-CCP or four-CCP C3b-binding segment is connected to a C-terminal transmembrane helix or a GPI anchor, which localizes the regulator upon a cell surface requiring protection from C3b promulgation. FH is not constrained to a membrane but relies on some or all of its C-terminal 16 CCPs for localization, and exploits the preponderance of polyanionic self-surface markers to selectively increase its residency time on self-surfaces. We investigated the path taken by CCPs 5-18, which connect N-terminal and C-terminal C3b-binding sites, to construct an experimentally derived model of the FH:C3b complex.
When the C3d of C3d:FH19–20 is superimposed on the TED of C3b, the FH1–4 C-terminus and FH19–20 N-terminus are close together. A bent-back FH structure would therefore be required were a single molecule to simultaneously occupy both sites on C3b (and interact with self-surface associated polyanions), as in our model (Fig. 5a). This is fully consistent with our SAXS-derived shape-envelopes of FH fragments presented here, as well as with existing SAXS, crystal and NMR-derived structures, and agrees with previous work showing that non-liganded FH could adopt a predominantly bent-back conformation 37,47-50. Our data are incompatible with an alternative model in which FH wraps around the C3b molecule. However, we cannot exclude the existence of a complex in which one FH molecule binds to two neighboring, closely packed, C3b molecules.
Previous studies suggested a weak additional C3b-binding site within FH CCPs 6–8 12,13. While no such interface exists within our model of the complex, uncertainties in reconstruction of the N-terminal half of FH (lacking structural information for FH4–5/FH5–6) means we cannot exclude CCP6/7-C3b contacts. Nonetheless, our model places CCP7 close to the self-surface where it could recognize additional GAG surface markers; it also places the structurally deviant module, CCP13 37 close to the self-surface, although the nature of any ligand for CCP13 remains undetermined.
In summary, we present an experimentally supported structural model for how a FH molecule engages bivalently with a C3b molecule and simultaneously with self-surface polyanionic markers. The model indicates that effective complement inhibitors, with therapeutic potential, might be constructed through flexible linkage of FH19–20, via a domain with appropriately apposed N and C termini, to FH1–4 or homologous segments of other complement regulators.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGEMENTS
Use of the Protein Production Facilities at the University of Edinburgh was supported by The Wellcome Trust, the Scottish University Life Sciences Alliance and the BBSRC. A.P.H was supported by Wellcome Trust (078780/Z/05/Z to D.U. and P.N.B.), B.S.B was supported by EastChem PhD studentship and a University of Edinburgh campaign small project grant, C.M.J was supported by EC FP6 (Marie Curie EST Fellowship, contract: MEST-CT-2005-020744).
APPENDIX
ONLINE METHODS
FH1–4, FH8–15, FH15–18, FH15–19 and FH18-20 and FH19–20 Protein Production
Recombinant wild-type forms of human FH1–4, FH8–15 and wild-type and mutant forms of FH19–20 were produced utilizing a Pichia pastoris expression system as previously described 12,16,31. Resulting recombinant forms of FH were secreted into the media and purified by successive ion exchange and size exclusion chromatography steps. Additional CCP15–18, CCP15–19 and CCP18–20 constructs of human FH were also engineered utilizing a FH codon-optimized gene and cloned into the Pichia pastoris pPICZαB expression vector (Invitrogen). Recombinant forms of these proteins were generated and purified as outlined above for FH1–4, FH8–15 and FH19–20.
Wild-type and mutant C3d production
Recombinant wild-type, E117A (prepro C3 numbering: E1110A), D122A (D1115A), E117A/D122A (E1110A/D1115A), E160A (E1153A) and I164A (I1157A) mutant forms of C3d, corresponding to residues 996–1306 of wild-type human C3d (C3d996-1306), and containing an additional C17A (C1011A) point mutation, were produced utilizing an Escherichia coli expression system as previously described 52,53. GST-tagged forms of this protein were obtained as described in the Supplementary Methods.
Analysis of wild-type and mutant FH binding by surface plasmon resonance (SPR)
Surface plasmon resonance was used to determine the binding affinity of wild-type and mutant forms of FH19–20 (D1119G, R1182S, W1183L, W1183R, T1184R, L1189F, L1189R, S1191L, S1191L-V1197A, V1197A, R1203S, R1210C, R1215Q), and wild-type FH15–18, FH15–19 and FH18–20 to amine-coupled and physiologically-immobilized forms of wild-type C3d and C3dg, respectively, as described in the Supplementary Methods. Binding analyses under identical solution conditions were also carried out utilizing immobilized recombinant mutant forms of C3d (E117A, (E1110A) D122A (D1115A), E117A D122A (E1110 D1115A), E160A (E1153A) and I164A (I1157A)), and wild-type and R1182S, L1189F, L1189R and R1215Q variant forms of FH19–20. Binding of C3b to FH15–19, and FH18-20 were carried out under identical conditions to those described above. To demonstrate that the GAG-binding site of FH CCP20 does not overlap with the FH19–20 binding site on C3d, SPR was also used to determine the binding affinity of wild-type FH19–20 to amine-coupled recombinant C3d in the presence of a 20-fold excess of the pentasaccharide GAG-surrogate, Fondaparinux (kindly donated by GlaxoSmithKline PLC, U.K) (see Supplementary Methods).
Crystallization and data collection
Crystals of C3d in complex with FH19–20 were grown at 17 °C by the vapor diffusion method from hanging drops. Drops contained an equal volume of a protein solution in which a complex had been formed with a 1:1 molar ration of C3d:FH19–20 in phosphate buffered saline, pH 7.4 (PBS: 137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4), with an equal volume of well solution. Well solution conditions for the C3d:FH19–20 complex were 0.1 M Tris-HCl, pH 9.0, 8% w/v PEG 8K. Prior to data collection, crystals were cryoprotected with well solution additionally containing 25% v/v glycerol (added stepwise), mounted in loops and flash frozen in liquid nitrogen at 77K. Additional crystals of the C3d:FH19–20 complex were also grown at pH 7.0 as described in the Supplementary Methods. Intensity data were collected (Phi scans were 1° over 360°) at the Diamond synchrotron radiation facility in Oxfordshire, United Kingdom on beamlines I02, I03 and I04. Data were then processed in the P1 space group with MOSFLM 54 and scaled with SCALA 55. The P1 space-group was confirmed using the program POINTLESS and there was no indication of non-crystallographic symmetry by self rotation function analysis 55.
C3d:FH19–20 Structure determination
The structure of the C3d:FH19–20 complex was solved by molecular replacement using the program PHASER 56, with the previously determined structure of FH (PDB accession code: 2G7I 43) and C3d (PDB accession code: 1C3D) 57) as search models. There was a clear molecular replacement solution, with three C3d and three FH monomers in the crystallographic asymmetric unit. The initial model was subjected to ten cycles of restrained refinement using the program REFMAC 58 resulting in R/Rfree values of 37.85 and 41.22, respectively. Module 20 of FH19–20 (Chain F) was removed at this stage of refinement as there was no clear interpretable electron density. Using the program COOT 59, alternative side chain conformers were constructed and additional residues were added to the N- and C-terminal. The model was then subjected to several rounds of restrained refinement, resulting in R/Rfree values of 32.4 and 35.8, respectively. With the improvement of phases, Fo-Fc electron density corresponding to module 20 of FH·19-20 (Chain F) appeared, allowing for manual building and completion of this module. Areas of disorder were carefully modeled into Fo-Fc electron density and the changes in R/Rfree values were used to assess final model quality. Water molecules were then added to the model using the program PHENIX and after several rounds of refinement the R/Rfree values converged to 17.06/20.59, respectively (Table 1). The geometry of the model was assessed using MolProbity 60 and the coordinates for the 2.1 Å structure have been deposited in the Protein Data Bank with the code 3OXU. Superimpositions of structures were performed using Pymol. Also see Supplementary Methods.
Glycosaminoglycan (GAG) preparation
Heparin-derived octasaccharides were prepared as outlined in the Supplementary Methods.
Hemolysis assay
Ability of recombinant wild-type and D1119G forms of FH19–20 to inhibit FH mediated protection of sheep erythrocytes was measured as outlined in the Supplementary Methods.
Nuclear Magnetic Resonance of the C3d:FH19–20 complex and FH19–20:GAG interactions
C3d for the NMR study was purchased from Complement Technology (Texas) and 15N-labeled FH19–20 was prepared as described previously 16. All [1H, 15N]-HSQC spectra were collected at 25 °C on a Bruker Avance 800 MHz NMR spectrometer equipped with a cryogenic TCI probe as described in the Supplementary Methods.
Small-angle X-ray scattering, ab initio shape determination and molecular modelling
Synchrotron radiation X-ray scattering data of recombinantly expressed-forms of FH1–4, FH8-15 and FH15-19 were collected, processed and these data together with existing three-dimensional FH structural data were utilized to generate a composite full-length FH:C3b model as set out in the Supplementary Methods.
Footnotes
Accession codes. Protein Data Bank: Coordinates and structure factors for the C3d:FH19–20 complex have been deposited under accession number 3OXU.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
REFERENCES
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Cordoba SR, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin. Exp. Immunol. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–6. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 4.Pangburn MK, Schreiber RD, Muller-Eberhard HJ. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein beta1H for cleavage of C3b and C4b in solution. J. Exp. Med. 1977;146:257–70. doi: 10.1084/jem.146.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiler JM, Daha MR, Austen KF, Fearon DT. Control of the amplification convertase of complement by the plasma protein beta1H. Proc. Natl. Acad. Sci. USA. 1976;73:3268–72. doi: 10.1073/pnas.73.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whaley K, Ruddy S. Modulation of the alternative complement pathways by beta 1 H globulin. J. Exp. Med. 1976;144:1147–63. doi: 10.1084/jem.144.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ripoche J, et al. Partial characterization of human complement factor H by protein and cDNA sequencing: homology with other complement and non-complement proteins. Biosci. Rep. 1986;6:65–72. doi: 10.1007/BF01145180. [DOI] [PubMed] [Google Scholar]
- 8.Meri S, Pangburn MK. Regulation of alternative pathway complement activation by glycosaminoglycans: specificity of the polyanion binding site on factor H. Biochem. Biophys. Res. Commun. 1994;198:52–9. doi: 10.1006/bbrc.1994.1008. [DOI] [PubMed] [Google Scholar]
- 9.Pangburn MK. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology. 2000;49:149–57. doi: 10.1016/s0162-3109(00)80300-8. [DOI] [PubMed] [Google Scholar]
- 10.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J. Immunol. 1995;155:348–56. [PubMed] [Google Scholar]
- 11.Pangburn MK. Cutting edge: localization of the host recognition functions of complement factor H at the carboxyl-terminal: implications for hemolytic uremic syndrome. J. Immunol. 2002;169:4702–6. doi: 10.4049/jimmunol.169.9.4702. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt CQ, et al. A new map of glycosaminoglycan and C3b binding sites on factor H. J. Immunol. 2008;181:2610–2619. doi: 10.4049/jimmunol.181.4.2610. [DOI] [PubMed] [Google Scholar]
- 13.Sharma AK, Pangburn MK. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc. Natl. Acad. Sci. USA. 1996;93:10996–1001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, et al. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat. Immunol. 2009;10:728–33. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buddles MR, Donne RL, Richards A, Goodship J, Goodship TH. Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2000;66:1721–2. doi: 10.1086/302877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert AP, Uhrin D, Lyon M, Pangburn MK, Barlow PN. Disease-associated sequence variations congregate in a polyanion recognition patch on human factor H revealed in three-dimensional structure. J. Biol. Chem. 2006;281:16512–16520. doi: 10.1074/jbc.M513611200. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Caballero D, et al. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2001;68:478–84. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira VP, Herbert AP, Hocking HG, Barlow PN, Pangburn MK. Critical role of the C-terminal domains of factor H in regulating complement activation at cell surfaces. J. Immunol. 2006;177:6308–16. doi: 10.4049/jimmunol.177.9.6308. [DOI] [PubMed] [Google Scholar]
- 19.Jokiranta TS, et al. Binding of complement factor H to endothelial cells is mediated by the carboxy-terminal glycosaminoglycan binding site. Am. J. Pathol. 2005;167:1173–81. doi: 10.1016/S0002-9440(10)61205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackmore TK, et al. Identification of the second heparin-binding domain in human complement factor H. J. Immunol. 1998;160:3342–8. [PubMed] [Google Scholar]
- 21.Blackmore TK, Sadlon TA, Ward HM, Lublin DM, Gordon DL. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J. Immunol. 1996;157:5422–7. [PubMed] [Google Scholar]
- 22.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7227–32. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehn BM. Gene discovery provides clues to cause of age-related macular degeneration. Jama. 2005;293:1841–5. doi: 10.1001/jama.293.15.1841. [DOI] [PubMed] [Google Scholar]
- 25.Zareparsi S, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am. J. Hum. Genet. 2005;77:149–53. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiesmann C, et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–20. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 27.Lian LY, Barsukov IL, Sutcliffe MJ, Sze KH, Roberts GC. Protein-ligand interactions: exchange processes and determination of ligand conformation and protein-ligand contacts. Methods Enzymol. 1994;239:657–700. doi: 10.1016/s0076-6879(94)39025-8. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo H, et al. Identification by NMR spectroscopy of residues at contact surfaces in large, slowly exchanging macromolecular complexes. J. Am. Chem. Soc. 1999;121:9903–9904. [Google Scholar]
- 29.Daughdrill GW, et al. Chemical shift changes provide evidence for overlapping single-stranded DNA- and XPA-binding sites on the 70 kDa subunit of human replication protein A. Nucleic Acids Res. 2003;31:4176–4183. doi: 10.1093/nar/gkg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAlister MSB, et al. NMR analysis of interacting soluble forms of the cell-cell recognition molecules CD2 and CD48. Biochemistry. 1996;35:5982–5991. doi: 10.1021/bi952756u. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira VP, et al. The Binding of Factor H to a Complex of Physiological Polyanions and C3b on Cells Is Impaired in Atypical Hemolytic Uremic Syndrome. J. Immunol. 2009;182:7009–7018. doi: 10.4049/jimmunol.0804031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehtinen MJ, Rops AL, Isenman DE, van der Vlag J, Jokiranta TS. Mutations of factor H impair regulation of surface-bound C3b by three mechanisms in atypical hemolytic uremic syndrome. J. Biol. Chem. 2009;284:15650–8. doi: 10.1074/jbc.M900814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morikis D, Lambris JD. The electrostatic nature of C3d-complement receptor 2 association. J. Immunol. 2004;172:7537–47. doi: 10.4049/jimmunol.172.12.7537. [DOI] [PubMed] [Google Scholar]
- 34.Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Geisbrecht BV, Lambris JD. Cutting edge: members of the Staphylococcus aureus extracellular fibrinogen-binding protein family inhibit the interaction of C3d with complement receptor 2. J. Immunol. 2008;181:7463–7. doi: 10.4049/jimmunol.181.11.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee A, Lehtinen MJ, Kajander T, Goldman A, Jokiranta TS. Both domain 19 and domain 20 of factor H are involved in binding to complement C3b and C3d. Mol. Immunol. 47:1686–91. doi: 10.1016/j.molimm.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Prosser BE, et al. Structural basis for complement factor H linked age-related macular degeneration. J. Exp. Med. 2007;204:2277–83. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt CQ, et al. The Central Portion of Factor H (Modules 10-15) Is Compact and Contains a Structurally Deviant CCP Module. J. Mol. Biol. 2010;395:105–122. doi: 10.1016/j.jmb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barlow PN, et al. Solution structure of a pair of complement modules by nuclear magnetic resonance. J. Mol. Biol. 1993;232:268–84. doi: 10.1006/jmbi.1993.1381. [DOI] [PubMed] [Google Scholar]
- 39.Barlow PN, et al. Solution structure of the fifth repeat of factor H: a second example of the complement control protein module. Biochemistry. 1992;31:3626–34. doi: 10.1021/bi00129a011. [DOI] [PubMed] [Google Scholar]
- 40.Szakonyi G, et al. Structure of complement receptor 2 in complex with its C3d ligand. Science. 2001;292:1725–8. doi: 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- 41.Hammel M, et al. A structural basis for complement inhibition by Staphylococcus aureus. Nat. Immunol. 2007;8:430–7. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, et al. Allosteric inhibition of complement function by a staphylococcal immune evasion protein. Proc. Natl. Acad. Sci. U. S. A. 2010 doi: 10.1073/pnas.1003750107. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jokiranta TS, et al. Structure of complement factor H carboxyl-terminus reveals molecular basis of atypical haemolytic uremic syndrome. Embo J. 2006;25:1784–94. doi: 10.1038/sj.emboj.7601052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders RE, et al. The interactive Factor H-atypical hemolytic uremic syndrome mutation database and website: update and integration of membrane cofactor protein and Factor I mutations with structural models. Hum. Mutat. 2007;28:222–34. doi: 10.1002/humu.20435. [DOI] [PubMed] [Google Scholar]
- 45.Miller EC, Roumenina L, Fremeaux-Bacchi V, Atkinson JP. Functional characterization of mutations in complement C3 that predispose to aHUS. Mol. Immunol. 2010;47:2291. [Google Scholar]
- 46.Fremeaux-Bacchi V, et al. Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood. 2008;112:4948–52. doi: 10.1182/blood-2008-01-133702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aslam M, Perkins SJ. Folded-back solution structure of monomeric factor H of human complement by synchrotron X-ray and neutron scattering, analytical ultracentrifugation and constrained molecular modelling. J. Mol. Biol. 2001;309:1117–38. doi: 10.1006/jmbi.2001.4720. [DOI] [PubMed] [Google Scholar]
- 48.Fernando AN, et al. Associative and structural properties of the region of complement factor H encompassing the Tyr402His disease-related polymorphism and its interactions with heparin. J. Mol. Biol. 2007;368:564–81. doi: 10.1016/j.jmb.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 49.Oppermann M, et al. The C-terminus of complement regulator Factor H mediates target recognition: evidence for a compact conformation of the native protein. Clin. Exp. Immunol. 2006;144:342–52. doi: 10.1111/j.1365-2249.2006.03071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okemefuna AI, et al. The regulatory SCR-1/5 and cell surface-binding SCR-16/20 fragments of factor H reveal partially folded-back solution structures and different self-associative properties. J. Mol. Biol. 2008;375:80–101. doi: 10.1016/j.jmb.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young KA, Chen XS, Holers VM, Hannan JP. Isolating the Epstein-Barr virus gp350/220 binding site on complement receptor type 2 (CR2/CD21) J. Biol. Chem. 2007;282:36614–25. doi: 10.1074/jbc.M706324200. [DOI] [PubMed] [Google Scholar]
- 53.Shaw CD, et al. Delineation of the Complement Receptor Type 2-C3d complex by site-directed mutagenesis and molecular docking. J. Mol. Biol. 2010;404:697–710. doi: 10.1016/j.jmb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. 1992. [Google Scholar]
- 55.Evans P. Scaling and assessment of data quality. Acta. Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 56.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–81. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 58.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.