Abstract
Aldehyde oxidase (AO; EC 1.2.3.1) activity was measured in seedlings of wild type or an auxin-overproducing mutant, superroot1 (sur1), of Arabidopsis thaliana. Activity staining for AO after native polyacrylamide gel electrophoresis separation of seedling extracts revealed that there were three major bands with AO activity (AO1–3) in wild-type and mutant seedlings. One of them (AO1) had a higher substrate preference for indole-3-aldehyde. This AO activity was significantly higher in sur1 mutant seedlings than in the wild type. The difference in activity was most apparent 7 d after germination, the same time required for the appearance of the remarkable sur1 phenotype, which includes epinastic cotyledons, elongated hypocotyls, and enhanced root development. Higher activity was observed in the root and hypocotyl region of the mutant seedlings. We also assayed the indole-3-acetaldehyde oxidase activity in extracts by high-performance liquid chromatography detection of indole-3-acetic acid (IAA). The activity was about 5 times higher in the extract of the sur1 seedlings, indicating that AO1 also has a substrate preference for abscisic aldehyde. Treatment of the wild-type seedlings with picloram or IAA caused no significant increase in AO1 activity. This result suggested that the higher activity of AO1 in sur1 mutant seedlings was not induced by IAA accumulation and, thus, strongly supports the possible role of AO1 in IAA biosynthesis in Arabidopsis seedlings.
AO (EC 1.2.3.1) has been extensively investigated in animals and microorganisms (Hall and Krenitsky, 1986; Yoshihara and Tatsumi, 1986). The enzyme has been implicated in the detoxification of various xenobiotics (Bauer and Howard, 1991; Stoddart and Levine, 1992; Hirao et al., 1994) and is thought to be involved in the biosynthesis of biologically important compounds such as retinoic acid (Tomita et al., 1993; Huang and Ichikawa, 1994). In plants much attention has been focused on the role of this enzyme in IAA biosynthesis, because it can catalyze the oxidation of IAAld to form IAA, a possible route for IAA synthesis in higher plants. In fact, some investigations have shown that an AO, tentatively called IAAld oxidase, may be involved in IAA synthesis (Rajagopal, 1971; Bower et al., 1978; Miyata et al., 1981; Koshiba and Matsuyama, 1993; Koshiba et al., 1996; Tsurusaki et al., 1997), but the actual function of the enzyme in IAA biosynthesis has not been definitively confirmed.
The characterization of mutants defective for the biosynthesis of phytohormones such as GA3, ethylene, or ABA has led to significant advances in the understanding of hormone biosynthetic pathways and of the involved enzymes (Davies, 1995). Unfortunately, no auxin-deficient mutants have been identified so far, most probably because such mutants would be lethal. Trp auxotrophic mutants have been used to investigate the IAA biosynthesis pathway in maize (Wright et al., 1991) and Arabidopsis (Normanly et al., 1993). These mutants, which contained very low amounts of Trp, accumulated high levels of IAA. These results indicate the presence of a Trp-independent IAA biosynthesis pathway. More recently, the Arabidopsis mutants superroot1 (sur1), rooty (rty), aberrant lateral root formation1 (alf1), and hookless 3 (hls3) were isolated independently (Boerjan et al., 1995; Celenza et al., 1995; King et al., 1995; Lehman et al., 1996). They are now known to be alleles of a single locus located on chromosome 2. These mutant seedlings have excess adventitious and lateral roots and contain increased levels of endogenous IAA. The auxin-overproducing mutants are useful for understanding how IAA biosynthesis is regulated and for identifying the enzymes that are involved (Kawaguchi and Syōno, 1996). Gopalraj et al. (1996) reported that the Sur1 gene encoded a protein that had great similarity to Tyr aminotransferases from animals. Several studies of plant aminotransferases have indicated that aspartate and aromatic amino acid aminotransferases have a wide substrate specificity, including not only Tyr but also Trp (Wightman and Forest, 1978). These facts suggest that the Sur1 gene may be involved in the metabolism of chorismic acid, which is a possible precursor of IAA in both Trp and non-Trp pathways or of Trp as an IAA precursor in the Trp pathway.
We have investigated AO activities in wild-type and sur1 mutant seedlings and found that one of the three AO isoforms detected by activity staining of native polyacrylamide gels was expressed at much higher levels in sur1 mutant seedlings. In the present work we studied the substrate preference and tissue specificity of the enzyme in relation to IAA overproduction in the sur1 mutant seedlings.
MATERIALS AND METHODS
Arabidopsis thaliana (Columbia ecotype) seeds of wild type and a sur1 (sur1–1) mutant were obtained as described by Boerjan et al. (1995). The seeds were aseptically sown on agar plates and germinated under 16 h of light and 8 h of dark at 22°C. Whole seedlings, or cotyledons (including first leaves), hypocotyls, and roots, were sampled as appropriate. The whole seedlings or excised tissues were washed with distilled water, the excess water was removed, and then the samples were immediately frozen in liquid N2 and stored at −80°C until use. To investigate the effect of picloram the wild-type seeds were sown on agar plates containing 5 μm picloram and germinated for 8 d as described above. IAA treatment was performed as follows: after 4 d of germination, the wild-type seedlings were transferred to IAA-containing (10 μm) agar plates and growth was continued under the same conditions. The seedlings were sampled on the 8th d after germination and the AO activity was determined.
Enzyme Extraction
The following procedure was used for the extraction of small samples (less than 100 mg fresh weight). Frozen samples were quickly homogenized in 1.5-mL Eppendorf tubes with 200 to 400 mL of extraction buffer containing 50 mm Tris-HCl buffer (pH 7.5), 1 mm EDTA, 1 μm sodium molybdate, 5 μm leupeptin, 10 μm FAD, 2 mm DTT, and Polyclar AT (0.2 g/g fresh weight, Wako Pure Chem, Osaka, Japan) with a Physcotron (equipment with a 4-mm-diameter knife, Nichion-Irika, Funabashi, Japan). After centrifugation of the homogenate at 15,000 rpm for 20 min, the supernatant was concentrated by filtration using a Centricon (model UFC-C3LGC, Millipore) to give a final volume of 50 to 100 μL. The concentrated solutions were used as crude enzyme preparations for native PAGE.
Larger samples (1–3 g) were ground to a powder with liquid N2 and homogenized in 10 mL of the extraction buffer and Polyclar AT (0.2 g/g fresh weight) with a mortar and pestle. After the sample was centrifuged at 12,000g for 15 min, the supernatant was fractionated with ammonium sulfate (0–60% saturation). The precipitate was dialyzed against 20 mm Tris-HCl buffer (pH 8.0) containing 1 mm EDTA, 1 μm sodium molybdate, 1 μm leupeptin, 10 μm FAD, and 2 mm DTT. The dialyzed sample was centrifuged at 20,000g for 20 min, and the supernatant was used for determining the substrate specificity after native PAGE and for assaying the capacity to oxidize IAld and IAAld to form ICA and IAA, respectively.
The protein concentration of samples was measured by using a protein assay kit (Bio-Rad).
Native PAGE and AO Activity Stain
Native PAGE was performed with a 7.5% acrylamide gel in a Laemmli buffer system (Laemmli, 1970) in the absence of SDS at 4°C. After electrophoresis, the gel was immersed in 0.1 m potassium phosphate buffer (pH 7.5) for 5 min, and then enzyme activity staining was developed in a mixture containing 0.1 m phosphate buffer (pH 7.5), 1 mm substrate, 0.1 mm phenazine methosulfate, and 0.4 mm 3(4,5-dimethylthiazolyl-2)2,5-diphenyltetrazolium bromide at 30°C in the dark for 30 to 60 min. After activity staining, the gels were scanned to quantify the relative intensity of AO activity bands using the computer software NIH Image 1.6.
Assay of Activity with IAld and IAAld
AO activity with IAld and IAAld as the substrates was assayed by determining the amount of corresponding acid, ICA and IAA, respectively, formed with a reversed-phase HPLC with an ODS C18 column as described previously (Koshiba et al., 1996). Reaction mixtures (100 μL) contained 10 to 50 μL of enzyme solution, 0.05 mm IAld or 0.1 mm IAAld, and 0.1 m potassium phosphate buffer (pH 7.5). The reaction was performed at 30°C for 30 min and stopped by adding 1 n HCl (and 2 m NaHSO4, when IAAld was used as a substrate) and methanol. The mixture was then centrifuged and a portion of the supernatant (100 μL) was subjected to HPLC. The amounts of ICA and IAA were determined from their peak areas.
Chemicals
IAAld was prepared from IAAld bisulfite (Sigma), according to the method described by Bower et al. (1978). (±)-ABAld was synthesized by active manganese dioxide oxidation of (±)-(2Z,4E)-abscisic alcohol in chloroform, followed by purification with TLC (Silica gel 60 PF254, Merck), as described by Yamamoto and Oritani (1996).
RESULTS
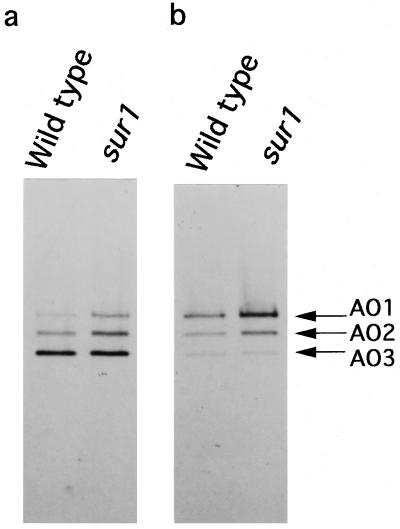
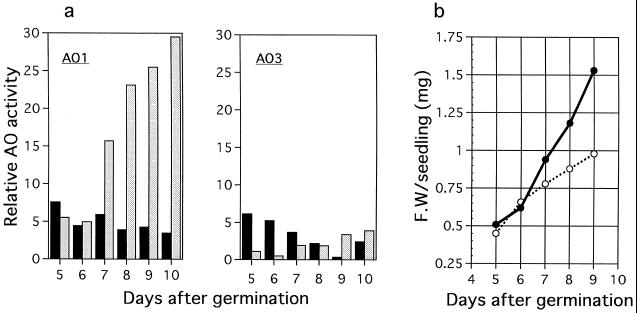
Crude extracts obtained from 8-d-old seedlings of wild-type and sur1 plants were subjected to native PAGE and then activity staining with benzaldehyde or IAld as the substrates (Fig. 1). Three bands (AO1, AO2, and AO3) of activity were detected and the intensity of the lower bands was stronger when benzaldehyde was the substrate, whereas the opposite result was observed when IAld was the substrate. This suggests that AO1 and AO3 have different substrate preferences related to their biological role. The changes in the intensity of these AO1 and AO3 activity bands were investigated in seedlings 5 to 10 d after germination (Fig. 2a). The intensity of AO2 is not shown, because the band of AO2 activity had intermediate intensities between the AO1 and AO3 bands. AO1 and AO3 had similar activities in wild-type seedlings, slightly decreasing over time during the period studied. However, in seedlings of the sur1 mutant the AO1 activity was remarkably higher than that in the wild type after 7 d of germination. A differential increase in fresh weight of the mutant seedlings compared with the wild type could first be noticed at 7 d (Fig. 2b). Mutant seedlings started exhibiting the typical phenotype with epinastic cotyledons, elongated hypocotyls, and enhanced root development at approximately this time. Approximately 6 d after germination, IAA overproduction was observed in the mutant seedlings (Boerjan et al., 1995).
Figure 1.
Pattern of AO activity bands from Arabidopsis seedlings after native PAGE. Ammonium sulfate-fractionated crude enzyme samples were obtained from 8-d-old seedlings of the wild type and the sur1 mutant. Gel electrophoresis and activity staining were performed as described in Methods using benzaldehyde (a) or IAld (b) as the substrates. Eighty micrograms of protein was loaded in each lane. The positions of the three bands, AO1, AO2, and AO3, are indicated with arrows.
Figure 2.
Changes in band intensities of AO activities (a) and in fresh weights (b) of Arabidopsis wild-type (shaded symbols) and sur1 (open symbols) seedlings during germination. After the AO activity developed on native gels using IAld, the relative intensity of the bands was calculated by using computer software (NIH Image 1.6). Each sample contained 30 μg of protein. Fresh weight (F.W) of the seedlings was measured by weighing 50 to 200 seedlings from each day, and the values for the average fresh weight per seedling are presented.
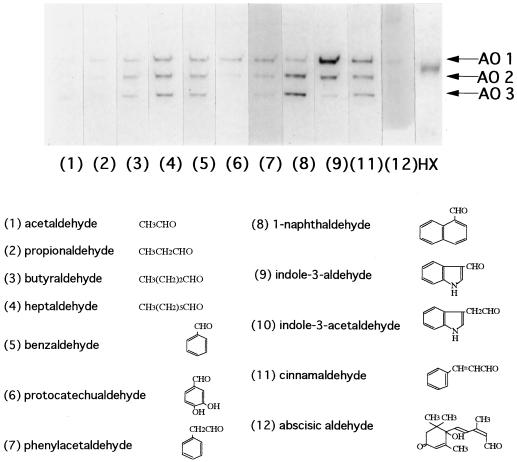
Using the enzyme preparation obtained by ammonium sulfate fractionation of extracts from 8-d-old sur1 mutant seedlings, we studied further the substrate preferences of the different AOs using 11 different aldehydes and hypoxanthine as the substrates for gel detection of the AO activities. The results with the structures of the tested substrates are summarized in Figure 3. AO1 activity showed a strong preference for IAld, and AO3 could efficiently oxidize 1-naphthaldehyde. AO2 had intermediate properties. When ABAld was used as a substrate, only AO1 showed a very low activity, and AO2 and AO3 had almost no activity (Fig. 3, lane 12). XD activity was detected at a position between AO1 and AO2, suggesting that the enzyme is different from the AOs (Fig. 3, lane HX).
Figure 3.
Arabidopsis AO activity after native PAGE with different substrates. Ammonium sulfate-fractionated enzyme solution obtained from 8-d-old sur1 mutant seedlings was loaded with 54 μg of protein in each lane. After native PAGE, the activity bands were developed separately with strips from each lane using 11 aldehydes (lanes 1–12, except for 10) and hypoxanthine (HX). The number at the bottom of each lane corresponds to the substrate used. The substrates are shown with their structure. IAAld (lane 10) could not be used as a substrate for the activity stain (Koshiba et al., 1996) but is added here so that the structure can be compared with the structure of the others.
Because the staining of native gels with IAAld became increasingly difficult (as mentioned previously; Koshiba et al., 1996), the level of IAAld oxidase activity was determined using HPLC to measure the amount of IAA formed by oxidation of IAAld. About 5 times higher activity was detected in sur1 mutant seedlings compared with wild-type seedlings (Table I). When IAld was used as a substrate, about 3.5-fold higher activity was found in the mutant seedlings. The difference in IAAld oxidase activity in mutant and wild-type seedlings corresponded well with the increase in the band intensity of AO1 in the 8-d-old seedlings (Fig. 2a), in which the intensity of AO1 was about 5 times higher in the sur1 mutant than in the wild type. This result also indicates that AO1 is able to oxidize IAld as well as IAAld, because no such difference in activity was observed for the AO3 (Figs. 1 and 3), which could account for this increased IAAld oxidase activity. AO2 could also contribute to the higher activity in sur1 mutant seedlings but to a lesser extent than AO1. We conclude from this study that AO1 has a preference for the aldehydes having an indole-ring structure and has higher activity in the IAA-overproducing sur1 mutant.
Table I.
AO activity of wild-type and surI mutant seedlings of Arabidopsis using IAld and IAAld as the substrates
| Substrate | Activity
|
|
|---|---|---|
| Wild type | sur1 | |
| pmol product min−1 mg−1 protein | ||
| IAld | 54.5 | 186.9 |
| IAAld | 22.2 | 119.5 |
The activity was assayed by determining the amounts of ICA and IAA formed during the reaction, as described in Methods. Two independent experiments each with two assays were carried out and one typical result is presented here.
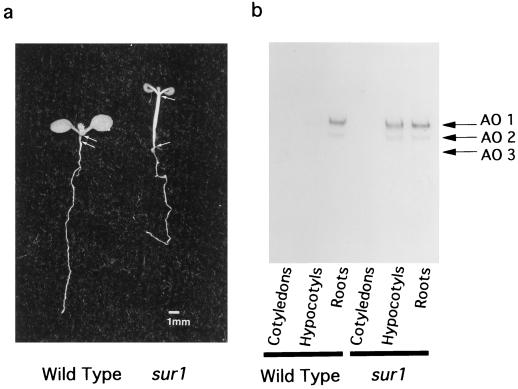
The organ-specific distribution of AOs was investigated in cotyledons (including first leaves), hypocotyls, or roots derived from 8-d-old wild-type and sur1 mutant seedlings (Fig. 4). Crude extracts from these tissues were subjected to native PAGE, and activity was developed using IAld as a substrate (Fig. 4b). In wild-type seedlings AO1 activity was detected almost only in roots. However, in sur1 mutant seedlings AO1 activity was detected not only in roots but also in hypocotyls, with the band in the hypocotyls having much stronger intensity than in the wild type. When IAld was used as a substrate, the AO3 band was too weak to be detectable, but when benzaldehyde was used as a substrate, AO3 was seen almost exclusively in the cotyledons of both wild-type and sur1 mutant seedlings (data not shown). Again, AO2 had intermediate activity. Thus, AO1 is highly expressed in sur1 mutant seedlings, especially in root and hypocotyl regions.
Figure 4.
Organ-specific distribution of AO activity detected after native PAGE. Cotyledons (including first leaves), hypocotyls, and roots were excised from 8-d-old seedlings of Arabidopsis wild type and the sur1 mutant. a, Picture of wild-type and sur1 seedlings, indicating the excised parts with white arrows. b, AO activity was developed using IAld as a substrate. Ten micrograms of protein was loaded in each lane.
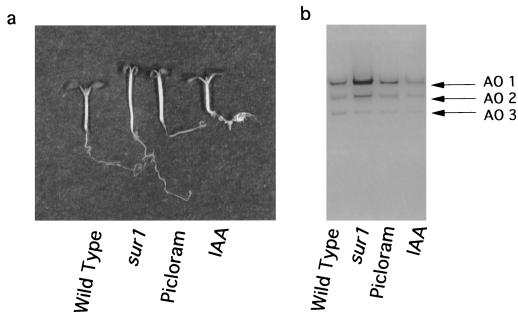
To determine whether the higher activity of AO1 in sur1 mutant seedlings is induced by a high level of endogenous IAA in the mutant seedlings, the activity of auxin-treated wild-type seedlings was investigated (Fig. 5). Whereas the phenotype of picloram-treated seedlings was very similar to that of sur1 seedlings, and the IAA treatment induced the formation of many roots (Fig. 5a), no significant increase in AO1 activity was detected in picloram- or IAA-treated wild-type seedlings (Fig. 5b).
Figure 5.
Effect of picloram and IAA on morphology of wild-type seedlings of Arabidopsis and AO activities in the seedlings. a, Picture of wild-type, sur1 mutant, picloram, and IAA-treated 8-d-old seedlings. b, Crude enzyme extracts were obtained from wild-type, sur1, picloram, and IAA-treated 8-d-old seedlings, and AO activity was developed using IAld as a substrate. Sixty micrograms of protein was loaded in each lane.
DISCUSSION
Plant AOs have been studied from several sources, including oat (Avena sativa) coleoptiles (Rajagopal, 1971), potato tubers (Rothe, 1974), cucumber (Cucumis sativus) seedlings (Bower et al., 1978), pea seedlings (Miyata et al., 1981), and maize (Zea mays L.) coleoptiles (Koshiba et al., 1996). All of the AOs had relatively wide substrate specificity, and after native PAGE, several isoforms with different mobilities were separated (Rothe, 1974; Koshiba et al., 1996). Among these, one of the maize AOs had affinity for IAld and IAAld, and AOs from oat and cucumber have been shown to oxidize IAAld to produce IAA. Thus, the enzyme has been presumed to be implicated in IAA biosynthesis, but no direct evidence for this has previously been reported in any plants.
In this study we detected three major bands of AO in Arabidopsis seedlings and one of them (AO1) clearly had a strong preference for IAld and IAAld. The activity is much higher in the seedlings of the sur1 mutant than in those of the wild type. The mutant develops excess adventitious and lateral roots and contains 4- to 14-fold higher levels of free IAA than that of the wild type (Boerjan et al., 1995). Therefore, it is likely that AO1, which has higher activity in this mutant, may be involved in IAA biosynthesis and contribute to the overproduction of IAA in the seedlings. However, alternative explanations are also possible; AO1 activity is induced by high levels of endogenous IAA in sur1 mutant seedlings, or the enzyme is mainly expressed in root primordia in sur1 mutant seedlings. If the latter is the case, the numerous roots generated in sur1 mutant seedlings would result in large amounts of the enzyme in mutant seedlings. But, since auxin treatments with either picloram or IAA did not cause any increase in AO1 activity (Fig. 5), these two possibilities could be eliminated. Thus, it is possible that AO1 has a role in IAA biosynthesis in Arabidopsis, at least at early developmental stages.
The results presented here also demonstrate that AO is expressed in an organ-specific manner. AO1 was highly expressed in roots of both wild-type and sur1 mutant plants and also specifically in the hypocotyl of the mutant seedlings (Fig. 4), whereas AO3 was detected almost exclusively in the cotyledon (and first-leaf) region. The fact that excised roots and hypocotyls of the mutant seedlings could grow on auxin-free agar medium clearly suggests that they must have the ability to synthesize IAA in situ (Boerjan et al., 1995). In addition, when a reporter construct made from the IAA-inducible GH3 promoter (Hagen et al., 1991) fused to the gene encoding β-glucuronidase was used to transform sur1 and wild-type seedlings, higher expression levels were detected in the lower region of the hypocotyl and roots of the transgenic sur1 mutant seedlings (Delarue, 1996). This is consistent with the above results. However, further experiments using molecular and genetic approaches are required to understand where and how IAA is synthesized.
Recently, the Rty gene, allelic to the Sur1 gene, was reported to encode a protein with high similarity to Tyr aminotransferases from human and rat (Gopalraj et al., 1996). This indicates that the Rty (Sur1) gene may play a role in modulating IAA levels. Gopalraj et al. (1996) proposed a model suggesting that the gene was involved in Tyr and Phe synthesis. The mutation, by blocking the synthesis of these amino acids, could cause accumulation of chorismic acid, which would then be available for the synthesis of Trp and IAA. Nonhebel et al. (1993) presented results suggesting that aromatic aminotransferases are involved in IAA biosynthesis because the enzyme could catalyze the formation of IPy from Trp. In fact, several reports have suggested that Trp aminotransferase was able to transaminate other aromatic amino acids, including Tyr (Truelsen, 1972; Noguchi and Hayashi, 1980; McQueen-Mason and Hamilton, 1989; Koshiba et al., 1993). Thus, another possible explanation is that the Rty (Sur1) gene is an aromatic aminotransferase and could be involved in IAA biosynthesis by transaminating Trp. In the mutant seedlings the production of IPy is blocked, resulting in the accretion of the last step reaction, from IAAld to IAA, by increasing IAAld oxidase activity. According to this hypothesis, the question of whether the different steps of a pathway producing IAAld, such as via tryptamine or indole-3-acetaldoxime, are also increased in the mutant seedlings remains to be further elucidated.
The physiological role of plant AO(s) has also been discussed in relation to ABA biosynthesis, because MoCo-deficient mutants of barley (Walker-Simmons et al., 1989), tobacco (Leydecker et al., 1995), tomato (Marin and Marion-Poll, 1997), and Arabidopsis leaves (Schwartz et al., 1997) lacked MoCo-containing AO and XD activities and these mutants had impaired ABA production. These findings suggest that ABAld oxidase is a Mo-containing AO. However, in the present study we could not detect any ABAld-specific AO activity (band) in the extracts of Arabidopsis seedlings.
If one of the different AOs is involved in IAA biosynthesis, the MoCo biosynthesis mutant plants must also be impaired in IAA production. However, the MoCo mutants exhibit no obvious IAA deficiency or IAA auxotrophy phenotype. One possible explanation could be that these mutants are leaky and small amounts of IAA are sufficient to promote normal growth. Another possibility is that several parallel pathways of IAA biosynthesis exist in plants, operating at different stages of development and/or in different organs or tissues (Normanly et al., 1995; Kawaguchi and Syono, 1996; Normanly, 1997). In fact, Michalczuk, et al. (1992) observed stage-specific operation of Trp and non-Trp pathways, and the presence of a non-Trp IAA biosynthesis pathway was suggested by the Arabidopsis alf3–1 mutant, which has a specific defect in the elongation of lateral roots (Celenza et al., 1995).
By using degenerate primers designed from deduced amino acid sequences of maize AO cDNAs (Sekimoto et al., 1997), we recently isolated three independent cDNA clones from Arabidopsis. Work is in progress to identify which of these cDNAs encodes AO1 and to characterize further their substrate specificity by heterologous expression and in vitro assay.
ACKNOWLEDGMENT
We thank Dr. Heather McKhann of the Institut National de la Recherche Agronomique (Versailles, France) for her critical reading of this manuscript.
Abbreviations:
- ABAld
abscisic aldehyde
- AO
aldehyde oxidase
- IAAld
indole-3-acetaldehyde
- IAld
indole-3-aldehyde
- ICA
indole-3-carboxylic acid
- IPy
indole-3-pyruvic acid
- MoCo
Mo cofactor
- XD
xanthine dehydrogenase (oxidase)
Footnotes
This research was supported in part by a Grant-in-Aid for Japan-France Joint Study on Development in Higher Plants from the Ministry of Education, Science, Sports, and Culture, Japan.
LITERATURE CITED
- Bauer SL, Howard PC. Kinetics and cofactor requirements for the nitroreductive metabolism of 1-nitropyrene and 3-nitrofluoranthene by rabbit liver aldehyde oxidase. Carcinogenesis. 1991;12:1545–1549. doi: 10.1093/carcin/12.9.1545. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower PJ, Brown HM, Purves WK. Cucumber seedling indoleacetaldehyde oxidase. Plant Physiol. 1978;61:107–110. doi: 10.1104/pp.61.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Gene Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Davies PJ. Plant Hormones. Physiology, Biochemistry and Molecular Biology, Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. [Google Scholar]
- Delarue M (1996) Approches génétique et physiologique du développment d'Arabidopsis thaliana: caracterisation des mutants cristal et superroot. Doctoral thesis. Paris University 6, Paris
- Gopalraj M, Tseng T-S, Olszewski N. The rooty gene of Arabidopsis encodes a protein with highest similarity to amino-transferases (abstract no. 469) Plant Physiol. 1996;111:S-114. [Google Scholar]
- Hagen G, Martin G, Li Y, Guilfoyle TJ. Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol. 1991;17:567–579. doi: 10.1007/BF00040658. [DOI] [PubMed] [Google Scholar]
- Hall WW, Krenitsky TA. Aldehyde oxidase from rabbit liver: specificity toward purines and their analogs. Arch Biochem Biophys. 1986;251:36–46. doi: 10.1016/0003-9861(86)90048-2. [DOI] [PubMed] [Google Scholar]
- Hirao Y, Kitamura S, Tatsumi K. Epoxide reductase activity of mammalian liver cytosols and aldehyde oxidase. Carcinogenesis. 1994;15:739–743. doi: 10.1093/carcin/15.4.739. [DOI] [PubMed] [Google Scholar]
- Huang D-Y, Ichikawa Y. Two different enzymes are primarily responsible for retinoic acid synthesis in rabbit liver cytosol. Biochem Biophys Res Commun. 1994;205:1278–1283. doi: 10.1006/bbrc.1994.2803. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Syōno K. The excessive production of indole-3-acetic acid and its significance in studies of the biosynthesis of this regulator of plant growth and development. Plant Cell Physiol. 1996;37:1043–1048. doi: 10.1093/oxfordjournals.pcp.a029051. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell. 1995;7:2023–2037. doi: 10.1105/tpc.7.12.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Matsuyama H. An in vitro system of indole-3-acetic acid formation from tryptophan in maize (Zea mays) coleoptile extracts. Plant Physiol. 1993;102:1319–1324. doi: 10.1104/pp.102.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Mito N, Miyakado M. l- and d-Tryptophan aminotransferases from maize coleoptiles. J Plant Res. 1993;106:25–29. [Google Scholar]
- Koshiba T, Saito E, Ono N, Yamamoto N, Sato M. Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from maize coleoptiles. Plant Physiol. 1996;110:781–789. doi: 10.1104/pp.110.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Leydecker M-T, Moureaux T, Kraepiel Y, Schnorr K, Caboche M. Molybdenum cofactor mutants, specifically impaired in xanthine dehydrogenase activity and abscisic acid biosynthesis, simultaneously overexpress nitrate reductase. Plant Physiol. 1995;107:1427–1431. doi: 10.1104/pp.107.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Marion-Poll A. Tomato flacca mutant is impaired in ABA aldehyde oxidase and xanthine dehydrogenase activities. Plant Physiol Biochem. 1997;35:369–372. [Google Scholar]
- McQueen-Mason SJ, Hamilton RH. The biosynthesis of indole-3-acetic acid from d-tryptophan in Alaska pea plastids. Plant Cell Physiol. 1989;30:999–1005. [Google Scholar]
- Michalczuk L, Ribnicky DM, Cooke TJ, Cohen JD. Regulation of indole-3-acetic acid biosynthetic pathways in carrot cell cultures. Plant Physiol. 1992;100:1346–1353. doi: 10.1104/pp.100.3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Suzuki Y, Kamisaka S, Masuda Y. Indole-3-acetaldehyde oxidase of pea seedlings. Physiol Plant. 1981;51:402–406. [Google Scholar]
- Noguchi T, Hayashi S. Peroxisomal localization and properties of tryptophan aminotransferase in plant leaves. J Biol Chem. 1980;255:2267–2269. [PubMed] [Google Scholar]
- Nonhebel HM, Cooney TP, Simpson R. The route, control and compartmentation of auxin synthesis. Aust J Plant Physiol. 1993;20:527–539. [Google Scholar]
- Normanly J. Auxin metabolism. Physiol Plant. 1997;100:431–442. [Google Scholar]
- Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA. 1993;90:10355–10359. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Slovin JP, Cohen JD. Rethinking auxin biosynthesis and metabolism. Plant Physiol. 1995;107:323–329. doi: 10.1104/pp.107.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R. Metabolism of indole-3-acetaldehyde. III. Some characteristics of the aldehyde oxidase of Avena coleoptiles. Physiol Plant. 1971;24:272–281. [Google Scholar]
- Rothe GM. Aldehyde oxidase isoenzymes (E.C.1.2.3.1) in potato tubers (Solanum tuberosum) Plant Cell Physiol. 1974;15:493–499. [Google Scholar]
- Schwartz SH, Léon-Kloosterziel KM, Koornneef M, Zeevaart JAD. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114:161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Dohmae N, Takio K, Kamiya Y, Koshiba T. Cloning and molecular characterization of plant aldehyde oxidase. J Biol Chem. 1997;272:15280–15285. doi: 10.1074/jbc.272.24.15280. [DOI] [PubMed] [Google Scholar]
- Stoddart AM, Levine WG. Azoreductase activity by purified rabbit liver aldehyde oxidase. Biochem Pharmacol. 1992;43:2227–2235. doi: 10.1016/0006-2952(92)90182-i. [DOI] [PubMed] [Google Scholar]
- Tomita S, Tsujita M, Ichikawa Y. Retinal oxidase is identical to aldehyde oxidase. FEBS Lett. 1993;336:272–274. doi: 10.1016/0014-5793(93)80818-f. [DOI] [PubMed] [Google Scholar]
- Truelsen TA. Indole-3-pyruvic acid as an intermediate in the conversion of tryptophan to indole-3-acetic acid. I. Some characteristics of tryptophan transaminase from mung bean seedlings. Physiol Plant. 1972;26:289–295. [Google Scholar]
- Tsurusaki K, Takeda K, Sakurai N. Conversion of indole-3-acetaldehyde to indole-3-acetic acid in cell-wall fraction of barley (Hordeum vulgare) seedlings. Plant Cell Physiol. 1997;38:268–273. [Google Scholar]
- Walker-Simmons M, Kudrna DA, Warner RL. Reduced accumulation of ABA during water stress in a molybdenum cofactor mutant of barley. Plant Physiol. 1989;90:728–733. doi: 10.1104/pp.90.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman F, Forest JC. Properties of plant aminotransferases. Phytochemistry. 1978;17:1455–1471. [Google Scholar]
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD. Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science. 1991;254:998–1000. doi: 10.1126/science.254.5034.998. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Oritani T. Stereoselectivity in the biosynthetic conversion of xanthoxin into abscisic acid. Planta. 1996;200:319–325. [Google Scholar]
- Yoshihara S, Tatsumi K. Kinetic and inhibition studies on reduction of diphenyl sulfoxide by guinea pig liver aldehyde oxidase. Arch Biochem Biophys. 1986;249:8–14. doi: 10.1016/0003-9861(86)90554-0. [DOI] [PubMed] [Google Scholar]