Abstract
A cDNA of pea (Pisum sativum L.) RbcS 3A, encoding a small subunit protein (S) of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), has been expressed in Arabidopsis thaliana under control of the cauliflower mosaic virus 35S promoter, and the transcript and mature S protein were detected. Specific antibodies revealed two protein spots for the four Arabidopsis S and one additional spot for pea S. Pea S in chimeric Rubisco amounted to 15 to 18% of all S, as judged by separation on two-dimensional isoelectric focusing/sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels from partially purified enzyme preparations and quantitation of silver-stained protein spots. The chimeric enzyme had 11 ± 1% fewer carbamylated sites and a 11 ± 1% lower carboxylase activity than wild-type Arabidopsis Rubisco. Whereas pea S expression, preprotein transport, and processing and assembly resulted in a stable holoenzyme, the chimeric enzyme was reproducibly catalytically less efficient. We suggest that the presence of, on average, one foreign S per holoenzyme is responsible for the altered activity. In addition, higher-plant Rubisco, unlike the cyanobacterial enzyme, seems to have evolved species-specific interactions between S and the large subunit protein that are involved in carbamylation of the active site.
In higher plants Rubisco (EC 4.1.1.39) is composed of eight L and eight S subunits, L8S8. Two active sites are formed by residues from two L in the L2 dimer (Gutteridge and Gatenby, 1987; Roy, 1989). The L are encoded by a chloroplast gene and are identical, whereas the S are derived from gene families (Dean et al., 1989; Silverthorne et al., 1990). We decided to examine whether and how the presence of a foreign S affected holoenzyme stability or carboxylation activity and chose to express a pea (Pisum sativum L.) S in Arabidopsis thaliana, a species with as few as four different small subunit genes (RbcS; Krebbers et al., 1988).
Crystallographic analysis of spinach Rubisco indicates extensive contacts at the S/L interface. S interact with two neighboring L and a third L dimerized to one of these neighbors (Knight et al., 1990). Thirteen S residues contact residues of α-helix 7 of one L and of α-helix 8 of a neighboring L. Both helices are close to active sites (Knight et al., 1990; Schneider et al., 1990; Taylor and Andersson, 1996). Recently, Taylor and Andersson (1996) proposed a model that assumes movement in one L being transmitted through S to catalytic site movement in a neighboring L.
Studies with cyanobacterial L8S8 have shown that foreign or mutagenized S influence catalysis. Altered S have been used to replace endogenous S in cyanobacterial Rubisco (Andrews and Lorimer, 1985; Goloubinoff et al., 1989; Smrcka et al., 1991; Read and Tabita, 1992). Removal of S from L8S8 or expression of cyanobacterial L in the absence of S generated soluble, carbamylated L, capable of less than 1% of the carboxylase activity of the holoenzyme (Andrews and Ballment, 1984; Andrews, 1988). The consequences of altered S sequences in higher-plant holoenzymes have, however, been impossible to analyze because higher-plant L or L8 cannot be manipulated in vitro (Gatenby, 1984; Gatenby et al., 1987). Removal of S leads to denaturation of the L8 core and higher-plant L expressed in bacterial systems is inactive. For higher-plant enzymes, distinguishing how the products of different RbcS genes might influence activity has received little attention, and it is unknown how isoforms of S might influence structural or kinetic properties of plant holoenzymes.
Work on RbcS has focused mainly on gene expression (Tobin, 1981; Fluhr and Chua, 1986; Silverthorne and Tobin, 1990; Silverthorne et al., 1990; Dedonder et al., 1993) and not on the S. It is unknown whether specific S affect holoenzyme structure or kinetic parameters; in fact, the minimal differences between different S in one species are generally considered irrelevant. Tobacco ecotypes, for example, which express distinct forms of S (Li et al., 1983), did not show significant biochemical differences in activity. In contrast, ferns grown under different light conditions contain two differentially expressed pools of S, the presence of which was correlated with changes in carboxylase activity (Eilenberg et al., 1991).
We chose Arabidopsis thaliana as a model to express a foreign S. This plant contains four RbcS genes, RbcS 1A, 1B, 2B, and 3B, all of which are expressed (Krebbers et al., 1988; Dedonder et al., 1993). These S share high sequence identity and have identical pIs. A cDNA of the pea RbcS 3A gene was introduced into Arabidopsis because its calculated pI was sufficiently different from the pIs of the endogenous S. The RbcS 3A gene encodes a protein that is 30% dissimilar compared with the Arabidopsis S (Fluhr and Chua, 1986; Krebbers et al., 1988). As an added advantage, overexpression of a new S driven by the cauliflower mosaic virus 35S promoter allowed for its light-independent expression (Benfey et al., 1990a, 1990b).
We report that this pea S in A. thaliana is stably incorporated into the holoenzyme at a ratio of 1 to 8 S. The carboxylase activity of the chimeric holoenzyme was reduced relative to wild-type Arabidopsis Rubisco by a small although reproducible amount, indicating that one of eight active sites was inoperative, seemingly because this site was not carbamylated in the chimeric holoenzyme.
MATERIALS AND METHODS
Plant Transformations and Plant Growth Conditions
The binary vector pBin19 (Bevan, 1984) was used for Agrobacterium tumefaciens-mediated plant transformations. A. tumefaciens LBA4404 (Ooms et al., 1982) was transformed either by triparental mating, using Escherichia coli XL1Blue as the donor strain and E. coli RK2013 as the Hfr strain, or by electroporating binary vectors directly into the host. Arabidopsis thaliana (ecotype Landsberg erecta) was transformed according to the method of Valvekens et al. (1988). Transformed plants were identified by kanamycin selection at 50 μg mL−1 kanamycin. Plants were grown for 5 weeks under 24 h of cool-white fluorescent light at 100 μmol m−2 s−1 PPFD at 20°C. Plants were treated with blue light for 12 h at 100 μmol m−2 s−1 at 20°C using fluorescent lights (F20T12BB, Phillips Lighting, Amsterdam, The Netherlands) emitting 95% of the light intensity in the range 425 and 475 nm.
RNA Isolation and Analysis
RNA was isolated from white- and blue-light-treated plants that were harvested simultaneously; leaves were frozen in liquid nitrogen and stored at −70°C. RNA extraction was performed by the DNA-extraction method of Gustincich et al. (1991), with the modification that the LiCl-precipitated RNA pellets were kept and resuspended in H2O. The RNA was separated on 4% (v/v) formaldehyde, 20 mm Mops, pH 8.0, 5 mm sodium acetate, 1 mm EDTA, and 1.1% (w/v) agarose gels for 4 h at 100 V. Gels were blotted onto membranes (Zetaprobe, Bio-Rad). Prehybridization and hybridization of an oligonucleotide probe specific for pea (Pisum sativum L.) RbcS 3A (5′-GCACTTTACTCGGCCACCATTGCT-3′) was performed according to the method of Berent et al. (1985).
Rubisco Enrichment
A. thaliana leaves, frozen at −70°C, were ground in the presence of extraction buffer (either 100 mm Tris, pH 6.8, 100 mm NaCl, and 20 mm EDTA or 50 mm Bicine, pH 8.0, 10 mm EDTA, and 1 mm DTT). Immediately prior to extraction 1 mm leupeptin and 0.8 mm PMSF were added to the buffer. The homogenate was then clarified at 12,000g for 10 min. The supernatant fluid was brought to 30% (w/v) ammonium sulfate and centrifuged again. The supernatant fraction was adjusted to 60% (w/v) ammonium sulfate and recentrifuged. Pellets were resuspended in 50 mm Bicine, pH 8.0, 2 mm EDTA, and 1 mm DTT. Samples were either purified by fast-protein liquid chromatography or precipitated by addition of PEG-3350. Fast-protein-liquid chromatography purification utilized an anion-exchange column (model PSC10-QM, Productive Column, Bps Separations, Natick, MA). Rubisco eluted at 0.3 m KCl in 50 mm Bicine, pH 8.0, and was collected and concentrated over a Centricon-100 concentrator (Amicon, Beverly, MA), exchanging the buffer with 50 mm Bicine, pH 8.0, 2 mm EDTA, 1 mm DTT, and 20% (w/v) Gly. Samples were stored in aliquots at −70°C. Otherwise, 18% (w/v) PEG-3350 precipitation and centrifugation at 12,000g followed ammonium sulfate fractionation. The supernatant fluid was collected and MgCl2 was added to 30 mm. The precipitate was collected, resuspended, and stored in 50 mm Bicine, pH 8.0, 2 mm EDTA, 1 mm DTT, and 20% (v/v) Gly at −70°C.
Native Gel Purification of Rubisco
Rubisco samples were treated as described above except that, following the 60% (w/v) ammonium sulfate precipitation and resuspension, samples were separated by 6% (w/v) PAGE in 1× Laemmli buffer without SDS under constant cooling at 10°C for 6 to 8 h. A thin strip down the length of the gel was removed for Coomassie blue staining, and the remaining gel was equilibrated in 10 mm Tris-HCl, pH 7.8, 0.5 mm DTT, and 1 mm EDTA. The 550-kD band of Rubisco was excised and cut into small fragments. Rubisco protein was electroeluted (model 1750, Sample Concentrator, Isco, Lincoln, NE) into 50 mm Bicine, pH 8.0, 2 mm EDTA, 1 mm DTT, and 20% (v/v) Gly.
2-D Gel Electrophoresis
Denaturing IEF was performed by a modified method of O'Farrel (1975) using wide-range (pH 3.0–10.0) and narrow-range (pH 5.0–7.0) ampholines (Serva, Paramus, NJ). Proteins were run from the anodic reservoir (0.5% [v/v] ethanolamine) to the cathodic reservoir (0.3% [w/v] citric acid). Ten to 15 μg of vacuum-dried, enriched Rubisco was suspended in IEF buffer (9.5 m urea, 0.5% [w/v] 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate [Chaps], 0.5% [w/v] DTE, and 2% [v/v] pH 3.0–10.0, and 8% [v/v] pH 5.0–7.0 ampholines) and loaded into an electrofocusing tube gel (model SE600–1.5, electrofocusing apparatus, Hoefer Scientific [San Francisco, CA]). IEF acrylamide tube gels were composed of 9.16 m urea, 4% (w/v) acrylamide, 0.1% (w/v) bisacrylamide, 1% (w/v) Chaps, and 1.7% (v/v) pH 3.0 to 10.0 ampholines. IEF gels were developed with decreasing current and a voltage ramp from 0.03 to 0.015 mA and from 50 to 400 V over 15 h. IEF gels were then equilibrated for 30 min in a 62 mm Tris base, pH 6.8, 2.3% (w/v) SDS, and 10% (v/v) glycerol. IEF gels were either stored at −70°C or separated by 15% (w/v) acrylamide SDS-PAGE.
Quantitation of Pea S Protein
Three 2-D gels each of two transgenic Arabidopsis lines (T7.3 and T7.5) were scanned using an Apple OneScanner and stored as pict files, which were analyzed using NIH image software (National Institutes of Health, Bethesda, MD) for quantitation. Percentage of pea S was determined as (pea S value/[Arabidopsis S value plus pea S value]).
Antibody Selection by Affinity Purification
Polyclonal antibodies were generated in rabbits against purified tobacco Rubisco. Crude serum (0.5 mL) was used in 50 mL of 1× TBS, 5% (w/v) low-fat dry milk, and 0.5% (v/v) Tween 20 against Rubisco S from pea blotted onto nitrocellulose membrane. Following incubation for 1 h at room temperature, membranes were washed three times in 1× TBS and 0.5% (v/v) Tween 20. Antibodies were removed from the membrane with 0.2 m Gly, pH 2.2, and neutralized with 1 m Tris, pH 8.8. IgG was either precipitated with 35% (w/v) ammonium sulfate or concentrated in 1× TBS.
Enzyme Assays
Ribulose-1,5-bisphosphate was synthesized by the method of Bahr and Jensen (1978). Enzyme concentration of enriched Rubisco from three sets of wild-type Arabidopsis, T7.3, and T7.5 plants was determined by using the extinction coefficient for tobacco (Nicotiana tabacum L.) Rubisco (McCurry et al., 1982). Concentrations for wild-type Arabidopsis Rubisco were verified by 2-carboxy-d-arabinitol 1,5-bisphosphate titration (Pierce et al., 1980), indicating a purity of approximately 85% for all preparations. Carboxylase activity was determined after activation in 50 mm Bicine, pH 8.0, 10 mm MgCl2, and 10 mm NaH14CO3 (2 Ci mol−1) for 20 min at 25°C (Lorimer et al., 1976). Activity assays were performed for enzymes in the same buffer following addition of 0.8 mm ribulose-1,5-bisphosphate at 25°C. Carbamylation (activation) assays were performed in the same manner as for activation assays with the modification that 10 mm NaH14CO3 (5 Ci mol−1) was used. 2-Carboxy-d-arabinitol 1,5-bisphosphate was then added to 0.6 mm for 2 h to trap activator CO2 (Pierce et al., 1980). The Rubisco samples were desalted with Econo-Pac 10DG columns (Bio-Rad), and the bound activator 14CO2 was quantified by liquid-scintillation counting. Sixty micrograms of Rubisco, as determined by A280, was used per assay. Calculations to determine the number of activator CO2-bound sites are based on the molecular mass of Arabidopsis S (14.7 kD; Krebbers et al., 1988) and L (52.9 kD; Zhu et al., 1997). The molecular mass of Arabidopsis Rubisco is 541 kD, resulting in 14.8 nmol sites mg−1 protein or 0.89 nmol sites per assay (Table I). This amount of wild-type Arabidopsis Rubisco had 0.91 nmol activator CO2-bound sites (Table I), equaling 8.2 sites in wild-type Rubisco, which was taken as 8 sites. The error may be from using the extinction coefficient of 0.7 for tobacco Rubisco (McCurry et al., 1982) or impurities. Assays were repeated six times for each Rubisco preparation.
Table I.
Carboxylase activity and carbamylation
| Parameter | Wild Type | T7.3 | T7.5 |
|---|---|---|---|
| Activitya | 1.71 ± 0.12b | 1.45 ± 0.11 | 1.51 ± 0.10 |
| Wild type activity | — | 85% | 88% |
| Activated sitesc | 0.91 ± 0.01 | 0.81 ± 0.01 | 0.80 ± 0.01 |
| Wild type Carbamylation | — | 89% | 88% |
| Average ACO2 sites/Rubiscod | 8 | 7 | 7 |
Carboxylase activity as μmol CO2 mg−1 protein min−1.
sds are based on three independently purified sets of Rubisco from the wild type, T7.3, and T7.5.
Carbamylation of Rubisco as nmol activator CO2 (ACO2) per 60 μg of protein.
Calculated average number of carbamylated sites per Rubisco (see Methods).
RESULTS
Pea RbcS 3A Construct
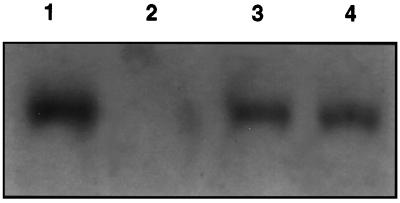
A pea RbcS 3A cDNA (Fig. 1) in the binary vector pBIN19 was introduced into A. thaliana by A. tumefaciens-mediated transformation. The RbcS 3A cDNA was expressed under control of the duplicated cauliflower mosaic virus 35S promoter. Thirty kanamycin-resistant plants were regenerated. T3 plants were screened to determine relative amounts of pea RbcS 3A transcript by RNA-blot analysis (data not shown). T7.3 and T7.5 were selected for a high amount of the transcript. Based on total RNA, T7.3 and T7.5 contained pea RbcS 3A at approximately 55% of that of RbcS transcripts found in pea leaves (Fig. 2).
Figure 1.
Pea RbcS3A gene construct. The gene construct is composed of a duplicated cauliflower mosaic virus 35S promoter (Dup. CaMV 35S) followed by the pea RbcS 3A transit peptide (TP) sequence and RbcS 3A cDNA, including the 3′ untranslated region.
Figure 2.
Pea RbcS 3A transcript in A. thaliana. Transcripts of pea RbcS 3A were determined by RNA-blot analysis using a probe, 14 nucleotides in length, specific for the pea RbcS 3A transcript. The probe is complementary to a pea RbcS 3A-specific sequence in the 3′ untranslated region. Lane 1, Pea RNA; lane 2, wild-type Arabidopsis RNA; lane 3, transgenic Arabidopsis (T7.3) RNA; and lane 4, transgenic Arabidopsis (T7.5) RNA. Ten micrograms of total RNA was loaded in each lane.
Protein Sequence Comparisons
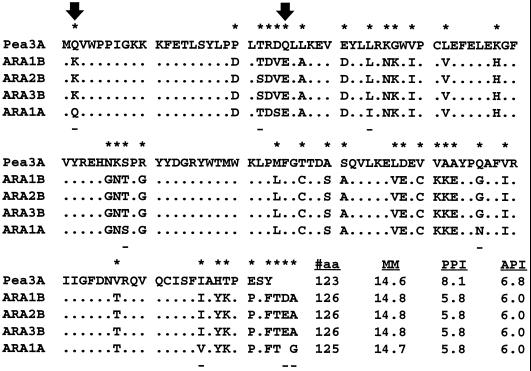
Comparisons of the polypeptides indicated that the pea and Arabidopsis S are different. Whereas the Arabidopsis S are difficult to discern from one another, molecular mass and pI distinguish the pea S-3A from Arabidopsis S (Fig. 3). Two Arabidopsis RbcS genes (RbcS 2B and RbcS 3B) encode identical proteins and cannot be distinguished. S encoded by RbcS 1B differs from those of RbcS 2B and 3B only in positions T-22 and D-125, which do not result in molecular mass or pI differences. Only the protein product from RbcS 1A should be discernible from other members of the family because it has a slightly lower molecular mass. It differs by seven amino acid substitutions and one deletion near its C terminus from the other S proteins (Krebbers et al., 1988).
Figure 3.
Comparison of mature pea and Arabidopsis (ARA) S polypeptide sequences. Dots represent identical residues. An asterisk above the deduced protein sequence indicates differences between pea S 3A (Fluhr and Chua, 1986) and Arabidopsis S (Krebbers et al., 1988). A dash below the sequences indicates differences among Arabidopsis S. Arrows identify residues that reportedly participate in ionic interactions at the S/L interface of spinach Rubisco (Knight et al., 1990; Schneider et al., 1990). Also shown are the length of the proteins (#aa), molecular mass (MM), predicted pI (PPI), and apparent pI (API).
2-D Gel Electrophoresis
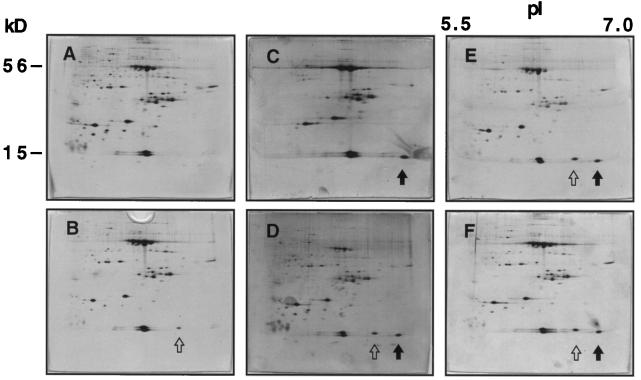
Rubisco protein was enriched to approximately 85% purity from populations of T7.3, T7.5, and wild-type Arabidopsis and separated on 2-D gels in the IEF range from pH 5.5 to 7.0. Silver-stained 2-D gels of Rubisco preparations from wild-type and transgenic Arabidopsis revealed Rubisco L near the top of the gel at 56 kD and putative Rubisco S near the bottom of the gel at 15 kD (Fig. 4). Rubisco L was distributed along the IEF gradient as four major spots, suggesting the existence of differentially modified L populations in Arabidopsis, possibly similar to observations in other systems (Houtz et al., 1989). The presence of two prominent protein products of the expected mass (14.7 and 14.8 kD) for endogenous Arabidopsis S is shown in Figure 4A. We think that the RbcS 1B, 2B, and 3B gene products migrated together, generating the spot of a molecular weight of 14,800, and that S from RbcS 1A produced the 14.7-kD spot (Fig. 4A). This would be consistent with transcription data, since all Arabidopsis RbcS genes are expressed in white light (Dedonder et al., 1993). Under blue-light conditions, a third spot that reacted with S antibodies (see below) appeared with an apparent pI of 6.4 (Fig. 4B). This suggested either up-regulation of an unidentified endogenous S, which we consider unlikely, or the posttranslational modification of an S present in white light.
Figure 4.
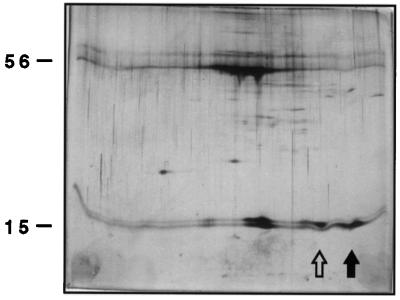
Separation of S proteins from enriched Rubisco samples. Silver-stained 2-D gels containing enriched Rubisco from white-light-grown plants (top) and blue-light-treated plants (bottom) are shown. L, resolved into four spots, migrate near the top of each gel at approximately 56 kD. S are resolved as multiple spots at approximately 15 kD. In A and B, wild-type Rubisco is shown. In B, an open arrow indicates the blue-light S. C and D show T7.3 Rubisco, which contains a new 15-kD spot (solid arrow). E and F present T7.5 Rubisco containing both the pea S spot (solid arrow) and the blue-light S spot (open arrow) under both light conditions.
A New 15-kD Protein Present in Transgenic Arabidopsis
An additional protein spot with a mass of about 15 kD was found in the transgenic plant lines T7.3 and T7.5 (Fig. 4, C–E). This protein was also detected when a construct that introduced a genomic version of the pea RbcS 3A was used (data not shown). The mass of this protein was consistent with the expected mass (14.6 kD) of the pea RbcS 3A gene product (Fig. 3). Wild-type pea S, which was used as a control, showed S separated into two spots, one of which co-migrated with the spot found in the transgenic Arabidopsis lines T7.3 and T7.5 expressing pea S 3A (data not shown; Getzoff, 1997).
In the transgenic line T7.3 the blue-light-dependent 15-kD protein appeared (Fig. 4, C and D), which was already seen in wild-type Rubisco (Fig. 4B). This putative blue-light Arabidopsis S appeared, however, in white light in several other independently transformed lines (represented by T7.5; Fig. 4E). Furthermore, blue light no longer increased the relative amount of this putative Arabidopsis S in pea S-expressing plants (Fig. 4F), and no additional S was expressed under blue light in T7.5. This suggested that the blue-light-enhanced putative S in wild-type Arabidopsis was identical to the protein expressed in white light in line T7.5. S from three independent Rubisco preparations from each line (T7.3, Fig. 4C; T7.5, Fig. 4E) were quantified after separation on 2-D gels. In Rubisco from line T7.3 pea S amounted to 18 ± 3% of the total S, and 15 ± 3% of all S was determined in line T7.5 Rubisco, i.e. pea S amounted on average to one of eight S in the chimeric holoenzymes.
All Four 15-kD Proteins Are S Proteins
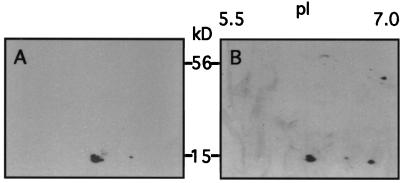
S-specific antibodies were used to ascertain the nature of the 15-kD spots. After separation of Rubisco proteins from blue-light-treated wild type and white-light-treated T7.5 on 2-D gels, gel blots were probed with anti-S antibodies. All putative Arabidopsis S, including the “blue-light” S (Fig. 5A), and putative pea S (Fig. 5B) were recognized, thus identifying each as S.
Figure 5.
Antibody detection of S from enriched Rubisco samples resolved in 2-D gels. S-specific antibodies were used to probe protein blots of 2-D gels. Chemiluminescence was used to produce fluorographs displaying S positions. A, Antibody detection of S in wild-type Arabidopsis treated with blue light. Three S spots are apparent. B, Antibody detection of S in T7.5 under white light. A fourth S is resolved. Also shown in B is an unidentified spot at 45 kD with a pI of 7, which is neither S nor its precursor.
Assembly of Pea S in the Arabidopsis Holoenzyme
To establish whether the pea S was assembled, the holoenzyme was further purified. The 30 to 60% (NH4)2SO4 fraction was separated on nondenaturing gels. The Rubisco complex of approximately 550 kD, identified by Coomassie blue staining of part of each gel, was excised and eluted, and the L and S subunits were separated on 2-D gels (Fig. 6). Pea S was present in chimeric holoenzymes, confirming assembly following chloroplast import and maturation of the precursor protein. When wild-type and recombinant holoenzymes were incubated with increasing concentrations of urea, no difference was observed in the concentration of urea that led to dissociation, as judged by the emergence of S from protein complexes on nondenaturing acrylamide gels (not shown).
Figure 6.
Purified transgenic Rubisco containing pea S. A silver-stained 2-D gel of T7.5 Rubisco, which was first fractionated with 30 to 60% saturated (NH4)2SO4 and then purified by separation on a 6% (w/v) acrylamide native gel. Rubisco for this 2-D gel was eluted from the native gel as a 550-kD band. All Arabidopsis 15-kD S, including the blue-light-induced S (open arrow), and the pea RbcS 3A S (solid arrow) are present.
Comparison of Wild-Type and Chimeric Rubisco Activity and Carbamylation
Biochemical characteristics of wild-type and chimeric Rubisco were examined. In three sets of independently purified enzymes, chimeric Rubisco consistently had a modest 12 to 15% reduction in total activity relative to the wild type (Table I). Further examination determined that the presence of pea S in Arabidopsis Rubisco was correlated with an equally modest decrease in the bound activator 14CO2. Table I shows nanomoles of activator 14CO2-labeled sites present in equal amounts (60 μg) of wild-type, T7.3, and T7.5 Rubisco. Wild-type Rubisco had 0.91 ± 0.01 nmol of carbamylated sites. In contrast, transgenic holoenzymes from lines T7.3 and T7.5 showed 0.81 ± 0.01 and 0.80 ± 0.01 nmol of carbamylated sites, respectively, indicating a decline of approximately 11% in activated sites.
DISCUSSION
All four A. thaliana RbcS genes are transcribed (Dedonder et al., 1993). If all transcripts are translated, the S derived from RbcS 2B and 3B are identical, and a third S, derived from RbcS 1B, is distinguished from these by two amino acid exchanges that do not alter mass or pI. Thus, three S could generate a single spot with a molecular mass of 14.8 kD on 2-D gels. S derived from RbcS 1A can be distinguished from the others by its mass (Figs. 3 and 4). Thus, the four A. thaliana S form two spots on 2-D gels, which was observed with S-specific antibodies. The introduced pea S, which formed a new spot, was identified immunologically and by its comigration with one of the authentic pea S proteins.
The appearance of an additional S in the wild type after blue-light treatment was unexpected. We think that this S is the product of posttranslational modification of one of the endogenous S and not the product of an unidentified fifth Arabidopsis RbcS gene. It is unknown which S may be modified and what type of modification might occur. Preliminary experiments provided no evidence for N-glycosylation or phosphorylation (data not shown), but phosphorylation of S has been reported in response to light/dark treatments (Foyer, 1985). The blue-light-induced wild-type S is present in some of the independently transformed Arabidopsis expressing pea S following blue and white light. We have no explanation for its appearance in white light but it seems possible that this S is modified in response to the presence of pea S.
Carboxylase activity of T7.3 and T7.5 was modestly but consistently lower (12–15%) than that of wild-type enzyme (Table I), which paralleled a similar decrease in carbamylation (about 11%) relative to the wild type. We will discuss arguments supporting that these altered characteristics may be due to an incompatible interaction between Arabidopsis L and pea S.
Pea S represents between 15 and 18% of the total S content in chimeric Rubisco, equivalent to one pea S per holoenzyme. We assume that the pea S are distributed equally among the chimeric holoenzymes, but we cannot exclude an unequal distribution. Attempts to increase the ratio of pea S to Arabidopsis S were made by including a second construct into the T7.3 and T7.5 lines. This was achieved by antisense expression of the Arabidopsis RbcS transit peptide and the Arabidopsis 5′ untranslated region. Plants were regenerated that contained more than 30% of pea S in relationship to the Arabidopsis S. When grown in tissue culture with added Suc, these plants developed chlorotic vasculature and we were unable to obtain sufficient material for biochemical analyses (Getzoff, 1997).
Incompatibility between the Arabidopsis L and pea S can be deduced from sequence comparisons in the context of crystallographic data. Arabidopsis and pea S differ in 39 residues (Fig. 3), many of which are oriented toward the solute phase (Knight et al., 1990) and are likely to be irrelevant to the discussion. Two residues in S, Q-2 and Q-25 (Fig. 3, arrows), have been shown to contact amino acids W-411 of α-helix 7 and E-433 of α-helix 8 in neighboring L of the spinach enzyme (Schneider et al., 1990). W-411 and E-433 are present in Arabidopsis and pea L. The residues Q-2 and Q-25 are conserved in pea S (Fluhr and Chua, 1986; Zurawski et al., 1986) but differ in Arabidopsis S (Krebbers et al., 1988). Residues K-2 (in Arabidopsis S1B, 2B, and 3B) and E-25 (in all Arabidopsis S) do not support interaction with W-411 and E-433 of L (Zurawski et al., 1981, 1986; Zhu et al., 1997). Apparently, Arabidopsis and pea Rubisco evolved different interactions at the S/L interface and, possibly, Arabidopsis may lack two S/L interactive pairs of residues.
Additional differences between Arabidopsis and pea S are G56N, N57K, T58S, and G60R (Fig. 3). These residues are located in a hairpin loop, protruding between two neighboring L into the central channel of the holoenzyme (Knight et al., 1990). Engineering of this loop has been shown to enable assembly of cyanobacterial S (in which the hairpin loop is missing) into the higher-plant holoenzyme (Wasmann et al., 1989; Flachmann and Bohnert, 1992). Arabidopsis RbcS 1A shares identity with pea at position 58 in this loop, whereas the substitutions at position 56 introduce a charge difference. Crystallographic analysis of spinach Rubisco has recently indicated that two isoforms of S are positioned in an ordered fashion in the holoenzyme. Shibata et al. (1996) could distinguish the two different spinach S based on the presence or absence of an ionic interaction between positions 56, in the hairpin loop of S, and E-259 of L. Although pea S residues allow for this interaction with E-259 in Arabidopsis L, Arabidopsis S residues do not. Three of the four positions in the hairpin loop that differ between Arabidopsis and pea S are conserved positions among Arabidopsis S (Fig. 3). Thus, pea S could introduce an interaction at N56 with Arabidopsis L at E-259, which is not possible between the homologous subunit proteins in Arabidopsis. The additional interaction could influence the active site. Additional experiments will be necessary to investigate whether such an interaction exists and whether this interaction causes changes in carbamylation or specific activity.
We demonstrated that heterologous expression, preprotein import, and stable assembly in the holoenzyme of a foreign S in a transgenic plant is possible. Unexpectedly, the presence of one foreign S in eight slightly affected carbamylation and carboxylation of the chimeric holoenzyme. It could be that the pea S formed contacts with the Arabidopsis L that were different from those in the homologous assembly. This suggests that, unlike in cyanobacterial L/S (Andrews and Ballment, 1984), higher-plant L may require S for activation. The three pea S residues at the S/L interface, which differ in charge from the Arabidopsis counterparts, are prime candidates to test whether they interfere with carbamylation. Expression of foreign S in transgenic plants, in combination with silencing or elimination of the endogenous S family members, will prove useful in determining the critical residues at the S/L interface. The results will have important implications for attempts at engineering or replacing subunits of Rubisco enzymes in higher plants.
ACKNOWLEDGMENTS
We wish to thank Dr. Don P. Bourque for the gift of purified tobacco holoenzyme, and we are thankful for the technical assistance given by Mrs. Wendy Chmara. We thank Dr. E. Krebbers for providing gene probes.
Abbreviations:
- L
large subunit protein(s)
- S
small subunit protein(s)
- 2-D gel
two-dimensional IEF/SDS-PAGE gel
Footnotes
The work was supported by the National Science Foundation (Biochemistry Program, 1991–1994), by Japan Tobacco, and in part by the Arizona Agricultural Experiment Station.
LITERATURE CITED
- Andrews TJ. Catalysis by cyanobacterial ribulose bisphosphate carboxylase large subunits in the complete absence of small subunits. J Biol Chem. 1988;263:12213–12219. [PubMed] [Google Scholar]
- Andrews TJ, Ballment B. Active-site carbamate formation and reaction intermediate-analog binding by ribulose bisphosphate carboxylase/oxygenase in the absence of its small subunits. Proc Natl Acad Sci USA. 1984;81:3660–3664. doi: 10.1073/pnas.81.12.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ, Lorimer GH. Catalytic properties of a hybrid between cyanobacterial large subunits and higher plant small subunits of ribulose bisphosphate carboxylase-oxygenase. J Biol Chem. 1985;260:4632–4636. [PubMed] [Google Scholar]
- Bahr JT, Jensen RG. Activation of ribulose bisphosphate carboxylase in intact chloroplast by CO2and light. Arch Biochem Biophys. 1978;185:39–48. doi: 10.1016/0003-9861(78)90141-8. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua N-H. Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 1990a;9:1677–1684. doi: 10.1002/j.1460-2075.1990.tb08291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua N-H. Combinational and synergistic properties of CaMV 35S enhancer subdomains. EMBO J. 1990b;9:1685–1696. doi: 10.1002/j.1460-2075.1990.tb08292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berent SL, Mahmoudi M, Torczynscki RM, Bragg PW, Bollon AP. Comparison of oligonucleotide and long DNA fragments as probes in DNA and RNA dot, southern, northern, colony and plaque hybridizations. Biotechniques. 1985;3:208–220. [Google Scholar]
- Bevan M. Binary Agrobacteriumvectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Pichersky E, Dunsmuir P. Structure, evolution, and regulation of RbcSgenes in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:415–439. [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E. ArabidopsisRbcS genes are differentially regulated by light. Plant Physiol. 1993;101:801–808. doi: 10.1104/pp.101.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilenberg H, Beer S, Gepstein S, Geva N, Orly T, Zilberstein A. Variability in ribulose-1,5-bisphosphate carboxylase/oxygenase small subunits and carboxylation activity in fern gametophytes grown under different light spectra. Plant Physiol. 1991;95:298–304. doi: 10.1104/pp.95.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachmann R, Bohnert HJ. Replacement of a conserved arginine in the assembly domain of ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit interferes with holoenzyme formation. J Biol Chem. 1992;267:10576–10582. [PubMed] [Google Scholar]
- Fluhr R, Chua N-H. Developmental regulation of two genes encoding ribulose-1,5-bisphosphate carboxylase small subunit in pea and transgenic petunia plants: phytochrome response and blue-light induction. Proc Natl Acad Sci USA. 1986;83:2358–2362. doi: 10.1073/pnas.83.8.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH. Stromal protein phosphorylation in spinach (Spinacia oleracea) Biochem J. 1985;231:97–103. doi: 10.1042/bj2310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby AA. The properties of the large subunit of maize ribulose bisphosphate carboxylase/oxygenase synthesized in Escherichia coli. Eur J Biochem. 1984;144:361–366. doi: 10.1111/j.1432-1033.1984.tb08472.x. [DOI] [PubMed] [Google Scholar]
- Gatenby AA, Van der Vies SM, Rothstein SJ. Coexpression of both the maize large and wheat small subunit genes of ribulose-bisphosphate carboxylase in E. coli. Eur J Biochem. 1987;168:227–231. doi: 10.1111/j.1432-1033.1987.tb13409.x. [DOI] [PubMed] [Google Scholar]
- Getzoff TP (1997) Introduction of a pea small subunit into Arabidopsis Rubisco: studies in kinetics and protein substitution. PhD thesis, University of Arizona, Tucson
- Goloubinoff P, Gatenby AA, Lorimer GH. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Manfioletti G, Del Sal G, Schneider C. A fast method for high quality genomic DNA extraction from whole human blood. Biotechniques. 1991;11:298–302. [PubMed] [Google Scholar]
- Gutteridge S, Gatenby AA. The molecular analysis of the assembly, structure, and function of Rubisco. Oxf Surv Plant Mol Cell Biol. 1987;4:95–135. [Google Scholar]
- Houtz RL, Stults JT, Mulligan RM, Tolbert NE. Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1989;86:1855–1859. doi: 10.1073/pnas.86.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Andersson I, Brandén CI. Crystallographic analysis of ribulose-1,5-bisphosphate carboxylase from spinach at 2.4 A resolution: subunit interactions and the active site. J Mol Biol. 1990;215:113–160. doi: 10.1016/S0022-2836(05)80100-7. [DOI] [PubMed] [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore A, Timko M. Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol. 1988;11:745–759. doi: 10.1007/BF00019515. [DOI] [PubMed] [Google Scholar]
- Li L, Sisson V, Kung S. Relationship between the kinetic properties and the small subunit composition of Nicotianaribulose-1,5-bisphosphate carboxylase. Plant Physiol. 1983;71:404–408. doi: 10.1104/pp.71.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer GH, Badger MR, Andrews TJ. The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. Equilibria, kinetics, a suggested mechanism, and physiological implications. Biochemistry. 1976;15:529–536. doi: 10.1021/bi00648a012. [DOI] [PubMed] [Google Scholar]
- McCurry SD, Gee R, Tolbert NE. Ribulose-1,5-bisphosphate carboxylase/oxygenase from spinach, tomato or tobacco leaves. Methods Enzymol. 1982;90:515–521. doi: 10.1016/s0076-6879(82)90178-1. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ooms G, Hooykaas PJ, Van Veen RJ, van Beelen P, Regensburg-Tuink TJ, Schilperoort RA. Octopine Ti-plasmid deletion mutants of Agrobacterium tumifacienswith emphasis on the right side of the T-region. Plasmid. 1982;7:15–29. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Pierce J, Tolbert NE, Barker R. Interaction of ribulose-1,5-bisphosphate carboxylase/oxygenase with transition-state analogs. Biochemistry. 1980;19:934–942. doi: 10.1021/bi00546a018. [DOI] [PubMed] [Google Scholar]
- Read BA, Tabita FR. Amino acid substitutions in the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase that influence catalytic activity of the holoenzyme. Biochemistry. 1992;31:519–525. doi: 10.1021/bi00117a031. [DOI] [PubMed] [Google Scholar]
- Roy H. Rubisco assembly: a model system for studying the mechanisms of chaperonin action. Plant Cell. 1989;1:1035–1042. doi: 10.1105/tpc.1.11.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Knight S, Andersson I, Brandén CI, Lindqvist Y, Lundqvist T. Comparison of the crystal structures of L2 and L8S8 Rubisco suggests a functional role for the small subunit. EMBO J. 1990;9:2045–2050. doi: 10.1002/j.1460-2075.1990.tb07371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Inoue T, Fukuhara K, Nagara Y, Ktagawa R, Harada S, Kasai N, Uemura K, Kata K, Yokota A and others. Orderly disposition of heterogeneous small subunits in d-ribulose-1,5-bisphosphate carboxylase/oxygenase from spinach. J Biol Chem. 1996;271:26449–26452. doi: 10.1074/jbc.271.43.26449. [DOI] [PubMed] [Google Scholar]
- Silverthorne J, Tobin EM. Post-transcriptional regulation of organ-specific expression of individual RbcS mRNA in Lemna gibba. Plant Cell. 1990;2:1181–1190. doi: 10.1105/tpc.2.12.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverthorne J, Wimpee CF, Yamada T, Rolfe SA, Tobin EM. Differential expression of individual genes encoding the small subunit of ribulose-1,5-bisphosphate carboxylase in Lemna gibba. Plant Mol Biol. 1990;15:49–58. doi: 10.1007/BF00017723. [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Ramage RT, Bohnert HJ, Jensen RG. Purification and characterization of large and small subunits of ribulose-1,5-bisphosphate carboxylase expressed separately in Escherichia coli. Arch Biochem Biophys. 1991;286:6–13. doi: 10.1016/0003-9861(91)90002-z. [DOI] [PubMed] [Google Scholar]
- Taylor T, Andersson I. Structural transitions during activation and ligand binding in hexadecameric Rubisco inferred from the crystal structure of the activated unliganded spinach enzyme. Nature Struct Biol. 1996;3:95–101. doi: 10.1038/nsb0196-95. [DOI] [PubMed] [Google Scholar]
- Tobin EM. Phytochrome-mediated regulation of messenger RNAs for the small subunit of ribulose-1,5-bisphosphate carboxylase and the light-harvesting chlorophyll a/b-protein in Lemna gibba. Plant Mol Biol. 1981;1:35–51. doi: 10.1007/BF00023012. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsevettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thalianaroot explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmann CC, Ramage RT, Bohnert HJ, Ostrem JA. Identification of an assembly domain in the small subunit of ribulose-1,5-bisphosphate carboxylase. Proc Natl Acad Sci USA. 1989;86:1198–1202. doi: 10.1073/pnas.86.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Jensen RG, Bohnert HJ. DNA sequence of ribulose-1,5-biphosphate carboxylase/oxygenase large subunit from Arabidopsis thaliana (accession no. U91966) (PGR 97-074) Plant Physiol. 1997;114:395. [Google Scholar]
- Zurawski G, Perrot B, Bottomley W, Whitfield PR. The structure of the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase from spinach chloroplast DNA. Nucleic Acids Res. 1981;9:3251–3270. doi: 10.1093/nar/9.14.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G, Whitfeld PR, Bottomley W. Sequence of the gene for the large subunit of ribulose-1,5-bisphosphate carboxylase from pea chloroplasts. Nucleic Acids Res. 1986;14:3975–3980. doi: 10.1093/nar/14.9.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]