Abstract
To investigate the proposed molecular characteristics of sugar-mediated repression of photosynthetic genes during plant acclimation to elevated CO2, we examined the relationship between the accumulation and metabolism of nonstructural carbohydrates and changes in ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) gene expression in leaves of Arabidopsis thaliana exposed to elevated CO2. Long-term growth of Arabidopsis at high CO2 (1000 μL L−1) resulted in a 2-fold increase in nonstructural carbohydrates, a large decrease in the expression of Rubisco protein and in the transcript of rbcL, the gene encoding the large subunit of Rubisco (approximately 35–40%), and an even greater decline in mRNA of rbcS, the gene encoding the small subunit (approximately 60%). This differential response of protein and mRNAs suggests that transcriptional/posttranscriptional processes and protein turnover may determine the final amount of leaf Rubisco protein at high CO2. Analysis of mRNA levels of individual rbcS genes indicated that reduction in total rbcS transcripts was caused by decreased expression of all four rbcS genes. Short-term transfer of Arabidopsis plants grown at ambient CO2 to high CO2 resulted in a decrease in total rbcS mRNA by d 6, whereas Rubisco content and rbcL mRNA decreased by d 9. Transfer to high CO2 reduced the maximum expression level of the primary rbcS genes (1A and, particularly, 3B) by limiting their normal pattern of accumulation through the night period. The decreased nighttime levels of rbcS mRNA were associated with a nocturnal increase in leaf hexoses. We suggest that prolonged nighttime hexose metabolism resulting from exposure to elevated CO2 affects rbcS transcript accumulation and, ultimately, the level of Rubisco protein.
Exposure of C3 plants to elevated CO2 frequently results in an immediate increase in the rate of CO2 assimilation; however, a reduction in photosynthetic capacity often occurs after prolonged periods (days to weeks) at elevated CO2 (for reviews, see Stitt, 1991; Griffin and Seemann, 1996). This down-regulation or acclimation of photosynthesis is generally accompanied by a large increase in leaf carbohydrates. On average, leaf soluble sugars increase by 52% and starch content increases by 160% (Long and Drake, 1992; Webber et al., 1994). Growth at elevated CO2 may also result in a large decline in Rubisco protein (up to 60%; Sage et al., 1989; Besford et al., 1990; Rowland-Bamford et al., 1991) and significant decreases in the transcript levels of genes encoding the small (rbcS) and large (rbcL) subunits of Rubisco (Nie et al., 1995a; Van Oosten and Besford, 1995). However, the metabolic signals and biochemical/molecular mechanisms underlying this acclimation to elevated CO2 are not well understood. Understanding the mechanisms that will ultimately determine the response of photosynthesis to the all-but-certain doubled atmospheric CO2 of the 21st century is a critical component in predicting the impact of global change on the earth's terrestrial ecosystems.
Sugars are known to influence many metabolic and cellular processes in both prokaryotes and eukaryotes, in part through modulation of gene expression (for reviews, see Sheen, 1994; Saier et al., 1995; Koch, 1996). To date, there is substantial evidence indicating that increased sugar levels can trigger repression of photosynthetic gene transcription. Using a transient expression system in maize protoplasts, Sheen (1990) showed that transcription of seven photosynthetic genes, including rbcS, is repressed by Glc and Fru. Furthermore, overexpression of a yeast invertase gene in the apoplast of tobacco leaves resulted in leaf hexose accumulation, bleached leaves, and stunted growth (von Schaewen et al., 1990). These transgenic plants also showed an inhibition of photosynthesis attributable to a decrease in the levels of several Calvin-cycle enzymes, including Rubisco. When detached spinach leaves were supplied with Glc through the transpiration stream, levels of rbcS mRNA decreased within hours (Krapp et al., 1993), and the amount of Rubisco protein declined 90% after 7 d (Krapp et al., 1991). Such sugar repression of photosynthetic genes appears to be widespread; this phenomenon has also been demonstrated to occur in an autotrophic cell culture of Chenopodium rubrum (Krapp et al., 1993), in photomixotrophic cultures and protoplasts of rapeseed (Harter et el., 1993), and in intact leaves/plants of Arabidopsis thaliana, tomato, potato, and wheat (Cheng et al., 1992; Heineke et al., 1994; Van Oosten and Besford, 1994; Jones et al., 1996; Dijkwel et al., 1997).
Repression of photosynthetic gene transcription by accumulated leaf soluble sugars is an attractive hypothesis to explain the acclimation responses of photosynthesis to elevated CO2. However, research on plant responses to high CO2 has largely focused on growth and physiological acclimation, with only a few studies addressing the effects of elevated CO2 on photosynthetic gene expression (e.g. Van Oosten et al., 1994; Van Oosten and Besford, 1995; Majeau and Coleman, 1996). Although these studies indicate that plants do modulate the levels of photosynthetic mRNAs in parallel with leaf carbohydrate status after exposure to high CO2, the link between sugar repression of gene expression and control of photosynthetic acclimation at elevated CO2 remains elusive.
All of the attributes of Arabidopsis that have made it a model experimental organism (e.g. the existence of many mutants, the small genome, the short generation time, and the large amount of genome information) for addressing a myriad of important questions in plant biology make it valuable for high-CO2 research. In Arabidopsis and other higher plants, rbcS mRNAs are encoded by a multigene family and their expression patterns can differ both quantitatively and qualitatively in response to light and development, and in different organs (for review, see Manzara and Gruissem, 1988; Dean et al., 1989). In Arabidopsis, the rbcS gene family consists of four members, namely 1A, 1B, 2B, and 3B (Krebbers et al., 1988). Dedonder et al. (1993) showed that the expression of individual Arabidopsis rbcS genes is differentially regulated by light of different quality and quantity. Whether other environmental factors such as elevated CO2 exert differential effects on the expression of individual rbcS genes in any species has not yet been determined.
Furthermore, in many species, such as Arabidopsis, grown in a light/dark photoperiod, rbcS mRNA exhibits a diurnal pattern of expression, with peak abundance occurring soon after dawn and minimum levels at the end of the light period (Pilgrim and McClung, 1993; this paper). This diurnal oscillation of rbcS mRNA occurs in an inverse time frame to the normal daytime accumulation and nighttime mobilization of leaf carbohydrates (e.g. Trethewey and ap Rees, 1994; Geiger et al., 1995). The response of such diurnal patterns after treatment of the plant with high CO2 can be a useful approach for evaluating carbohydrate regulation of photosynthetic gene expression (e.g. Nie et al., 1995a).
In this study we have examined the accumulation of leaf carbohydrates and changes in Rubisco expression (both protein and transcripts) during exposure of Arabidopsis to elevated CO2 (both long-term growth and short-term transfer). We have also closely examined the impact of elevated CO2 on the diurnal expression of rbcS gene family members in relation to leaf carbohydrate metabolism. These data provide insight into the regulation of photosynthetic gene expression by elevated sugar levels and on the control points of Rubisco synthesis (e.g. transcription, mRNA stability, translation, and protein turnover) during plant acclimation to high CO2.
MATERIALS AND METHODS
Plants of Arabidopsis thaliana (L.) Heynh. ecotype Columbia were germinated and grown five plants per 1-L pot in growth chambers at either 360 μL L−1 CO2 (ambient conditions) or 1000 μL L−1 CO2 (high-CO2 conditions), a 10-h photoperiod, a 21/18°C thermoperiod, 80% RH, and an irradiance of 400 μmol quanta m−2 s−1. To avoid growth-chamber effects, two chambers per treatment were used in replicate experiments. Plants were watered with one-fourth-strength Hoagland solution twice weekly.
For long-term growth experiments, plants were grown continuously for 40 d at ambient or high CO2. At this stage, plants grown at high CO2 were 3 to 4 d farther along developmentally than those grown at ambient CO2, as judged by subsequent bolting. Thus, high-CO2-grown plants on average may have been 2 d more advanced developmentally. For transfer experiments, 30-d-old ambient-CO2-grown plants were transferred to high CO2 for up to 12 d, and no accelerated development was apparent after the transfer. Diurnal tissue sampling of ambient control or high-CO2-treated plants began on the 6th d after transfer from ambient to high CO2. Five plants of each treatment were harvested at the indicated times and leaves were pooled for analysis. Shoots of Arabidopsis were harvested and frozen in liquid N2. Stems and petioles were removed from the samples before leaf analyses.
Biochemical Measurements
Leaf Rubisco content was measured by binding [14C]2-carboxyarabinatol-1,5-bisphosphate, followed by immunoprecipitation (Evans and Seemann, 1984). For carbohydrate measurements, samples were extracted in hot ethanol and processed as described by Moore et al. (1997). Starch in residual material was autoclaved and hydrolyzed as described by Schulze et al. (1991). All sugars were measured using high-performance anion exchange–pulsed-amperometric detection and a CarboPac PA1 column (Dionex, Sunnyvale, CA) under conditions described previously for parsley (Moore et al., 1997).
RNA Isolation and Northern-Blot Analysis
Total leaf RNA was isolated as described previously (Cheng and Seemann, 1998), except that the RNA pellet was dissolved in 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA after LiCl precipitation, and no alcohol precipitation was performed thereafter. Carbohydrate or protein contamination of the RNA preparation was evaluated by measuring A230, A260, and A280, and the A260 values were used for quantitation.
Total RNA was denatured, size fractionated by electrophoresis in a Mops-formaldehyde-1.4% agarose gel, transferred to a nylon membrane (Schleicher & Schuell), and cross-linked to the membrane by UV irradiation (Stratalinker, Stratagene) (Sambrook et al., 1989). Before hybridization, the nylon membrane was stained with methylene blue to check RNA integrity and ensure equal loading of RNA amounts. cDNAs used as probes were a 750-bp SalI/NotI fragment of Arabidopsis rbcS (no. 11C1T7P) and a 1.6-kb SalI/NotI fragment of Arabidopsis α-tubulin (no. 32C11T7) obtained from the Arabidopsis Biological Research Center (Ohio State University, Columbus), and a 1.2-kb plastid BamHI/EcoRI fragment of tobacco rbcL (Shinozaki and Sugiura, 1982). These DNA fragments were labeled with [α-32P]dCTP by random priming (Prime-a-Gene, Promega).
Hybridization was carried out at 42°C in a buffer containing 6× SSC buffer (1× SSC is 150 mm NaCl and 15 mm sodium citrate, pH 7.0), 50% (v/v) formamide, 5× Denhardt's solution (1× Denhardt's solution is 0.02% PVP, 0.02% Ficoll, and 0.02% BSA), 50 mm sodium phosphate, pH 7.0, 0.2% SDS (w/v), and 100 μg mL−1 denatured salmon-sperm DNA. After 16 h of hybridization the blots were washed twice for 5 min at room temperature in 2× SSC/0.1% SDS, and then twice for 10 min in 0.2× SSC/0.1% SDS at 60°C (for rbcS mRNA) or in 0.5× SSC/0.1% SDS at 60°C (for rbcL and tubulin mRNAs). In each blot a dilution series of an RNA sample was included to ensure that 32P-labeled probes were in excess. The hybridizing DNA probe was removed by incubating the blots in 50 mm Tris-HCl, pH 8.0, 60% formamide, and 1% SDS at 75°C for 1 h, and the blots were reprobed. Hybridization signals were quantified with a phosphor imager (Bio-Rad) to determine the relative amount of RNA present in each lane.
The expression of individual members of the Arabidopsis rbcS gene family was determined by using gene-specific oligonucleotide probes (Dedonder et al., 1993). The sequences of the probes were 5′-TTTTGAGGTTTACACAAAAG-3′ (1A), 5′-CGGATAGTCAACATTGAAT-3′ (1B), 5′-AGAATAATCAACGCTGAATAT-3′ (2B), and 5′-AGATAATTCATAAGAATGTT-3′ (3B). These synthetic oligonucleotides are complementary to the 3′ untranslated regions of the rbcS mRNAs (Krebbers et al., 1988). Ten micrograms of total RNA from each sample was processed as described as above, and the hybridization conditions were the same as reported by Dedonder et al. (1993).
RESULTS
Effects of Long-Term Growth at Elevated CO2
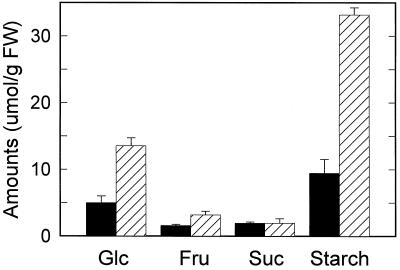
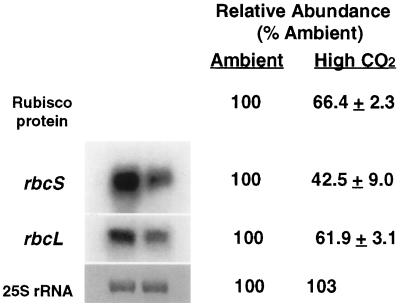
Glc, Fru, Suc, and starch were the predominant nonstructural leaf sugars in both ambient- and high-CO2-grown Arabidopsis. Long-term growth of Arabidopsis at elevated CO2 resulted in a 2-fold or greater increase in Glc and Fru and a 3.5-fold increase in starch, whereas Suc levels remained relatively constant (Fig. 1). Growth of Arabidopsis at high CO2 caused an approximately 34% reduction in Rubisco protein content and an approximately 38% decrease in rbcL mRNA (Fig. 2). However, the abundance of total rbcS transcript decreased nearly 60% at elevated CO2 relative to that at ambient CO2. Notably, the decrease in Rubisco protein content was consistently similar in magnitude to the decrease in rbcL mRNA but not to that in rbcS mRNA. Although growth at high CO2 was slightly accelerated relative to that at ambient CO2 (see Methods), the large decreases in Rubisco protein and subunit transcript levels were not attributable to accelerated development, but primarily to the effects of high CO2 (S.-H. Cheng and J.R. Seeman, unpublished data).
Figure 1.
Effects of long-term growth at elevated CO2 on nonstructural carbohydrate content in leaves of Arabidopsis. Values represent means ± sd (n = 3) for plants collected at midday. Starch is expressed as micromoles of Glc equivalents. Total sugar amounts were 19.9 and 53.8 μmol hexose equivalents g−1 fresh weight in ambient-CO2-grown (black bars) and high-CO2-grown (hatched bars) plants, respectively. Leaf chlorophyll was about 1.25 mg g−1 fresh weight in plants grown under both conditions. FW, Fresh weight.
Figure 2.
Effects of long-term growth at elevated CO2 on Rubisco content and rbcS and rbcL transcript abundance. Absolute amounts of leaf Rubisco were 33.7 and 22.3 nmol g−1 fresh weight for ambient- and high-CO2-grown plants, respectively. One microgram of total RNA per lane was used for northern-blot analysis. rRNA was used as an internal control to ensure equal loading of the lanes; 25S rRNA is shown stained with methylene blue. Relative hybridization signals quantified by phosphor imaging were expressed as a percentage of the values obtained from the ambient control. Values are means ± sd from three filters each, with RNA samples extracted from two replicate experiments. Leaves were collected at midday.
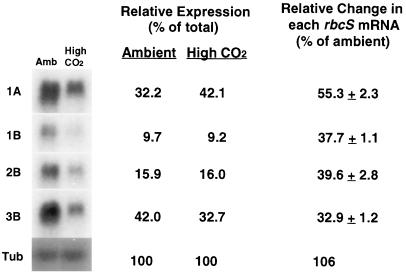
In Arabidopsis, rbcS transcripts are encoded by four different genes, 1A, 1B, 2B, and 3B (Krebbers et al., 1988). Although the coding regions of Arabidopsis rbcS genes have a high degree of homology with one another, the 3′ untranslated regions are sufficiently divergent to allow their use as gene-specific probes to examine individual rbcS mRNA expression (Krebbers et al., 1988; Dedonder et al., 1993). Under both CO2 growth conditions, the levels of 1A and 3B gene mRNA accounted for about 75% of the total rbcS transcript pool, and the 1B and 2B genes accounted for approximately 25% of the total (Fig. 3). Although total rbcS mRNAs decreased by approximately 60% in plants grown at elevated CO2, individual rbcS genes were down-regulated to somewhat different degrees. High CO2 caused more than a 60% decrease in the transcript abundance of 1B, 2B, and 3B genes, with a smaller reduction (45%) in 1A mRNA levels. The internal control gene α-tubulin was relatively unaffected by the CO2 growth conditions.
Figure 3.
Effects of long-term growth at elevated CO2 on the relative abundance of rbcS gene members. Ten micrograms of total RNA per lane was used for northern-blot analysis. Gene-specific probes were labeled to similar specific activities (approximately 5 × 107 dpm pmol−1), and equal amounts of radioactivity of each probe were used. Blots were exposed for the same length of time. The relative amounts of individual rbcS mRNA were expressed as a percentage of the sum of hybridizing signals from each of the rbcS members. Relative changes attributable to CO2 treatments were expressed using the ambient values of each rbcS gene as 100%. One of the blots was stripped and hybridized to an internal control gene (α-tubulin [Tub]). Numbers represent means ± sd from two filters each, with RNA samples extracted from two replicate experiments. Leaves were collected at midday.
Effects of Short-Term Transfer from Ambient to Elevated CO2
Rapid Down-Regulation of rbcS mRNA Relative to Rubisco Protein
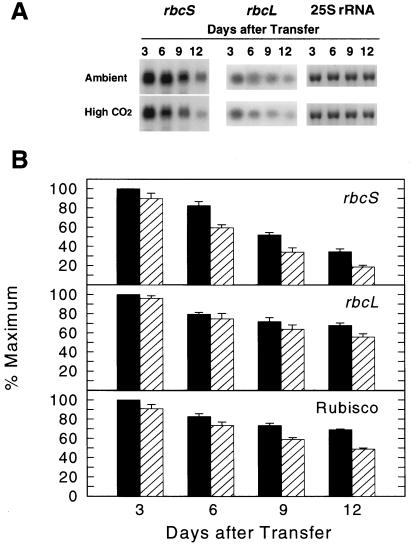
In an initial experiment, plants grown at ambient CO2 were transferred to high CO2 for up to 12 d, and leaves were collected at the beginning of the light period on each indicated sampling day for Rubisco protein and transcript measurements (Fig. 4). The time of day for sampling was selected based on a previous finding that the level of total rbcS mRNAs in Arabidopsis grown in a light/dark photoperiod displays a diurnal pattern, with peak abundance occurring soon after dawn (Pilgrim and McClung, 1993). In this experiment plants from both CO2 conditions showed an age-dependent decline in the levels of rbcS and rbcL mRNA and in Rubisco protein. Nonetheless, a distinct CO2 effect was evident. Total rbcS transcript abundance decreased significantly by d 6 (approximately 30%), with an additional decline thereafter. There was only a small effect of elevated CO2 on rbcL mRNA levels (an 18% decline by d 12). A significant CO2-induced decrease in Rubisco protein was observed on d 9 and 12 after the transfer to elevated CO2 (20 and 29%, respectively; Fig. 4B).
Figure 4.
Time course of relative abundance of Rubisco protein and total rbcS and rbcL transcripts in ambient-CO2-grown plants transferred to elevated CO2 for 12 d. Plants were transferred at the beginning of the light period on d 1, and leaves from 10 plants were collected on the indicated days at the beginning of the light period. A, Northern-blot analysis using 1 μg of total RNA as shown on the representative blot. 25S rRNA is shown stained with methylene blue. B, Leaf Rubisco content and quantified relative amounts of rbcS and rbcL mRNAs from ambient-CO2-grown (black bars) and high-CO2-grown (hatched bars) plants. Values are means ± sd from three separate protein extractions or from three replicate filters, with RNA extracted from one sample collection.
Dampening of Diurnal Oscillation of rbcS mRNA at Elevated CO2
Because a significant decrease of rbcS transcript abundance occurred by d 6 (Fig. 4), we chose this treatment time for a more detailed analysis of the diurnal expression patterns of rbcS and rbcL mRNAs and total leaf carbohydrate accumulation after high-CO2 treatments. The level of total rbcS mRNAs oscillated from a maximum at the beginning of the light period to a minimum at the end of the light period, with an ongoing accumulation throughout the dark period that reestablished the maximum early-morning level (Fig. 5). The change in transcript levels during the day was about 3-fold more than that in ambient-grown plants.
Figure 5.
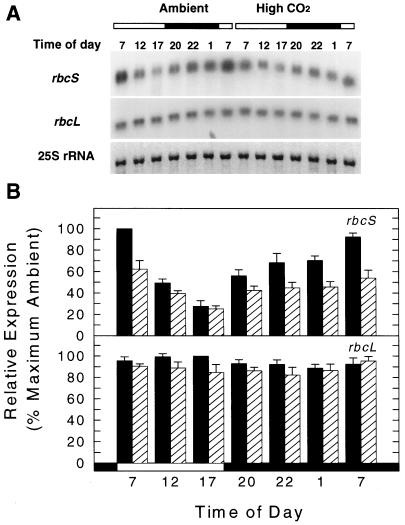
Effects of transfer to elevated CO2 on the relative abundance of rbcS and rbcL transcripts during a light/dark cycle. Ambient-CO2-grown plants were transferred to elevated CO2 at the beginning of d 1, and plants were collected through the 24-h period on d 6 of exposure. A, Northern-blot analysis of rbcS and rbcL expression using 1 μg of total RNA per lane. 25S rRNA is shown stained with methylene blue. Bars above blots and at the bottom of the graph in B indicate the light regime (the filled area indicates dark, the open area indicates light). B, Quantified signals of total rbcS and rbcL transcripts from ambient-CO2-grown (black bars) and high-CO2-grown (hatched bars) plants. The hybridizing signal at 7:00 am from ambient-CO2-grown plants was defined as 100%. The diurnal transfer experiment was done twice. Values are means ± sd from four filters (two filters for each replicate).
A similar diurnal pattern of total rbcS mRNA level occurred in plants transferred to elevated CO2 (Fig. 5), with maximum and minimum values also observed at the beginning and end of the light period, respectively. However, the maximum level was reduced about 40% relative to ambient CO2 (comparable to Fig. 4). Treatment differences in rbcS mRNA levels diminished from the beginning of the light period such that by the end of the light period (5:00 pm), virtually no difference in transcript abundance was found between CO2 treatments. However, the nighttime accumulation of rbcS mRNA at high CO2 was much less than what occurred at ambient CO2, thereby reducing the maximum accumulation level at the beginning of the light period. In contrast, rbcL transcript levels from both CO2 conditions remained approximately constant throughout the 24-h light/dark cycle, with rbcL mRNA amounts at elevated CO2 decreasing only slightly relative to amounts in the ambient treatment (Fig. 5).
Differential Sensitivity of rbcS Genes to Elevated CO2 during the Diurnal Cycle
Because individual rbcS genes displayed differential sensitivities to long-term growth at high CO2 (Fig. 3), we examined the effects of short-term elevated CO2 on the diurnal expression of specific rbcS genes. In ambient-CO2-grown plants, the patterns of diurnal oscillation of mRNA with respect to the timing of peak accumulation and the amplitude of the fluctuation were not identical among rbcS genes (Fig. 6). Similar to the oscillation patterns observed for total rbcS mRNAs, the expression of rbcS 1A and 3B mRNA (which constitute 75% of the total) was maximum at the beginning of the light period and lowest at the end. However, the relative magnitudes of these fluctuations and the rate of recovery during the dark were different in these two rbcS genes. 1A mRNA had a greater amplitude and slower recovery than did 3B mRNA. In contrast, the peak transcript abundance of both the 1B and 2B rbcS genes occurred before the beginning of the light period (5 and 8 h into the dark, respectively). Also, the minimum levels of mRNA accumulation occurred in the middle of the day for the 1B gene, but at the end of the light period for the 2B gene. Evidently, Arabidopsis rbcS gene members are expressed differently during the diurnal cycle.
Figure 6.
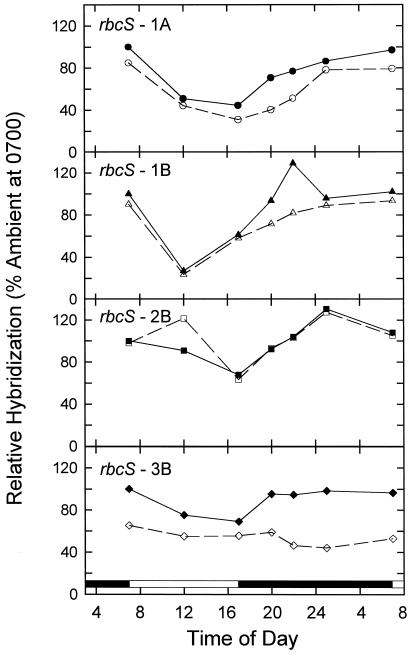
Effects of transfer to elevated CO2 on the diurnal oscillation of different rbcS transcripts. Ambient-CO2-grown plants were transferred to elevated CO2 at the beginning of d 1, and plants were collected through the 24-h period on d 6 of exposure. Ten micrograms of total RNA was used for northern-blot analysis. The hybridizing signal for each gene at 7:00 am from ambient-CO2-grown plants was defined as 100%. Values are means ± sd from three filters with total RNA extracted from one sample collection. Filled symbols, Ambient-CO2-grown plants; open symbols, high-CO2-treated plants.
Transfer to high CO2 on d 6 resulted in differential effects on the diurnal fluctuations of individual rbcS mRNAs (Fig. 6). Among the four rbcS genes, the 3B gene was most affected by the high-CO2 treatment. The 3B transcript levels in high-CO2-grown plants were as much as 60% lower than those in ambient plants throughout the light/dark cycle. In contrast to its normal nighttime recovery under ambient conditions, 3B transcript levels remained low throughout the night after transfer to high CO2. Elevated CO2 also reduced 1A mRNA levels somewhat throughout the light/dark cycle, but substantially less than occurred with 3B. 1A mRNA levels did increase during the night, but at a slower rate than at ambient CO2. Transfer to high CO2 had a minimal effect on the 2B diurnal levels except for an apparent stimulation at midday. Likewise, 1B mRNA levels were not much affected by high CO2 except that they were lower during the initial 5 h of darkness. After 6 d at high CO2, the relative expression of 1A, 1B, 2B, and 3B were about 35, 12, 23, and 30%, respectively, at the beginning of the light period (calculated from Figs. 3 and 6).
Diurnal Pattern of Leaf Sugar Accumulation
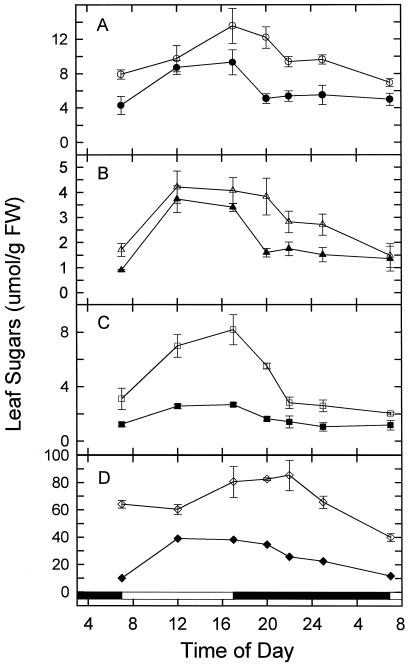
In both control plants and plants transferred to high CO2, leaf Glc, Fru, Suc, and starch were at a minimum at the beginning of the light period, accumulated throughout the day, and then decreased to different extents during the night (Fig. 7). This pattern of leaf carbohydrate accumulation was generally inverse to that for rbcS mRNA. Overall, high-CO2-treated plants had higher levels of all four sugars than did ambient-CO2-grown plants throughout the light/dark cycle. Moreover, the nighttime metabolism of all four sugars was delayed in plants transferred to high CO2. For example, the level of leaf hexoses was high and relatively constant during the first 4 h of darkness in treated plants, whereas the normal situation is that their levels have already declined to minimum values after 4 h of darkness. These data indicate that exposure to elevated CO2 not only resulted in a 2-fold increase in total nonstructural leaf carbohydrate content, but also caused a significant change in nighttime carbohydrate metabolism.
DISCUSSION
We report here the first detailed characterization of photosynthetic acclimation to elevated atmospheric CO2 in Arabidopsis at both the biochemical and molecular levels. Growth of Arabidopsis at high CO2 resulted in a 2-fold accumulation of nonstructural leaf carbohydrates and a substantial decrease in Rubisco protein content (Figs. 1 and 2), similar to that found in other species (e.g. wheat [Nie et al., 1995a, 1995b]; tomato [Van Oosten and Besford, 1995]; and pea [Majeau and Coleman, 1996]). Although such a decrease in Rubisco protein during plant growth at high CO2 is well documented (e.g. Sage et al., 1989; Stitt, 1991), the mechanism(s) that controls Rubisco expression at high CO2 has yet to be identified. Control of Rubisco synthesis is known to occur at the transcriptional, posttranscriptional (e.g. mRNA stability), translational, and/or posttranslational levels, depending on developmental factors and environmental stimuli (Deng and Gruissem, 1987; Berry et al., 1988; Shirley and Meagher, 1990; Wanner and Gruissem, 1991; Winder et al., 1992).
In Arabidopsis grown at high CO2, the levels of Rubisco protein and rbcL mRNA were reduced about 40%, whereas rbcS mRNA was reduced to an even greater extent (approximately 60%; Fig. 2). There are only a few reports to date on the influence of high- or low-CO2 growth conditions on the expression of both Rubisco protein and subunit transcript levels. In wheat grown at elevated CO2, both rbcS and rbcL transcripts decreased equally, with the same level of decline also observed in Rubisco protein (newly mature third leaves in chamber plants [Webber et al., 1994]; flag leaves in field plants [Nie et al., 1995b]). These results suggest that the regulation of transcriptional and/or posttranscriptional processes (e.g. mRNA stability) could determine the level of Rubisco protein at elevated CO2.
In tomato transferred to elevated CO2 for 22 d, the levels of Rubisco protein and subunit transcripts decreased to different extents relative to control plants, suggesting a different type of posttranscriptional regulation of protein content (Van Oosten and Besford, 1995). In Chlamydomonas reinhardtii grown under low-CO2 conditions, the steady-state levels of both rbcS and rbcL transcripts were not affected, but Rubisco protein content was found to decline rapidly (Winder et al., 1992). This was not because of an increased rate of protein degradation, but rather because of the inhibition of translation of both rbcS and rbcL mRNAs. Growth of pea at low levels of CO2 resulted in decreased rbcS mRNA, but had no effect on total Rubisco activity (i.e. fully activated enzyme; Majeau and Coleman, 1996).
The nature of the control of Rubisco content in Arabidopsis grown at high CO2 is intriguing because the coordination between rbcS and rbcL transcript levels was altered, as was the expression of protein content relative to subunit transcripts. The synthesis of Rubisco requires coordinated expression between the nucleus and the chloroplast genomes, but nuclear-encoded photosynthetic genes are generally more readily repressed by accumulated carbohydrates than are chloroplast-encoded genes (e.g. Van Oosten and Besford, 1994; Van Oosten et al., 1994). A decreased level of rbcS mRNA relative to rbcL mRNA has been observed during leaf senescence in bean (Bate et al., 1991), during severe water stress in tomato (Bartholomew et al., 1991), and with high-CO2 growth conditions in tomato (Van Oosten and Besford, 1994). Such differential transcript expression also has been observed in tobacco rbcS antisense plants, but reduced rbcS mRNA largely corresponded to reduced Rubisco content (Rodermel et al., 1988). Of particular significance was the finding that leaf rbcL mRNA in the rbcS antisense plants was not efficiently translated (Rodermel et al., 1996), a situation that may also occur in Arabidopsis at high CO2. During normal leaf development, Rubisco subunit transcripts and protein content are all coordinately expressed (Jiang and Rodermel, 1995). Growth of Arabidopsis at high CO2 may disrupt the homeostatic control of Rubisco protein and transcript expression.
We suggest that control of Rubisco content in Arabidopsis grown at high CO2 may have three primary components: (a) an inhibitory signal may repress levels of rbcS mRNA more than rbcL mRNA by differential effects on transcriptional activity and/or message stability. Transcription and mRNA stability are both important mechanisms for Glc repression of α-amylase in rice (Sheu et al., 1994); (b) leaf rbcL mRNA may not be as efficiently translated as is rbcS mRNA, or large subunit protein may simply accumulate in excess of small subunit protein, as apparently occurs in tomato (Van Oosten and Besford, 1995), or be degraded (although the latter has not been shown to occur in any system [Rodermel et al., 1996]); and (c) Rubisco protein turnover may be altered such that protein levels are not repressed to an extent comparable to rbcS mRNA. That Rubisco protein may be longer-lived was also suggested in a study of wheat leaves of intermediate age grown at high CO2 (Webber et al., 1994).
In higher plants, rbcS mRNAs are encoded by a multigene family (Dean et al., 1989). Pilgrim and McClung (1993) showed that the diurnal expression of total leaf rbcS mRNAs in Arabidopsis is under the control of a circadian clock, but transcriptional activities are not involved in regulating the clock. We have extended this observation by demonstrating that individual rbcS gene members in Arabidopsis also generally show the same diurnal expression pattern (Fig. 6) as was previously reported for total rbcS mRNAs (Pilgrim and McClung, 1993). However, the expression of 1B mRNA occurred out of phase relative to expression of the other three genes. This result was not detected in the diurnal analysis of total rbcS mRNAs (Fig. 5) (Pilgrim and McClung, 1993) because 1B mRNA normally constitutes only about 10% of the total leaf rbcS mRNAs.
In this study we have also shown that individual rbcS genes in Arabidopsis are expressed differently during long-term growth at high CO2 or after a short-term transfer to high CO2. During growth at high CO2, the 1A gene was the least repressed, whereas the other three genes were repressed by a similarly increased magnitude (Fig. 3). Transfer to high CO2 did not significantly affect the diurnal expression pattern of the individual genes, but did affect the magnitude of the expression levels of some of the genes (Fig. 6). After transfer to high CO2, the 3B gene mRNA was the most reduced, the 1A mRNA was moderately affected, and the 1B and 2B mRNAs were minimally affected. Because the 1A and 3B genes contribute the largest portion of the rbcS mRNA pool, their reduced expression at high CO2 resulted in a 40% reduction in the maximum level of total rbcS mRNA by d 6, and ultimately resulted in a 20% reduction in Rubisco content by d 9 (Figs. 4 and 5).
Several investigators (e.g. Krapp et al., 1993; Sheen, 1994; Van Oosten et al., 1994) have proposed that the increased metabolism of accumulated soluble sugars at high CO2 may trigger a repression of photosynthetic gene transcription. This repression is thought to be mediated by hexose metabolism via cytosolic hexokinase (Jang and Sheen, 1994; Jang et al., 1997), but we have limited knowledge of the suggested biochemical role of hexokinase as a sugar sensor. For example, the presumed signal of hexokinase, hexoses, are found to be almost exclusively located in the vacuole of leaf mesophyll cells during the daytime in several species grown at ambient or high CO2 (Heineke et al., 1994; Moore et al., 1997). Although our data and those of others indicate a correlation between increased leaf hexose levels and decreased rbcS mRNA in plants exposed to high CO2, one possibility is that hexokinase-mediated gene repression may occur in leaves at night rather than during the day.
In Arabidopsis we found that the normal nighttime recovery of rbcS transcript levels was greatly reduced after transfer of the plant to high CO2 (Fig. 5). We also found that leaf hexose levels were unusually high during the early hours of darkness (Fig. 7). In most species, including Arabidopsis, leaf carbohydrates accumulate during the day and are mobilized at night (Trethewey and ap Rees, 1994; Geiger et al., 1995). We hypothesize that elevated nighttime cytosolic hexose concentrations resulting from high CO2 are sensed by hexokinase, and trigger a repression response that results in decreased rbcS transcript levels. The fact that transgenic tobacco overexpressing a yeast invertase gene had reduced photosynthetic rates and no diurnal turnover of leaf carbohydrates is consistent with this hypothesis (von Schaewen et al., 1990).
Figure 7.
Effects of transfer to elevated CO2 on the diurnal accumulation of nonstructural leaf carbohydrates: A, Glc; B, Fru; C, Suc; and D, starch. Ambient-CO2-grown plants were transferred to elevated CO2 at the beginning of d 1, and plants were collected through the 24-h period on d 6 of exposure. Values represent means ± sd from three extractions. Starch is expressed as micromoles of Glc equivalents. Total maximum sugar amounts were 55.7 and 108.1 μmol hexose equivalents g−1 fresh weight in ambient-CO2-grown and high-CO2-transferred plants, respectively. Filled symbols, Ambient-CO2-grown plants; open symbols, high-CO2-treated plants. FW, Fresh weight.
An alternative possibility for hexose sensing in the absence of any detectable cytosolic hexoses at any time during the day could involve futile cycling of Suc between the cytosol and vacuole and/or between the cytosol and apoplast (Foyer, 1987; Huber, 1989; Stitt et al., 1990). Exposure to elevated CO2 may result in increased carbon flux to Suc. Under sink-limited conditions this may result in increased hexose accumulation from vacuolar or apoplastic hydrolysis by acid invertase (e.g. Goldschmidt and Huber, 1992). These hexoses may then be transported to the cytosol, phosphorylated by hexokinase, and reassimilated into Suc. Such carbohydrate cycling could occur rapidly but with no substantial increase in cytosolic hexoses or other cellular metabolites, which is analogous to Suc cycling through Suc synthase (Geigenberger and Stitt, 1991). With Suc cycling under high CO2, hexokinase then would have to function as a flux sensor. One effect of such metabolism is that species with lower leaf acid invertase activity may be less susceptible to down-regulation of photosynthesis when grown at high CO2.
In summary, our results support the hypothesis that increased leaf carbohydrates in response to elevated CO2 may signal the down-regulation of photosynthesis through modulation of photosynthetic genes such as rbcS, and that hexokinase-mediated repression of gene expression may occur at night. However, results presented in this report also suggest that sugar-induced repression of photosynthetic gene transcription alone cannot totally explain the decrease of Rubisco protein at elevated CO2. Rather, high-CO2-mediated changes in Rubisco expression are very complex and may involve multiple controls at the levels of transcription, mRNA stability, translation, and/or protein turnover. Therefore, it is important to determine specific transcription, translation, and protein turnover rates to better understand the mechanisms controlling photosynthetic responses to elevated CO2.
ACKNOWLEDGMENTS
We are grateful to Shanti Rawat and Therese Charlet for their technical assistance. We thank Dr. Steve Rodermel (Iowa State University, Ames) for kindly providing the cDNA of tobacco rbcL. Oligonucleotides used in this report were supplied by the Oligonucleotide Synthesis Core Facility at the University of Nevada, Reno.
Footnotes
This work was supported by National Science Foundation grant no. 1940424 to J.R.S. and S.-H.C.
LITERATURE CITED
- Bartholomew DM, Bartley GE, Scolnik PA. Abscisic acid control of rbcS and cab transcription in tomato leaves. Plant Physiol. 1991;96:291–296. doi: 10.1104/pp.96.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate NJ, Rothstein SJ, Thompson JE. Expression of nuclear and chloroplast photosynthesis-specific genes during leaf senescence. J Exp Bot. 1991;42:801–811. [Google Scholar]
- Berry OJ, Carr JP, Klessing DF. mRNAs encoding ribulose-1,5-bisphosphate carboxylase remain bound to polysome but are not translated in amaranth seedlings transferred to darkness. Proc Natl Acad Sci USA. 1988;85:4190–4194. doi: 10.1073/pnas.85.12.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besford RT, Ludwig LJ, Withers AC. The green house effect: acclimation of tomato plants growing in high CO2: photosynthesis and ribulose-1,5-bisphosphate carboxylase protein. J Exp Bot. 1990;41:925–931. [Google Scholar]
- Cheng C-L, Acedo GN, Christinsin M, Conkling MA. Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA. 1992;89:1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-H, Seemann JR (1998) Extraction and purification of RNA from plant tissue enriched in polysaccharides. In R Rapley, DL Manning, eds, RNA Isolation and Characterization Protocols: Methods in Molecular Biology, Vol 86. Humana Press, Totowa, NJ (in press) [DOI] [PubMed]
- Dean C, Dunsmuir P, Bedbrook J. Structure, evolution, and regulation of rbcS genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:415–439. [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E. Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol. 1993;101:801–808. doi: 10.1104/pp.101.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X-W, Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987;49:379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser, Weisbeek PJ, Chua N-H, Smeekens SCM. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR, Seemann JR. Differences between wheat genotypes in specific activity of RuBP carboxylase and the relationship to photosynthesis. Plant Physiol. 1984;74:759–765. doi: 10.1104/pp.74.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH. The basis for source-sink interaction in leaves. Plant Physiol Biochem. 1987;25:649–657. [Google Scholar]
- Geigenberger P, Stitt M. A “futile” cycle of sucrose synthesis and degradation is involved in regulating partitioning between sucrose, starch and respiration in cotyledons of germinating Ricinus communis L. seedlings when phloem transport is inhibited. Planta. 1991;185:81–90. doi: 10.1007/BF00194518. [DOI] [PubMed] [Google Scholar]
- Geiger DR, Shieh WJ, Yu XM. Photosynthetic carbon metabolism and translocation in wild-type and starch-deficient mutant Nicotiana sylvestris L. Plant Physiol. 1995;107:507–514. doi: 10.1104/pp.107.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt EE, Huber SC. Regulation of photosynthesis by end-product accumulation in leaves of plants storing starch, sucrose, and hexose sugars. Plant Physiol. 1992;99:1443–1448. doi: 10.1104/pp.99.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin KL, Seemann JR. Plants, CO2 and photosynthesis in the 21st century. Chemistry and Biology. 1996;3:245–254. doi: 10.1016/s1074-5521(96)90104-0. [DOI] [PubMed] [Google Scholar]
- Harter K, Talke-Messerer C, Barz W, Schafer E. Light- and sucrose-dependent gene expression in photomixotrophic cell suspension cultures and protoplasts of rape (Brassica napus L.) Plant J. 1993;4:507–516. [Google Scholar]
- Heineke E, Wildenberger K, Sonnewald U, Willmitzer L, Heldt HW. Accumulation of hexoses in leaf vacuoles: studies with transgenic tobacco plants expressing yeast-derived invertase in the cytosol, vacuole or apoplasm. Planta. 1994;194:29–33. [Google Scholar]
- Huber SC. Biochemical mechanism for regulation of sucrose accumulation in leaves during photosynthesis. Plant Physiol. 1989;91:656–662. doi: 10.1104/pp.91.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Sheen J. Sugar sensing in higher plants. Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C-Z, Rodermel SR. Regulation of photosynthesis during leaf development in rbcS antisense DNA mutants of tobacco. Plant Physiol. 1995;107:215–224. doi: 10.1104/pp.107.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PG, Lloyd JC, Raines CA. Glucose feeding of intact wheat plants repressed the expression of a number of Calvin cycle genes. Plant Cell Environ. 1996;19:231–236. [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Krapp A, Hofmann B, Shafer C, Stitt M. Regulation of the expression of rbcS and other photosynthetic genes by carbohydrates: a mechanism for the ‘sink’ regulation of photosynthesis. Plant J. 1993;3:817–828. [Google Scholar]
- Krapp A, Quick WP, Stitt M. Ribulose-1,5-bisphosphate carboxylase-oxygenase, other photosynthetic enzymes and chlorophyll decrease when glucose is supplied to mature spinach leaves via the transpiration stream. Planta. 1991;186:58–69. doi: 10.1007/BF00201498. [DOI] [PubMed] [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP. Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol. 1988;11:745–759. doi: 10.1007/BF00019515. [DOI] [PubMed] [Google Scholar]
- Long SP, Drake BG. Photosynthetic CO2 assimilation and rising atmospheric CO2 concentrations. In: Baker NR, Thomas H, editors. Crop Photosynthesis: Spatial and Temporal Determinants. Amsterdam: Elsevier; 1992. pp. 69–95. [Google Scholar]
- Majeau N, Coleman JR. Effects of CO2 concentration on carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase expression in pea. Plant Physiol. 1996;112:569–574. doi: 10.1104/pp.112.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzara T, Gruissem W. Organization and expression of the genes encoding ribulose-1,5-bisphosphate carboxylase in higher plants. Photosynth Res. 1988;16:117–139. doi: 10.1007/BF00039489. [DOI] [PubMed] [Google Scholar]
- Moore Bd, Palmquist DE, Seemann JR. Influence of plant growth at high CO2 concentrations on leaf content of ribulose-1,5-bisphosphate carboxylase/oxygenase and intracellular distribution of soluble carbohydrates in tobacco, snapdragon, and parsley. Plant Physiol. 1997;115:241–248. doi: 10.1104/pp.115.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie GY, Hendrix DL, Webber AN, Kimball BA, Long SP. Increased accumulation of carbohydrates and decreased photosynthetic gene transcript levels in wheat grown at an elevated CO2 concentration in the field. Plant Physiol. 1995a;108:975–983. doi: 10.1104/pp.108.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie GY, Long SP, Garcia RL, Kimball BA, Lamorte RL, Pinter PJ, Wall GW, Webber AN. Effects of free-air CO2 enrichment on the development of the photosynthetic apparatus in wheat, as indicated by changes in leaf proteins. Plant Cell Environ. 1995b;18:855–864. [Google Scholar]
- Pilgrim ML, McClung CR. Differential involvement of the circadian clock in the expression of genes required for ribulose-1,5-bisphosphate carboxylase/oxygenase synthesis, assembly, and activation in Arabidopsis thaliana. Plant Physiol. 1993;103:553–564. doi: 10.1104/pp.103.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S, Abbott MS, Bogorad L. Nuclear-organelle interactions: nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell. 1988;55:673–681. doi: 10.1016/0092-8674(88)90226-7. [DOI] [PubMed] [Google Scholar]
- Rodermel S, Haley J, Jiang C-Z, Tsai C-H, Bogorad L. A mechanism for intergenomic integration: abundance of ribulose-1,5-bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA. 1996;93:3881–3885. doi: 10.1073/pnas.93.9.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland-Bamford AJ, Baker JT, Allen LH, Jr, Bowes G. Acclimation of rice to changing atmospheric CO2 on growth, photosynthesis and water relations of salt marsh grass species. Aquat Bot. 1991;39:45–55. [Google Scholar]
- Sage RF, Sharkey TD, Seemann JR. Acclimation of photosynthesis to elevated CO2 in five C3 species. Plant Physiol. 1989;89:590–596. doi: 10.1104/pp.89.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J-J. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schulze W, Stitt M, Schulze E-D, Heuhaus HE, Fichtner K. A quantification of the significance of assimilatory starch for growth of Arabidopsis thaliana L. Heynh. Plant Physiol. 1991;95:890–895. doi: 10.1104/pp.95.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Feedback control of gene expression. Photosynth Res. 1994;39:427–438. doi: 10.1007/BF00014596. [DOI] [PubMed] [Google Scholar]
- Sheu J-J, Jan S-P, Lee H-T, Yu S-M. Control of transcription and mRNA turnover as mechanisms for metabolic repression of alpha-amylase gene expression. Plant J. 1994;5:655–664. [Google Scholar]
- Shinozaki K, Sugiura M. The nucleotide sequence of the tobacco chloroplast gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Gene. 1982;20:91–102. doi: 10.1016/0378-1119(82)90090-7. [DOI] [PubMed] [Google Scholar]
- Shirley BW, Meagher RB. A potential role for RNA turnover in the light regulation of plant gene expression: ribulose-1,5-bisphosphate carboxylase small subunit in soybean. Nucleic Acids Res. 1990;18:3377–3386. doi: 10.1093/nar/18.11.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. Rising CO2 levels and their potential significance for carbon flow in photosynthetic cells. Plant Cell Environ. 1991;14:741–762. [Google Scholar]
- Stitt M, von Schaewen A, Willmitzer L. “Sink” regulation of photosynthetic metabolism in transgenic tobacco plants expressing yeast invertase in their cell wall involves a decrease of the Calvin-cycle enzymes and an increase of glycolytic enzymes. Planta. 1990;183:40–50. doi: 10.1007/BF00197565. [DOI] [PubMed] [Google Scholar]
- Trethewey RN, ap Rees T. The role of the hexose transporter in the chloroplasts of Arabidopsis thaliana L. Planta. 1994;195:168–174. [Google Scholar]
- Van Oosten J-J, Besford RT. Sugar feeding mimic effect of acclimation to high CO2 rapid down regulation of rubisco small subunit transcripts but not to the large subunit transcripts. J Plant Physiol. 1994;143:306–312. [Google Scholar]
- Van Oosten J-J, Besford RT. Some relationships between the gas exchange, biochemistry and molecular biology of photosynthesis during leaf development of tomato plants after transfer to different carbon dioxide concentrations. Plant Cell Environ. 1995;18:1253–1266. [Google Scholar]
- Van Oosten J-J, Wilkins S, Besford RT. Regulation of the expression of photosynthetic nuclear genes by high CO2 is mimicked by carbohydrates: a mechanism for the acclimation of photosynthesis to high CO2. Plant Cell Environ. 1994;17:913–923. [Google Scholar]
- von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. Expression of a yeast-derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbohydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO J. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Gruissem W. Expression dynamics of the tomato rbcS gene family during development. Plant Cell. 1991;3:1289–1303. doi: 10.1105/tpc.3.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber AN, Nie GY, Long SP. Effects of rising CO2 concentration on expression of photosynthetic proteins. Photosynth Res. 1994;39:413–425. doi: 10.1007/BF00014595. [DOI] [PubMed] [Google Scholar]
- Winder TL, Anderson JC, Spalding MH. Translational regulation of the large and small subunits of ribulose bisphosphate carboxylase/oxygenase during induction of the CO2 concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 1992;98:1409–1414. doi: 10.1104/pp.98.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]