Abstract
Antiretroviral therapy (ART) has evolved considerably over the last three decades. From the early days of monotherapy with high toxicities and pill burdens, through to larger pill burdens and more potent combination therapies, and finally, from 2005 and beyond where we now have the choice of low pill burdens and once-daily therapies. More convenient and less toxic regimens are also becoming available, even in resource-poor settings. An understanding of the individual variation in response to ART, both efficacy and toxicity, has evolved over this time. The strong association of the major histocompatibility class I allele HLA-B*5701 and abacavir hypersensitivity, and its translation and use in routine HIV clinical practice as a predictive marker with 100% negative predictive value, has been a success story and a notable example of the challenges and triumphs in bringing pharmacogenetics to the clinic. In real clinical practice, however, it is going to be the exception rather than the rule that individual biomarkers will definitively guide patient therapy. The need for individualized approaches to ART has been further increased by the importance of non-AIDS comorbidities in HIV clinical practice. In the future, the ideal utilization of the individualized approach to ART will likely consist of a combined approach using a combination of knowledge of drug, virus, and host (pharmacogenetic and pharmacoecologic [factors in the individual’s environment that may be dynamic over time]) information to guide the truly personalized prescription. This review will focus on our knowledge of the pharmacogenetics of the efficacy and toxicity of currently available antiretroviral agents and the current and potential utility of such information and approaches in present and future HIV clinical care.
Keywords: HIV, pharmacogenetics, pharmacogenomics, personalized medicine, antiretroviral
Introduction and current antiretroviral use
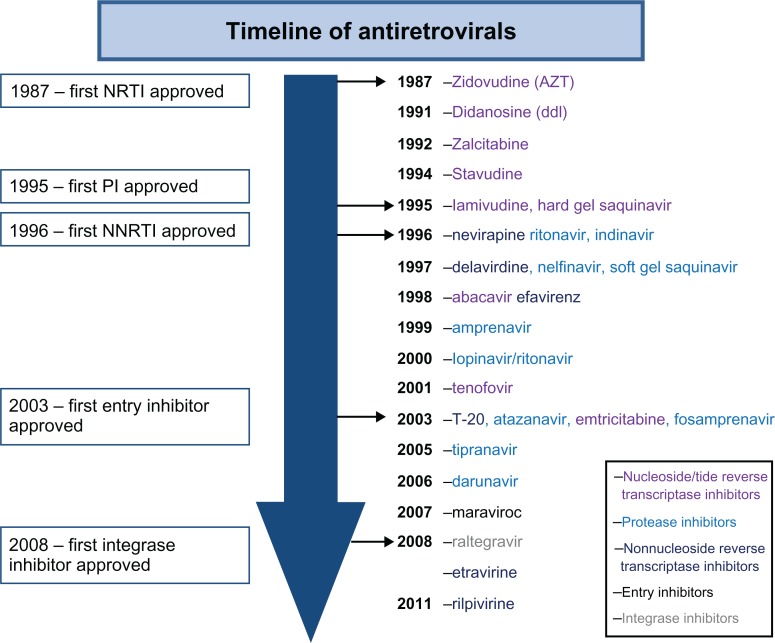
Since the recognition of AIDS as a clinical syndrome 30 years ago there have been 26 drugs approved for the HIV treatment (Figure 1). Early treatment of HIV was burdened with the inconvenience and intolerance of high-dose drugs such as zidovudine in the absence of improved monitoring with viral load and resistance testing. This type of treatment evolved into sequential monotherapy and finally in the mid-1990s into the true era of highly active antiretroviral therapy (HAART), when combination antiretroviral therapy (ART) with three or more agents became the norm. These early HAART years were marked by regimens that had high pill burdens and toxicities and, although efficacious, were hard to adhere to and tolerate. Early fixed-dose combination with newer nucleoside reverse transcriptase inhibitors (NRTIs) led to smaller pill burdens, and these later-generation NRTIs also had fewer toxicities. This evolved to the development of once-daily drugs with fewer pill burdens and toxicities across the NRTI, non-nucleoside reverse transcriptase inhibitor (NNRTI), and protease inhibitor classes (Figure 1).
Figure 1.
Timeline of antiretrovirals.
Abbreviations:
NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
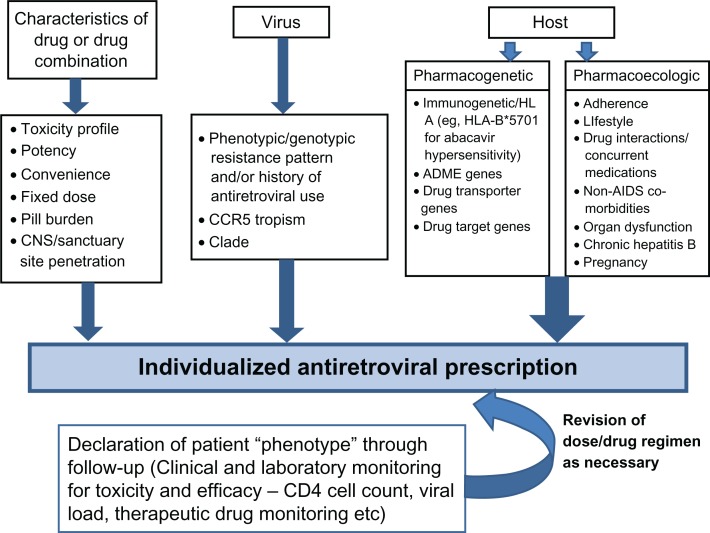
HIV is a chronic proinflammatory illness and, if left untreated, results in chronic CD4+ T-cell count depletion. The decision on when to start antiretroviral drugs has traditionally been based on the degree of immunosuppression, and evidence-based medicine unequivocally supports the initiation of therapy with CD4 T-cell counts of 350/μL or lower.1–4 An increasing body of observational literature supports much earlier initiation of ART, and this has been supported by increasing knowledge of the basic science of inflammation in relation to chronic HIV infection (Table 1).1–4 The evolution of less toxic and more convenient antiretroviral agents (Figure 1) and the current landscape of HIV treatment guidelines (Tables 1 and 2) have driven and demonstrate the significant shift toward earlier treatment of HIV, respectively. Introduction of newer, more convenient, and highly tolerable drugs near the end of this past decade has led to more treatment options for HIV. In the developed world, in particular, we currently have the luxury of individualizing ART based on specific characteristics of the drug, virus, and host (Figure 2). Characteristics of the drug that may play into the decision of treating a specific individual will include its toxicity profile, convenience, pill burden, and penetration into the central nervous system (CNS) and other sanctuary sites. Characteristics of the virus influencing drug choice include the viral resistance pattern demonstrated on genotypic and/or phenotypic testing and the history of previous antiretroviral use in treatment-experienced patients. More recently, C-C chemokine receptor type 5 (CCR5) antagonists such as maraviroc have been developed, which inhibit a host chemokine receptor utilized by a proportion of HIV, particularly early in the disease. Genotypic and phenotypic tropism assays are utilized to ascertain whether HIV and the population of viruses in a specific patient are likely to be CCR5 tropic and respond to treatment with these agents.5,6
Table 1.
Comparison of recommendations on when to start, based on CD4+ cell count
| CD4 <200/μL | CD4 200–350/μL | CD4 350–500/μL | CD4 >500/μL | |
|---|---|---|---|---|
| Department of Health and Human Services 20111 | ✓ | ✓ | ✓ | ✓ (optional) |
| International AIDS Society 20102 | ✓ | ✓ | ✓ | Consider |
| European AIDS Clinical Society 20093 | ✓ | ✓ | Select only | Deferral |
| World Health Organization 20104 | ✓ | ✓ | ✕ | ✕ |
Abbreviation: AIDS, acquired immune deficiency syndrome.
Table 2.
What to start for HIV treatment naive patients – DHHS guidelines 2011
| Preferred regimens | EFV or | }+ TDF/FTC |
|---|---|---|
| ATV)/r or | ||
| DRV/r or | ||
| RAL | ||
| Pregnant Women: LPV/r (twice daily)++ ZDV/3TC | ||
| Alternative regimens* | EFV + ABC or ZDV/3TC | |
| NVP + ZDV/3TC | ||
| ATV/r + (ABC or ZDV)/3TC | ||
| FPV/r (once or twice daily) + either (ABC or ZDV)/3TC or TDF/FTC | ||
| Acceptable regimens | EFV + ddi + (3TC or FTC) | |
| ATV + (ABC or ZDV)/3TC | ||
| MVC + ZDV/3TC | ||
| Data lacking | MVC + either ABC/3TC or TDF/FTC | |
| RAL + (ABC/ZDV) /3TC | ||
| DRV/r or SQV/r + (ABC or ZDV)/3TC | ||
as of August 2011, rilpivirine, a second generation non-nucleoside reverse transcriptase inhibitor has now been approved for second-line use in combination ART in treatment naive patient1
Once daily LPV/r (800/200 mg) is not recommended in pregnancy. Based on published pharmacokinetic studies it is generally recommended that the dose of LPV/r be increased to 600 mg/150 mg p.o. bid for the second and third trimesters.
Abbreviations: EFV, efavirenz; ATV, atazanavir; r, ritonavir; DRV, darunavir; TDF, tenofovir; FTC, emtricitibine; RAL, raltegravir; LPV, lopinavir; ZDV, zidovudine; 3TC, laminudine; ABC, abacavir; NVP, nevirapine; FPV, fosamprenavir; TDF, tenofovir; ddi, didanosine; MVC, maraviroc; RAL, raltegravir; SQV, saquinavir; DHHS, Department of Health and Human Services.
Figure 2.
Individualized antiretroviral prescription.
Abbreviation: CNS, central nervous system; CCR5, C-C Chemokine receptor type 5; AIDS, acquired immune deficiency syndrome; HLA, human leukocyte antigen; ADME, absorption distribution, metabolism and excretion.
Finally, characteristics of the host are very important in the individualization of ART and comprise both pharmacoecologic (factors in the individual’s environment that may be dynamic over time) and pharmacogenetic factors.7 Pharmacoecologic factors that influence our day-to-day prescribing in the individual include factors relating to lifestyle and adherence of the patient, drug interactions, underlying organ dysfunction such as hepatitis or renal disease, or pregnancy. Chronic hepatitis B infection is an important consideration because specific antiretroviral drugs such as lamivudine (2′,3′-dideoxy-3′-thiacytidine), emtricitabine (FTC), and tenofovir have activity against hepatitis B.
Non-AIDS comorbidities such as vascular disease, end-organ dysfunction, and cancer are playing an increasing role in the decision to individualize therapy. Antiretroviral drugs with benign metabolic profiles, for instance, are likely to be easier to use in an aging population either with or at risk for vascular disease. A more fixed aspect of the host that relates to individualization of ART is pharmacogenetic factors that potentially affect drug efficacy and toxicity. These include genes that may affect the overall disposition and activity of a drug, such as those related to absorption, distribution, metabolism, and excretion; drug transport; or the drug target itself. These may also include immune response or human leukocyte antigen (HLA) genes that appear to be particularly important in specific types of immunologically mediated drug toxicities. The discovery of a strong association between the major histocompatiblity complex (MHC) class I allele HLA-B*5701 and abacavir hypersensitivity and the subsequent widespread utilization of this marker in routine HIV clinical practice is a major example of the translation of pharmacogenetics in the individualization of ART.8–10 Many other examples exist that have not currently been or will not be amenable to clinical translation. The remainder of this review article will focus on pharmacogenetic aspects of variation in antiretroviral drug response and the prospects for utilization of this knowledge in individualizing ART.
Pharmacogenetics of antiretroviral drugs
Pharmacokinetic and efficacy correlations
Amongst antiretroviral drugs, the NNRTIs and protease inhibitors, in particular, are associated with significant interindividual variation in drug exposure, which is largely driven by genetic differences in drug transporters, nuclear receptors, and drug-metabolizing enzymes. Although the pharmacogenetics of these drugs is becoming well established and pharmacokinetic/pharmacogenetic relationships are established for some drugs, reports directly correlating genetic differences in absorption, distribution, metabolism, and excretion and drug transporter genes with treatment efficacy and response have been inconsistent. There are currently no large published studies that have directly examined the use of pharmacogenetics to guide antiretroviral dose. As such, although some have advocated using approaches combining therapeutic drug monitoring with pharmacogenetics, pharmacogenetics of the individual patient is generally not widely used in clinical practice in an attempt to improve treatment efficacy.11
Non-nucleoside reverse transcriptase inhibitors
Nevirapine and efavirenz are still two of the most common NNRTIs in clinical use. For efavirenz, toxicity and efficacy pharmacokinetic relationships have been established.12 There is wide interindividual variability in nevirapine and efavirenz plasma exposure, explained to a great extent for efavirenz by cytochrome P450 2B6 (CYP2B6), the main isoenzyme involved in the major metabolic pathway 8-hydroxylation (and to a lesser extent in 7-hydroxylation). For the gene CYP2B6, more than 30 alleles have been identified, and several of these result in a complete or partial loss of function. Depending on the alleles present, the CYP2B6 genotype may be described as extensive, intermediate, or slow metabolizing, and in the situation where CYP2B6 is impaired, alternative metabolic genes may then have increased importance.13–17 In addition to CYP2B6, which is responsible for 77%–92% of efavirenz clearance, efavirenz is metabolized to a lesser degree by CYP3A4/5, CYP2A6, and UDP-glucuronosyl-transferase-2B7 (UGT2B7) enzymes.18 Efavirenz is also known to induce CYP2B6, CYP3A4, P-glycoprotein, and some UGT1 enzymes.
The most extensively studied single nucleotide polymorphism (SNP) in relation to both nevirapine and efavirenz metabolism is CYP2B6 516G>T, which marks the slow metabolizing haplotypes containing alleles CYP2B6*6,*7, *9, and *13. These alleles result in a pronounced decrease in CYP2B6 expression and activity, as well as a low rate of efavirenz 8-hydroxylation.19,20 The 516 T allele is found at a significantly higher frequency in Sub-Saharan Africans (45.5%) and African Americans (46.7%) as compared with Hispanic (27.3%), European (21.4%), and Asian (17.4%) populations.21–23 CYP2B6 516G>T homozygosity is significantly associated with greater plasma and intracellular exposure to efavirenz and greater plasma exposure to nevirapine across a variety of ethnicities and in both adults and children.24–37 Although many have associated CYP2B6 polymorphisms with nevirapine metabolism, concluding similar effects for both efavirenz and nevirapine metabolism,30,34 several conflicting studies are reported.38,39 In general, it appears that the effect of slow metabolizer CYP2B6 alleles on nevirapine area under the plasma drug concentration versus time curve (AUC) is less extreme.
A less frequent CYP2B6 polymorphism, 983T>C, also predicts plasma efavirenz pharmacokinetics and, in fact, improves the predictive ability of CYP2B 516G>T.25,27,40 This SNP characterizes haplotypes containing the slow metabolizing alleles CYP2B6*16 and *18. Further, slow metabolizer alleles for CYP2B6 are genetically linked to the SNPs 136A>6 (CYP2B*11A, *11B), 1172T>A (CYP2B*15A, *15B), 499C>G, 516G>T, 785A>G (CYPB*26), 593T>C (CYP2B*27), and 1132C>T (CYP2B*28).24,41,42 Furthermore, a hybrid allele, CYP2B6*29, carries two amino acid substitutions, Q172H and M198T, associated with impaired enzyme function, and carriers show increased AUC when compared with extensive metabolizer genotypes.42,43 Carriers of the allele CYPA6*17 (marked by 76A>T, 83A>G, 85C>A, 86G>C, 933C>T) show a 1.8 times higher median efavirenz concentration (P = 0.017) compared with non-carriers.15 In a study that fully resequenced CYP2B6 and genotyped 15 relevant SNPs in 169 participants, 45% carried a loss/diminished function allele. Median AUC levels were more than doubled in individuals homozygous for a loss/diminished function allele compared with carriers and noncarriers.42 Another study that characterized CYP2B6 haplotypes with three SNPs (rs10403955, rs2279345, and rs8192719) found an association between carriers of four to six of these three alleles and the risk of high (>4000 μg/L) efavirenz plasma concentrations.44
In general, it appears that slow metabolizer genotypes benefit from decreased drug doses. However, there is some evidence that such genotypes may confer some virologic benefit. In the case of efavirenz, it may be that the drug itself was approved at a higher than needed dose for most of the population. The ENCORE (Evaluation of Novel Concepts in Optimization of Antiretroviral Efficacy) study is a randomized clinical trial that is currently examining the efficacy of the standard 600 mg daily of efavirenz versus 400 mg daily, and has a genetic substudy. Reduced clearance of nevirapine with the CYP2B6-516-T/T genotype was accompanied by a beneficial increase in CD4+ T cell from baseline to week 24 in one study.31 More recently, slow efavirenz metabolizers with the 516/983 genotypes displayed decreased virologic failure in African ethnicities.25 One explanation may be that sustained high efavirenz concentrations during brief periods of nonadherence may allow continued control of HIV-1 replication. This later finding was not seen across all ethnicities and thus requires replication in other cohorts. In all, drug metabolism should be considered together with CD4+ count when considering dose adjustments.
When CYP2B6 function is impaired, the relevance of accessory metabolic pathways utilizing CYP2A6 and CYP3A4/5 in limiting drug accumulation may increase.13,14 Without consideration for CYP2B6 function, the data regarding CYP2A6 and CYP3A4/5 are conflicting. When considered in isolation, a weak association between the promoter SNP CYP3A4*B (–392A>G) and CYP3A5*3 (6986A>G) and plasma efavirenz exposure was suggested in an analysis of 154 participants.33 However, subsequent studies that have examined these SNPs and, in addition, CYP3A4*2 have found that although these splice defects reduce enzyme activity, there is no influence on the metabolism of efavirenz or nevirapine.28,30,39,45–47 Subsequent studies that have considered the dual effect of CYP2B6 have shown that the CYP2A6 slow metabolizer variant carriers (*9 or *17) and CYP3A4/A5 accessory pathways appear to influence efavirenz elimination independent of CYP2B6.13–15 The cumulative influence of CYP2B6, CYP2A6, and CYP3A4_rs4646437 allelic variants on clearance implies a critical 90% decrease of efavirenz elimination in triple-Hom-loss of function (LOF) individuals.13 A similar study examining efavirenz concentrations in 169 patients in the Swiss HIV cohort found that the effect of the CYP2A6*7, *9, and/or *17 alleles was observed only in patients with CYP2B6 slow metabolizer genotypes.14 In this group, carriers of one or two CYP2A6 diminished (DOF)/LOF alleles, when compared with CYP2A6 extensive metabolizers, showed decreased metabolites of 8-hydroxylation efavirenz and 7-hydroxylation together with increased metabolites of N-glucuronidation of efavirenz.14 These results are consistent with the role of CYP2A6 as an alternative pathway in efavirenz metabolism, and the role of N-glucuronidation in the setting of multiple DOF/LOF alleles in the main and accessory hydroxylating pathways.14
The UDP-glucuronsyltransferases (UGT1A1 and UGT2B7) are a diverse group of enzymes that transform small lipophilic molecules into water-soluble, excretable metabolites. UDP-glucuronosyltransferase (UGT) 2B7 is the main UGT isoform responsible for the efavirenz glucuronidation, which may increase in importance in the background of impaired CYP function as mentioned.14,18 Another study that examined efavirenz plasma concentration in relation to UGT1B27, CYP2B6, and CYP2A6 genotypes found UGT2B7*1a carrier status to be an independent predictor for efavirenz concentration and accounted for 10.1% of the total variance in the drug metabolism in Ghanaian patients.16
In an in vitro heterologous system, conjugation of efavirenz by UGTB27 was examined together with atazanavir metabolism, another drug also known to be glucuronidated by UGTB27. Each of the drugs was found to be inhibited by the other, and this raises another level of complexity in the genotype/phenotype relationship of drug metabolism.18 Similarly, the presence of concurrent therapies may also affect the function of key CYP enzymes in the metabolism of efavirenz. Tenofovir, a nucleotide analog reverse transcriptase inhibitor, exacerbates the effect on efavirenz AUC in LOF/DOF homozygotes.42 Rifampicin is a known inducer of the P450 enzymes CYP2B6, CYP3A, CYP2C19, CYP2C8, CYP2C9, and CYP1A2, and despite its well-known interaction potential it is commonly used in the treatment of HIV/AIDS patients coinfected with tuberculosis.48 As a CYP2B6 enzyme inducer, rifampin coadministration would be expected to enhance efavirenz clearance and reduce plasma levels. However, a recent study found that patients with CYP2B6 516T/T slow metabolizing genotype receiving concomitant antituberculous therapy have higher efavirenz plasma concentrations than those volunteers with the same genotype but not receiving antituberculous therapy.49 This paradoxical effect of antituberculous therapy may be due to increased susceptibility of the CYP2B6 516G>T variant to inhibition by one (or more) of the antituberculous drugs as compared with the reference CYP2B6 enzyme. Alternatively, efavirenz metabolism may be inhibited by effects on the non-CYP2B6 accessory pathways, leading to higher efavirenz concentrations.49 Further studies are needed to determine the mechanism.
The multidrug resistance 1 (MDR1) gene encodes an adenosine triphosphate (ATP)-binding cassette (ABC) transporter, P-glycoprotein, which is present in the intestine, liver, kidney, and blood-brain barrier and on T lymphocytes. The role of this transporter in the response to efavirenz-containing regimens is controversial, as efavirenz does not appear to be a substrate for P-glycoprotein.50 Two studies have suggested that MDR1 3435C>T predicts more favorable virologic responses to efavirenz and nelfinavir treatment with the MDR1 T/T genotype, correlating with a greater rise in CD4 cell count after 6 months and the best recovery of naive CD4 cells.45,47 Contrasting studies looking at CD4 count have reported a marginal association with MDR1 1236 but not at MDR1 2677 or MDR1 3435, or, in fact, a lower increase in CD4 cell count after efavirenz therapy initiation.26,51,52 However, others have found no correlation between MDR1 SNPs and virologic success or immunologic failure.52,53 Similarly, there are conflicting findings with the SNP MDR1 2677G>T, which is in linkage disequilibrium with MDR1 3434C>T and was originally reported to be associated with decreased efavirenz resistance.33 A supporting study found virologic failure was associated with a two-locus interaction between MDR1 2677G>T and CYP2B6 516G>T, which also correlated with higher efavirenz AUC24 values. More recently, a novel variant allele in MDR1 4036A>G was a major predictor of efavirenz plasma exposure in a healthy Ugandan population after single-dose administration. Efavirenz relative bioavailability was 26% higher in subjects homozygous for the SNP.41 Although both MDR1 (2677 G>T/A, 3435 C>T) genotypes have not been associated with any effect on nevirapine pharmacokinetics or plasma concentrations,28,30,39 nevirapine cerebrospinal fluid:plasma ratios are higher in children with the MDR1-3435-C/T or -T/T genotypes in comparison with those with the MDR1-3435-C/C genotype.31
Etravirine is a newer second-generation NNRTI that shows efficacy in patients with primary NNRTI mutations.54 It is primarily hepatically metabolized by CYP3A4, CYP2C9, and CYP2C19. It is also an inhibitor of CYP2C9 and CYP2C19 and an inducer of CYP3A4.54 Carriers of CYP2C9*3 and CYP2C19*2 have lower etravirine clearance. However, there has been no established impact on clinical outcome.55
Protease inhibitors
Although several studies have associated drug concentration/pharmacogenetic relationships for various HIV protease inhibitors, these have been inconsistent, many have not been replicated, and pharmacogenetic efficacy relationships have yet to be established. The most commonly used protease inhibitors in HIV clinical practice that are combined with ritonavir as a boosting agent are atazanavir, darunavir, and lopinavir. CYP3A4 is predominantly involved in the metabolism of all HIV protease inhibitors. Protease inhibitors are also CYP3A inhibitors, with ritonavir being the most potent, and hence it is routinely used as a booster to increase plasma exposure of other protease inhibitors that are paired with it. Protease inhibitors are also substrates for the drug transporter P-glycoprotein. The pregnane X receptor (PXR) may explain the dual influence of CYP3A4 and MDR1 on the protease inhibitors such as atazanavir. PXR expression is correlated with CYP3A4 in liver in the absence of enzyme inducers. The PXR SNP 63396C→T alters PXR expression and CYP3A4 activity in vitro. Homozygosity for the T allele for PXR 63396 is associated with higher atazanavir clearance, likely mediated through an effect on hepatic PXR expression and therefore expression of its target genes (eg, CYP3A4, solute carrier organic anion transporter family member 1B1 (SLCO1B1), and MDR1), which are known to be involved in atazanavir clearance.56,57 A recent study examining PXR, CYP3A4, and CYP2B6 polymorphisms in a cohort of 1013 Africans found that darunavir, fosamprenavir, lopinavir, nelfinavir, and tipranavir increased CYP3A4 and/or CYP2B6 promoter activity, some through constitutive receptor but mainly through PXR. Addition of low-dose ritonavir enhanced levels of CYP promoter activity for several protease inhibitors. Some PXR variants displayed lower fosamprenavir- and lopinavir-induced CYP3A4 promoter activity than the PXR reference sequence, whereas efavirenz and nelfinavir induction was unchanged. The presence of PXR polymorphisms can therefore alter the induction of CYP3A4 and CYP2B6 promoter activity, potentially adding to the unpredictable nature of antiretroviral drug interactions.58 Studies have suggested a correlation between efficacy and toxicity and lopinavir/ritonavir exposure. More recent studies have associated a polymorphism in the organic anion-transporting polypeptide 1B1 (OATP1B1) hepatic influx drug transporter gene SCLO1B1 521 T>C with increased exposure to lopinavir and a polymorphism in intron 2 (rs4149032) of SLCO1B1 with a lower lopinavir Cmin.59,60 Another pharmacokinetic/pharmacogenetic modelling study suggested that 5% of lopinavir variability could be explained by genetic factors.61
Integrase and CCR5 inhibitors
Raltegravir is the only HIV integrase inhibitor currently approved for clinical use in the treatment of naive and experienced patients. Raltegravir is metabolized by UGT1A1. Although early studies that looked at an association between the UGT1A1*28/*28 genotype and raltegravir plasma concentrations showed higher concentrations in those with the UGT1A1*28/*28 genotype, these were not thought to be clinically significant.62 Maraviroc is a CCR5 receptor antagonist that works in patients who have a CCR5 trophic HIV virus. Maraviroc is also a substrate for CYP3A4. Previous studies have shown no difference in virologic response to maraviroc-based antiretroviral according to CCR5 Δ32 genotype in 982 patients genotyped in the MOTIVATE (Maraviroc versus Optimized Therapy in Viremic Antiretroviral Treatment-experienced Patients) 1 and 2 studies.63
Pharmacogenetics of antiretroviral drug toxicity
Drug toxicities mediated through a primary immunologic mechanism
Adverse drug reactions can be clinically classified into type A reactions that are predictable based on the pharmacologic action of the drug and tend to be dose-dependent, and type B reactions that are less dependent on dose, are not predictable based on the pharmacology of the drug, and are thought to be largely genetically mediated. Although type B reactions comprise only 20% or less of all adverse drug reactions, they are often associated with a very high morbidity and mortality. Many type B reactions are also immunologically mediated. Immunologically mediated drug reactions have traditionally been categorized according to the classification of Gell and Coombs type I–IV. Most antiretroviral drugs have been associated with Gel-Coombs type IV reactions or delayed hypersensitivity. Delayed hypersensitivity reactions more commonly consist of a mild to moderate skin rash occurring >72 hours after drug initiation. More severe reactions include drug hypersensitivity reactions consisting of fever, rash, and internal organ involvement; internal organ involvement alone; and severe skin syndromes such as Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN). Antiretroviral drugs most commonly associated with delayed skin rashes, delayed drug hypersensitivity syndromes, drug-induced hypersensitivity syndrome/drug reaction with eosinophilia and systemic symptoms, and hepatitis include abacavir, NNRTIs (nevirapine, efavirenz), and fosamprenavir. More recently, associations between the MHC and drug hypersensitivity syndromes have been described for abacavir and nevirapine.
Abacavir
Abacavir is a guanosine nucleoside analog acting as an HIV reverse transcriptase inhibitor that has been approved for clinical use since 1998 in combination with other antiretroviral drugs. The major treatment-limiting toxicity of abacavir is a drug hypersensitivity syndrome, which consists of fever, malaise, gastrointestinal symptoms, and later, skin rash, typically occurring in the second week after treatment initiation in approximately 5% of those initiating the drug. Most of the morbidity and mortality of abacavir hypersensitivity occurs on rechallenge, which is contraindicated in those who have previously experienced such symptoms in association with abacavir.
First clues to a potential genetic association with abacavir hypersensitivity came with postmarketing experience showing a lower frequency of abacavir hypersensitivity in black and Asian populations.64 A case report also documented abacavir hypersensitivity in a father and daughter. In 2002, two groups independently described a strong association between the HLA class I allele, HLA-B*5701, and abacavir hypersensitivity syndrome.65 The major barrier to the early application of HLA-B*5701 as a screening test to predict and prevent abacavir hypersensitivity was the apparent less than 100% negative predicative value in these early studies and subsequent case-control studies that included non-white populations.66 It was later discovered that this imperfect and apparent less than 100% negative predictive value was not real but was driven by the high false-positive clinical diagnosis of abacavir hypersensitivity in these early studies. Highlighting this, randomized double-blind clinical trials that included abacavir also consistently showed that between 2% and 7% of patients in the nonabacavir arms were given a diagnosis of abacavir hypersensitivity.67–70 Abacavir skin patch testing was later successfully used to define the true phenotype of immunologically mediated abacavir hypersensitivity.71–73 Two major clinical trials, the PREDICT-1 (Prospective Randomized Evaluation of DNA Screening in a Clinical Trial) and SHAPE (Study of Hypersensitivity to Abacavir and Pharmacogenetic Evaluation) studies, utilized skin patch testing as immunologic confirmation of abacavir hypersensitivity in their study design.74,75 The PREDICT-1 study was a randomized double-blind controlled study, randomizing patients to receive real-time HLA-B*5701 screening to exclude abacavir in those who tested positive on initiation of abacavir and clinical monitoring with later HLA-B*5701. The study was the first of its type to study the effectiveness of a genetic test to reduce a specific drug toxicity. It enrolled almost 2000 patients of predominant Caucasian ethnicity and demonstrated that HLA-B*5701 screening eliminated immunologically confirmed abacavir hypersensitivity.74 The SHAPE study, which was a case-control study of black and white patients in the US, demonstrated that 100% of both white and black patch test-positive patients with a clinical history consistent with abacavir hypersensitivity carried HLA-B*5701.75 Collectively, the PREDICT-1 and SHAPE studies hence provided strong evidence for the clinical utility of HLA-B*5701 to prevent abacavir hypersensitivity, generalizable across race.
In addition, several observational studies have convincingly shown the benefits of HLA-B*5701 screening to eliminate both true immunologically mediated abacavir hypersensitivity and false-positive clinical diagnosis.76–82 Convergent with evidence supporting the clinical utility of HLA-B*5701 as a screening test to prevent abacavir hypersensitivity was a body of basic science that suggested HLA-B*5701 was necessary but not sufficient for the development of abacavir hypersensitivity.83,84 In addition, feasible molecular techniques with rapid turnaround times were developed that were cost-effective alternatives to sequence-based typing.85–89 An HLA-B*5701-specific quality assurance program was validated and is currently actively administered by the Asia-Pacific Histocompatibility and Immunogenetics Association.90 HLA-B*5701 testing prior to initiation of abacavir ART is currently recommended as part of major HIV treatment guidelines and represents a success story in the translation of a pharmacogenetic testing to the routine clinic setting and provides a translational roadmap in this regard.
Nevirapine
Nevirapine is an NNRTI used in combination ART that has an excellent long-term toxicity profile. The major treatment-limiting toxicity of nevirapine is a rash, hypersensitivity syndrome, and hepatitis that typically occurs within 2–8 weeks of treatment initiation in approximately 5% of those starting the drug.91 SJS/TEN are rare and life-threatening severe skin syndromes that occur in 0.3% of those starting nevirapine within 8 weeks of treatment initiation.91 Unlike abacavir hypersensitivity, which was associated with a single HLA class I allele, different class I and class II associations have been associated with nevirapine rash, hypersensitivity, and hepatitis. In 2005, a population-based study from Western Australia was the first to associate HLA-DRB1*0101 with nevirapine rash-associated hepatitis in the setting of CD4%≥25.92 Subsequently, other groups have published class I HLA association with nevirapine hypersensitivity such as HLA-B*1402 and HLA-Cw8 in a Sardinian population,93 HLA-Cw8 in a Japanese population,94 and HLA-B*3505 in association with nevirapine rash and hypersensitivity in a Thai population.95 In an updated analysis of the Western Australian cohort study, the initial association of HLA-DRB1*0101 and CD4%≥25 was again confirmed with nevirapine rash-associated hepatitis, but an additional class I association between HLA-B*3501 and nevirapine hypersensitivity with rash became apparent.96 This analysis also suggested that ethnicity and phenotype are important and, in particular, that HLA-DRB1*0101 is associated with a hepatitis phenotype in Caucasians, whereas HLA-B*35 is associated with a rash phenotype across populations.
Yuan et al97 also recently published data from the Boerhringer-Ingelheim protocol 1100.1452, which also underscored the phenotype and ethnicity specific nature of nevirapine rash, hypersensitivity, and hepatitis. This study was a case-control format that enrolled 889 patients across 76 sites comprising 276 cases and 587 controls with CD4 cell counts ≥150 cells/μL at the start of treatment. This study used only two-digit HLA typing but similarly associated HLA-DRB1*01 with hepatitis in white, HLA-B*35 and HLA-Cw*04 with rash in Asian, and HLA-Cw*04 with rash in white and black populations.97 In addition, the CYP2B6 slow metabolizer genotype 516 G>T appeared to associate with rash rather than hepatitis. Previous studies had also associated the drug transporter genetic polymorphism MDR1 3435 C>T with a decreased risk of nevirapine hepatotoxicity, and another African study associated CYP3A5 6986 A>G and MDR1 3435 C>T with increases in liver enzymes in relation to nevirapine.98–100 Another recently presented case-control analysis involved examining high-resolution HLA class I and II typing in South Africans initiating nevirapine as part of protocol FTC–302. This study associated HLA-DRB1*0102, a class I allele with sequence homology to HLA-DRB1*0101, and nevirapine-associated hepatotoxicity.101 Another study that used HLA two-digit typing showed an association between both efavirenz rash, nevirapine hypersensitivity, and HLA-DRB1*01.102 Collectively, these nevirapine studies imply that different metabolic, immunologic, and genetic factors contribute to nevirapine hypersensitivity across different populations. Although it will further our understanding of the pathogenesis of nevirapine hypersensitivity syndromes, it seems unlikely at this point in time, given the diversity of associations and their complexity across populations, that HLA or other genetic tests could be feasibly applied in the clinical setting to prevent nevirapine rash, hepatitis, and hypersensitivity. For severe skin reactions such as SJS/TEN associated with nevirapine, a specific HLA association has yet to be defined, although genetic factors appear likely, with a case report of nevirapine SJS/TEN occurring in two family members.
Drug reactions not thought to be exclusively immunologically mediated
Nucleoside/nucleotide reverse transcriptase inhibitors
Mitochondrial toxicities: peripheral neuropathy
Peripheral neuropathy in the setting of ART has been particularly associated with the so-called d drugs, including the dideoxy-NRTIs (dNRTIs [ddI, ddC]) and the thymidine analog d4T (ddC>d4T>ddI).103 The risk tends to be higher in small females, when d4T and ddI are used together, and with advanced age and HIV disease. After 1 year of d-NRTI treatment, the incidence is estimated to be 10%–25%. In comparison with HIV-associated peripheral neuropathy, the quality of pain is similar with symmetric painful dysesthesias and sensory loss. However, the onset of ART-induced peripheral neuropathy tends to be abrupt and will be partially alleviated by ceasing the offending ARTs. Mounting evidence has suggested that drug-induced mitochondrial toxicity and inflammatory damage to the axons of peripheral nerves play a large part in the pathophysiology of ART-induced peripheral neuropathy. Candidate gene approaches have hence focused on genes related to mitochondrial function, NRTI metabolism, inflammatory response, apoptosis, and other factors where genetic variation could plausibly affect tendency to peripheral neuropathy, such as nutrient and antioxidant genes. Limited information currently exists on drug metabolism of NRTIs, as intracellular metabolite that correlates with efficacy and toxicity are difficult to measure. External factors that may increase concentrations of NRTI triphosphates such as hydroxyurea and ribavirin have been shown to increase the risk of NRTI-associated peripheral neuropathy and potentially other mitochondrial toxicities. Women, who have a higher risk of ART-associated peripheral neuropathy and other mitochondrial toxicities such as lipoatrophy and lactic acidosis, have also been shown to have significantly higher intracellular NRTI triphosphate concentrations. Mitochondrial DNA (mtDNA) is classified according to specific patterns of polymorphisms called haplogroups. In earlier studies, a common European mitochondrial haplogroup, haplogroup T (7028 C>T, 10398 G>A, and 13368 G>A) was associated with neuropathy during treatment in a case-control study from the ACTG 384.104 A follow-on study showed that another variant at complex I, subunit ND2, 4917A>G, which has been associated with neurodegenerative diseases, was associated with ART-associated peripheral neuropathy in the white population.105 Iron is essential for mitochondrial function, and in subjects receiving d4T/ddI in ACTG 384, common polymorphisms in the hemochromatosis gene (HFE) 845 G>A and 187 C>G have also been associated with a decreased risk of peripheral neuropathy.106 Because HFE is linked to the class I HLA-A3 allele, it needs to be ascertained that this effect is not due to linkage disequilibrium. Adding weight to the inflammatory pathogenesis of ART-associated peripheral neuropathy have been recent studies associating cytokine polymorphisms such as tumor necrosis factor-α (TNF-α) 1031*2 and markers of the 8.1 ancestral haplotype (HLA-A1/B8/DR3) such as TNF-α308*2 and B(0,+)-type amino acid transporter 1 (BAT1)*2.107,108 Interleukin 12B (IL12B)*2 genotype was over-represented in subjects who did not develop peripheral neuropathy.108 A small study attempted to combine patient demographics with these cytokine polymorphisms into a predictive model for peripheral neuropathy.109
Mitochondrial toxicities: lipoatrophy
Lipoatrophy is a largely irreversible mitochondrial toxicity that occurs largely in association with the thymidine analogs d4T>AZT. Like other mitochondrial toxicities, it is postulated that the toxicity is mediated by NRTI inhibition of DNA polymerase-γ in the host, which leads to depletion of mtDNA. In relation to other mitochondrial toxicities, a genetic variant of DNA polymerase-γ relating to 14% of functional activity was reported in a Thai patient with lactic acidosis related to d4T.110 With regards to lipoatrophy, risk is related to duration of treatment and is higher in females of lower body mass, suggesting a dose relationship. Previous fat biopsy studies have substantiated the early depletion of mtDNA preceding clinical disease and have highlighted the impressive active inflammatory response present in the fat in this disease. A promoter polymorphism in the TNF-α gene (TNF-238 A) has been associated with more rapid progression of disease.111,112 Mitochondrial haplogroup H was associated with increased thymidine analog-associated limb lipoatrophy and the HFE 187 C>G with less peripheral lipoatrophy.113,114 In another study, polymorphisms in IL-1β 3954 C>T were also associated with protection against lipoatrophy associated with thymidine analogs.115 In a separate study from ACTG 5005s and 384, an SNP in the resistin gene 7640369 C>T was associated positively with lipoatrophy.116 More recently, in a Thai case-control study, HLA-B*4001 was associated with moderate to severe lipoatrophy, defined by anthropomorphic measurements, questionnaire, and dual-emission x-ray absorptiometry scan, with a negative predictive value of 54%.117 These results would need to be verified. In particular, it ascertained that this was not due to linkage disequilibrium. In contemporary HIV practice in the developed world, new cases of lipoatrophy are being avoided through prevention and lack of use of thymidine analogs in first-line therapy. Challenges still exist in the developing world, where the demographics may independently put patients at risk for thymidine analog-associated mitochondrial toxicities. In addition, there is a significant burden of patients with past exposure to thymidine analogs.
Renal toxicity
Tenofovir disoproxil fumarate is the orally bioavailable prodrug of tenofovir that is an acyclic nucleotide reverse transcriptase inhibitor that is available in fixed-dose combination with emtricitibine and efavirenz. Tenofovir is eliminated unchanged in the kidney and has been associated with various presentations of nephrotoxicity, including proximal renal tubular dysfunction with and without decreased renal function. Although serious events occur in <1% of patients, approximately 2% will develop a significant increase in creatinine.118,119 Demographic factors that have been associated with tenofovir nephrotoxicity have included lower body weight, pre-existing renal dysfunction, medical comorbidities, concurrent nephrotoxic medications, advanced HIV, and advanced age. Some studies have also suggested a higher risk of developing renal impairment when tenofovir is used in combination with ritonavir-boosted protease inhibitors.120 Currently, the exact mechanism by which tenofovir causes renal toxicity is unclear and has been postulated to be mitochondrial toxicity similar to the related nucleotide analogs adefovir and cidofovir or direct interference with tubular function. Around 20%–30% of tenofovir is actively transported into renal proximal tubular cells by organic anion transporters hOAT1 (encoded by the solute carrier family 22 [SLC22] A6 gene) and OAT3. The membrane transporter multidrug resistance-associated protein (MRP) 4 encoded by the ATP-binding cassette sub-family C (ABC) C4 gene is primarily responsible for secreting the drug into the tubular lumen and into the urine, and the ABCC4 3463 variant has been associated with intracellular concentrations 35% higher than wildtype. Although MRP2 has not been shown to have a clear role in tenofovir excretion, two studies have shown associations between polymorphisms in the ABCC2 gene, which encodes MRP2 and tenofovir proximal tubulopathy, and the exact mechanism of this remains to be determined.121,122 One case-control study associated a specific CATC haplotype (polymorphisms-24, 1249, 3563, and 3972) in ABCC2 and proximal tubular disease, whereas another cross-sectional study, which used a different definition of renal toxicity, associated tenofovir tubular damage with the ABBC2-24C allele but not the CATC high-risk haplotype. Despite the fact that MRP4 is the primary transporter involved in the excretion of tenofovir, studies of genetic variation in association with tenofovir renal toxicity have been inconsistent, with one study suggesting an association with the polymorphism ABCC4 669 C>T in those with tenofovir renal damage but other studies looking at this and a larger number of SNPs not confirming this association.123 Polymorphisms in other transporter genes such as SLC22A6 and P-glycoprotein (coded by the ABCB1 gene) have shown no association with tenofovir clearance, levels, or renal damage. Recent evidence has suggested a role for ABCC10 (MRP7), a renal efflux transporter functionally similar to ABCC2, in the transport of tenofovir. Two ABCC10 SNPs (rs9349256 and rs2125739) and their haplotype were more commonly represented in 19 cases with tenofovir renal disease versus 96 tenofovir-tolerant controls without renal disease.124 Collectively, these findings are too preliminary to be used predictively in the clinic setting, and more studies using standardized definitions of renal toxicity and looking at generalizability across different populations will be needed to further clarify the pharmacogenetics of tenofovir toxicity.
Protease inhibitors
Unconjugated hyperbilirubinemia
Atazanavir and indinavir have commonly been associated with unconjugated hyperbilirubinemia due to inhibition of bilirubin binding to uridine diphosphate gluronosyl-transferase 1A1 (UGT1A1), and hence the inhibition and clearance of bilirubin.125 In view of dosing and toxicity considerations, indinavir is not commonly used in clinical practice. For atazanavir, it has been shown that there is a direct correlation between unconjugated hyperbilirubinemia and atazanavir plasma levels.126,127 Patients who have received indinavir and atazanavir are more likely to develop unconjugated hyperbilirubinemia if they have Gilbert syndrome, a genetic defect resulting from the promoter polymorphism in the TAT region of UGT1A1 (UGT1A1*28), which is independently associated with reduced bilirubin-conjugating activity.128 Polymorphisms in this promoter region are most important in the unconjugated hyperbilirubinemia associated with atazanavir in Caucasians. However, other polymorphisms such as the UGT1A1*6 (211 G>A) may be more important in the Asian population.129–131 Studies from clinical trials and clinical practice have documented that between 20% and 50% of patients exposed to atazanavir will develop unconjugated hyperbilirubinemia, of which up to 6% will be clinically apparent. Efavirenz has been shown to reduce unconjugated hyperbilirubinemia by its inducing effect on UGT1A1.132 Atazanavir plasma levels may also be affected by the P-glycoprotein drug transporter gene MDR1, with patients with the MDR1 3435 C>T polymorphism showing higher atazanavir plasma levels.133 Although unconjugated hyperbilirubinemia has no direct clinical consequences in adults, the effects of clinical jaundice may be cosmetically disturbing, and some studies have suggested a correlation between genetics and early treatment discontinuation of atazanavir in treatment-naive patients.134 There are currently, however, no studies that directly support the use of genetic screening in the routine clinical use of atazanavir.
Metabolic disturbances
Candidate gene studies in HIV-infected patients have focused on genetic variation in apolipoprotein E (APOE) and the gene cluster APOA1/C3/A4/A5 due to associations between polymorphisms in this group with hyperlipidemia in the general population. Some of these same SNPs have been examined in HIV-infected populations to add insights into the contribution of drugs and genetics to the dyslipidemia that is commonly seen in this population. Polymorphisms in APOA5 have been implicated, in that individuals not carrying the APOA5 *1/*1 haplotype (113IT>C, 64 G>C) have been shown to be at risk for low high-density lipoprotein cholesterol and hypertriglyceridemia in association with ritonavir.135,136 In this Swiss cohort predominantly Caucasian study, which longitudinally followed 438 HIV-infected patients for a median of 4.8 years across different ART regimens, SNPs in ABCA1, APOC3, APOE, APOA5, and cholesteryl ester transfer protein (CETP) aversely affected plasma triglyceride, and APOA5 and CETP were associated with low high-density lipoprotein cholesterol. The Swiss cohort group further validated the potential contribution of 42 SNPs, 33 of which had been reported in HIV-negative genome-wide association studies to dyslipidemia in 745 HIV-infected subjects.136 Taiwanese patients carrying APOA5 553 G>T had a higher risk of protease inhibitor-associated hypertriglyceridemia.137 Carriage of APOE ε4 has been associated with hypertriglyceridemia in HIV-positive patients not taking ART.138 Carriers of other variants of APOE also appear to be at risk for ritonavir-associated hypertriglyceridemia, and this risk appears to be enhanced in association with APOC3 variants.139,140 Collectively, these findings indicate that many genetic factors common to the general population are associated with hyperlipidemia in HIV and particularly during protease inhibitor therapy. Larger studies are needed to investigate associations between these gene variants, ART, and other vascular and metabolic events in HIV-infected individuals.
Non-nucleoside reverse transcriptase inhibitors
Neuropsychiatric disturbances
Up to two-thirds or more of patients initiating efavirenz will experience some degree of CNS side effects, which can be characterized by the full spectrum of minor sleep disturbances and vivid dreams through to major depression and anxiety. Most patients report amelioration of these symptoms by 2–4 weeks of treatment. However, up to 10% of patients have persistent symptoms that lead to efavirenz discontinuation.141 Not all studies have shown an association between efavirenz plasma levels and neuropsychiatric side effects.142 Many studies have confirmed associations between CYP2B6 variants, including CYP2B6 516 G>T and efavirenz exposure, and some have also suggested an association between CYP2B6 variants and early CNS toxicity.16,20,21,24–44 A recent study suggested an increased association with plasma efavirenz exposure and CNS events in white patients and a composite CYP2B6 516 G>T and 983 T>C genotype.25 This same genotype was associated with decreased virologic failure in black patients. A recent study also associated efavirenz plasma concentrations with a polymorphism 540 C>T in the constitutive androstone receptor (CAR), which appears to be important in the regulation of CYP2B6.143 A recent cohort study of patients who had initiated efavirenz therapy showed the CAR C/C genotype and the CYP2B6 516 T/T genotype to be independently associated with efavirenz early treatment discontinuation, with patients who carried both polymorphisms having a higher rate of discontinuation.143 More recent studies indicated that a high proportion of patients with at-risk genotypes have both high efavirenz exposure and CNS side effects, suggesting that combined genotyping-therapeutic drug monitoring (TDM) approaches may be useful to guide efavirenz therapy, particularly in higher-risk populations.11 Gatanaga et al24 looked at approaches combining genotyping and TDM. They showed consistently high efavirenz exposure in the CYP2B6 516 G>T variants *6/*6 and *6/*26. With daily dose reductions to 400 mg (n = 11) and 200 mg (n = 7), they were able to maintain virologic suppression and improve CNS side effects in >70% of patients.
Implications for clinical use: patient considerations
With a number of different combination regimens available for treatment of the HIV patient, individualized approaches to therapy have become both feasible and necessary in many cases. Individualized approaches to ART leading to the personalized antiretroviral prescription are driven not only by host factors but also by specific attributes of the virus and drugs being prescribed (Figure 2). Host factors that are increasingly important in driving a personalized approach to HIV therapy include pharmacoecologic factors relevant to individual patients such as lifestyle and adherence issues, food and drug-drug interactions, pregnancy, and underlying organ dysfunction, as well as hepatitis B/C coinfection (Figure 2). The increasing burden of non-AIDS morbidities, trends to initiate ART earlier, and the long period of time an individual patient would ideally spend on a single regimen also drive the need for individualized approaches to ART to ensure long-term success. Early approaches to individualization of ART included TDM, which was particularly pertinent to early protease inhibitor-based regimens because of the high interindividual variability of drug levels and the utility of increasing drug exposure in treatment-experienced patients. Although early studies of TDM showed promise, TDM of protease inhibitors was not shown to be beneficial in one randomized study of treatment-experienced patients (ACTG5146), where a normalized ratio of the protease inhibitor concentration in relation to its 50% inhibitory concentration (IC50) was used to dose adjust patients and the primary endpoint was virologic response at 20 weeks.144,145 However, it was noted in this study that TDM appeared to be more beneficial in patients with less viral resistance, suggesting that the specific population selected may have been important, and that TDM may benefit select populations of treatment-experienced patients.145 Nonetheless, measuring drug concentrations has remained a useful tool in clinical practice for individualizing patient therapy, particularly in cases of complex drug–drug interactions, in verifying patient compliance, and in other states, such as later stages of pregnancy where pharmacokinetics of certain protease inhibitors such as lopinavir/ritonavir may be altered and exposure reduced.146,147 Although there has been an explosion of pharmacogenetic/pharmacogenomic literature relevant to the field of HIV, it is anticipated that most of this knowledge will not be used to develop specific pharmacogenetic tests that are relevant to the treatment of the individual patient. In view of this continued influx of new literature, the website http://www.hiv-pharmacogenomics.org/ represents a useful reference for the reader, where tables of the pharmacogenetics of ART with reference to the pharmacokinetics, pharmacodynamics, reproducibility, and clinical relevance of pharmacogenetic associations are regularly updated.148
Although few pharmacogenetic associations will be directly translated into clinical practice, such knowledge has significantly furthered our understanding of the pathogenesis of host virus interactions, drug disposition, drug interactions, drug efficacy, and toxicity. Such knowledge also has future implications, such as to aid in the development of new drug targets and identify ways for screening drugs and drug candidates for toxicity in the premarketing period of drug development. Several steps are required to bring a pharmacogenetic test from discovery through to translation as a pharmacogenetic test in clinical practice.149 Factors influencing the success of translation include encompassing the right combination of characteristics of the test, the drug, the drug toxicity, the availability, or the ability to generate high levels of evidence and pharmacoeconomic data to support the test and having the right clinical and laboratory environment and infrastructure, including appropriate quality assurance programs. Robust partnerships between academia and pharma are also helpful in this process.
Finally, the actual pharmacogenetic test report needs to be easy to interpret and implemented by clinicians, and it must be readily understood and acted upon by both the healthcare system and patients. HLA-B*5701 testing for abacavir is one example of a test that has translated from discovery into a guideline-based routine HIV clinical practice, and even with the aforementioned factors being put into place and a simple and interpretable reporting scheme, this test took approximately 6 years from discovery to clinical implementation. Given that drug toxicities are still the most common reason why treatment-naive HIV patients discontinue treatment in the first year of therapy, it seems logical that pharmacogenetics could help identify the right drug regimen for the right patient. A recent observational cohort study that examined the association of pharmacogenetics with the time to treatment discontinuation during the first year of ART found that discontinuation rates were significantly higher for efavirenz- and atazanavir-based regimens in patients carrying high-risk genetic markers that had been previously shown to correlate with toxicity or pharmacokinetics of these drugs.134
Although it is intriguing that pharmacogenetics could inform more intelligent antiretroviral prescribing, other than for HLA-B*5701 and abacavir, prospective approaches to treatment using pharmacogenetic information to initiate ART have not been studied. Cost-effectiveness is uncertain, and the clinical, laboratory, and reporting infrastructure to implement such programs would be currently lacking at most centers. IL28B is a gene encoding for interferon-λ, a cytokine with antiviral activity. Three independent genome-wide association studies published in 2009 associated variation around the IL28B gene with treatment response to pegylated interferon and ribavirin for genotype 1 hepatitis C infection.150–152 HIV/hepatitis C virus coinfected patients with genotype 1 virus typically have a poor treatment response to pegylated interferon and ribavirin therapy. However, IL28B genotype also appears to affect treatment response in this population, even in those who have failed prior treatment, and may be useful to identify patients for whom early treatment would be warranted.153,154
Conclusions and future perspectives
Individualization of ART represents an important component of day-to-day HIV practice, given the diversity of drugs available, viral factors, and increasing diversity in HIV treatment populations. Over the last 5–10 years, a vast amount of published literature has been generated in the fields of HIV and antiretroviral pharmacogenetics and pharmacogenomics. Most of these discoveries, although not readily applicable to the clinic, will continue to guide our understanding of disease processes and antiretroviral efficacy and toxicity. Host pharmacogenetics currently play a minor role in our approach to individualization of drug therapy, although HLA-B*5701 screening for abacavir hypersensitivity represents an unusual example of a test that has been successfully translated from discovery to widespread clinical practice, and provides a general translational roadmap for other pharmacogenetics tests.
Footnotes
Disclosure
EP is an advisor and has received funding from Viiv, Janssen, Merck, and Gilead Australia.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services. 2011:1–66. Available from: http://www.aidsinfo.nih.gov/ContentFiles/Adultand-AdolescentGL.pdf. Accessed September 4, 2011. [Google Scholar]
- 2.Thompson M, Aberg J, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA. 2010;304(3):321–333. doi: 10.1001/jama.2010.1004. [DOI] [PubMed] [Google Scholar]
- 3.European AIDS Clinical Society Guidelines . 2009. Clinical Management and treatment of HIV-infected Adults in Europe. Version 5–4. Available from: www.eacs.eu. Accessed October 17, 2011. [Google Scholar]
- 4.World Health Organization Antiretroviral therapy for HIV infection in adults and adolescents. Recommendations for a public health approach: 2010 Revision. Available from: http://www.who.int/hiv/pub/arv/adult2010/en/index.html. Accessed October 17, 2011. [PubMed]
- 5.Swenson MC, Moores A, Low AJ, et al. Improved detection of CXCR4-using HIV by V3 genotyping: application of population based and “deep” sequencing to plasma RNA and proviral DNA. J Acquir Immune Defic Syndr. 2010;54(5):506–510. doi: 10.1097/QAI.0b013e3181d0558f. [DOI] [PubMed] [Google Scholar]
- 6.Parra J, Portilla J, Pulido F, et al. Clinical utility of maraviroc. Clin Drug Investig. 2011;31(8):527–542. doi: 10.2165/11590700-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 7.Phillips E, Mallal S. Personalizing antiretroviral therapy: is it a reality? Personalized Medicine. 2009;6(4):393–408. doi: 10.2217/pme.09.12. [DOI] [PubMed] [Google Scholar]
- 8.Mallal S, Nolan D, Witt C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359(9308):727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 9.Hetherington S, Hughes AR, Mosteller M, et al. Genetic variation in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359(9312):1121–1122. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]
- 10.Phillips E, Mallal S. Successful translation of pharmacogenetics into the clinic: the abacavir example. Mol Diagn Ther. 2009;13(1):1–9. doi: 10.1007/BF03256308. [DOI] [PubMed] [Google Scholar]
- 11.Figueroa SC, de Gatta MF, Garcia LH, et al. The convergence of therapeutic drug monitoring and pharmacogenetic testing to optimize efavirenz therapy. Ther Drug Monit. 2010;32(5):579–585. doi: 10.1097/FTD.0b013e3181f0634c. [DOI] [PubMed] [Google Scholar]
- 12.Marzolini C, Telenti A, Decosterd LA, Greub G, Bioliaz J, Buclin T. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1 infected patients. AIDS. 2001;15(1):71–75. doi: 10.1097/00002030-200101050-00011. [DOI] [PubMed] [Google Scholar]
- 13.Arab-alameddine M, Di Iulio J, Buclin T, et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1 infected individuals. Clin Pharmcol Ther. 2009;85(5):485–494. doi: 10.1038/clpt.2008.271. [DOI] [PubMed] [Google Scholar]
- 14.Di Iulio J, Fayet A, Arab-Alameddine M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogen Genomics. 2009;19(4):300–309. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 15.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. CYP2B6(c.516 G-->T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67(4):427–436. doi: 10.1111/j.1365-2125.2009.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS. 2009;23(16):2101–2106. doi: 10.1097/QAD.0b013e3283319908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogburn ET, Jones DR, Masters AR, Xu C, Guo Y, Desta Z. Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 2A6 as the principal catalyst of efavirenz 7-hydroxylaiton. Drug Metab Dispos. 2010;38(7):1218–1229. doi: 10.1124/dmd.109.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belanger AS, Caron P, Harvey M, Zimmerman PA, Mehlotra RK, Guillemette C. Glururonidation of the antiretroviral drug efavirenz by UGT2B7 and an in vitro investigation of drug-drug interaction with zidovudine. Drug Metab Dispos. 2009;37(9):1793–1796. doi: 10.1124/dmd.109.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desta A, Saussele T, Ward B, et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics. 2007;8(6):547–558. doi: 10.2217/14622416.8.6.547. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, Hollenberg PF. Polymorphic variants of Cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome P450 reductase complex. J Pharmacol Exp Ther. 2001;338(3):803–809. doi: 10.1124/jpet.111.183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyakutira C, Roshammar D, Chigutsa E, et al. High prevalence of the CYP2B6 516G-->T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol. 2008;64(4):357–365. doi: 10.1007/s00228-007-0412-3. [DOI] [PubMed] [Google Scholar]
- 22.Mehlotra RK, Bockarie MJ, Zimmerman PA. CYP2B6 983T>C polymorphism is prevalent in West Africa but absent in Papua New Guinea: implications for HIV/AIDS treatment. Br J Clin Pharmacol. 2007;64(3):391–395. doi: 10.1111/j.1365-2125.2007.02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakhmanina NY, van den Anker JN. Efavirenz in the therapy of HIV infection. Expert Opin Metab Drug Metab Toxicol. 2010;6(1):95–103. doi: 10.1517/17425250903483207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatanaga H, Hayashida T, Tsuchiya K, et al. Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6*6 and *26. Clin Infect Dis. 2007;45(9):1230–1237. doi: 10.1086/522175. [DOI] [PubMed] [Google Scholar]
- 25.Ribaudo HJ, Liu H, Schwab M, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202(5):717–722. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elens L, Vandercam B, Yombi JC, Lison D, Wallemacq P, Haufroid V. Influence of host genetic factors on efavirenz plasma and intracellular pharmacokinetics in HIV-1 infected patients. Pharmacogenomics. 2010;11(9):1223–1234. doi: 10.2217/pgs.10.94. [DOI] [PubMed] [Google Scholar]
- 27.Wyen C, Hendra H, Vogel M, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61(4):914–918. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penzak SR, Kabuye G, Mugyenyi P, et al. Cytochrome P450 2B6 (CYP2B6) G516T influences nevirapine plasma concentrations in HIV-infected patients in Uganda. HIV Med. 2007;8(2):86–91. doi: 10.1111/j.1468-1293.2007.00432.x. [DOI] [PubMed] [Google Scholar]
- 29.Leger P, Dillingham R, Beauhamais CA, et al. CYP2B6 variants and plasma efavirenz concentrations during antiretroviral therapy in Port-au- Prince, Haiti. J Infect Dis. 2009;200(6):955–964. doi: 10.1086/605126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahungu T, Smith C, Turner F, et al. Cytochrome P450 2B6 516G-->T is associated with plasma concentrations of nevirapine at both 200 mg twice daily and 400 mg once daily in an ethnically diverse population. HIV Med. 2009;10(5):310–317. doi: 10.1111/j.1468-1293.2008.00689.x. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh A, Sarles E, Capparelli E, et al. CYP2B6 genetic variants are associated with nevirapine pharmacokinetics and clinical response in HIV-1-infected children. AIDS. 2007;21(16):2191–2199. doi: 10.1097/QAD.0b013e3282ef9695. [DOI] [PubMed] [Google Scholar]
- 32.Ribaudo HJ, Haas DW, Tierney C, et al. Pharmacogenetics of plasma efavirenz exposure after treatment discontinuation: an Adult AIDS Clinical Trials Group Study. Clin Infect Dis. 2006;42(3):401–407. doi: 10.1086/499364. [DOI] [PubMed] [Google Scholar]
- 33.Haas DW, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous sytem side effects: an Adult AIDS Clinical Trials Group Study. AIDS. 2004;18(18):2391–4000. [PubMed] [Google Scholar]
- 34.Ramachandran G, Hemanth Kumar AK, Rajasekaran S, et al. CYP2B6 G516T polymorphism but not rifampin coadministration influences steady-state pharmacokinetics of efavirenz in human immunodefiency virus-infected patients in South India. Antimicrob Agents Chemother. 2009;53(3):863–868. doi: 10.1128/AAC.00899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotger M, Colombo S, Furrer H, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogent Genomics. 2005;15(1):1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Ramachandran G, Ramesh K, Hemanth Kumar AK, et al. Association of high T allele frequence of CYP2B6 G516T polymorphism among ethnic south Indian HIV-infected patients with elevated plasma efavirenz and nevirapine. J Antimicrob Chemother. 2009;63(4):841–843. doi: 10.1093/jac/dkp033. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya K, Gatanaga H, Tachikawa N, et al. Homozygous CYP2B6*6(Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319(4):1322–1326. doi: 10.1016/j.bbrc.2004.05.116. [DOI] [PubMed] [Google Scholar]
- 38.Haas DW, Gebretsadik T, Mayo G, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis. 2009;199(6):872–880. doi: 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Sun J, Ma Q, et al. CYP2B6 polymorphism and nonnucleoside reverse transcriptase inhibitor plasma concentrations in Chinese HIV-infected patients. Ther Drug Monit. 2010;32(5):573–578. doi: 10.1097/FTD.0b013e3181ea953c. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Sonnerborg A, Rane A, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenetic Genomics. 2006;16(3):191–198. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 41.Mukonzo JK, Roshammar D, Waako P, et al. A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br J Clin Pharmacol. 2009;68(5):690–696. doi: 10.1111/j.1365-2125.2009.03516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rotger M, Tegude H, Colombo S, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81(4):557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 43.Rotger M, Saumoy M, Zhang K, et al. Partial deletion of CYP2B6 owing to unequal crossover with CYP2B7. Pharmacogenet Genomics. 2007;17(10):885–890. doi: 10.1097/FPC.0b013e3282ef5cd1. [DOI] [PubMed] [Google Scholar]
- 44.Carr DF, la Porte CJ, Mirmohamed M, Owen A, Cortes CP. Haplotype structure of CYP2B6 and association with plasma efavirenz concentrations in a Chilean HIV cohort. J Antimicrob Chemother. 2010;65(9):1889–1893. doi: 10.1093/jac/dkq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas DW, Smeaton LM, Shafer RW, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult Aids Clinical Trials Group Study. J Infect Dis. 2005;192(11):1931–1942. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 46.Motsinger AA, Ritchie MD, Shafer RW, et al. Multilocus genetic interactions and response to efavirenz-containing regimens: an adult AIDS clinical trials group study. Pharmacogent Genomics. 2006;16(11):837–845. doi: 10.1097/01.fpc.0000230413.97596.fa. [DOI] [PubMed] [Google Scholar]
- 47.Fellay J, Marzolini C, Meaden ER, et al. Response to antiretroviral treatment in HIV-1 infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet. 2002;259(9300):30–36. doi: 10.1016/S0140-6736(02)07276-8. [DOI] [PubMed] [Google Scholar]
- 48.Rekic D, Roshammar D, Mukonzo J, Ashton M. In silico prediction of efavirenz and rifampicin drug-drug interaction considering weight and CYP2B6 phenotype. Br J Clin Pharmacol. 2011;71(4):536–543. doi: 10.1111/j.1365-2125.2010.03883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawara A, Lartey M, Sagoe KW, Court MH. Paradoxically elevated efavirenz concentrations in HIV/tuberculosis co-infected patients with CYP2B6 516TT genotype on rifampin-containing antituberculous therapy. AIDS. 2011;25(3):388–390. doi: 10.1097/QAD.0b013e3283427e05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marzolini C, Paus E, Buclin T, Kim RB. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther. 2004;75(1):13–33. doi: 10.1016/j.clpt.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Zhu D, Taguchi-Nakamura H, Goto M, et al. Influence of single-nucleotide polymorphisms in the multidrug resistance-1 gene on the cellular exposure of nelfinavir and its clinical implication for highly active antiretroviral therapy. Antivir Ther. 2004;9(6):929–935. [PubMed] [Google Scholar]
- 52.Winzer R, Langmann P, Zilly M, et al. No influence of the P-glycoprotein genotype (MDR1 C3435T) on plasma levels of lopinavir and efavirenz during antiretroviral treatment. Eur J Med Res. 2003;8(12):531–534. [PubMed] [Google Scholar]
- 53.Brumme ZL, Dong WW, Chan KJ, et al. Influence of polymorphisms within the CX3CR1 and MDR-1 genes on initial antiretroviral therapy response. AIDS. 2003;17(2):201–208. doi: 10.1097/00002030-200301240-00010. [DOI] [PubMed] [Google Scholar]
- 54.Adams J, Patel N, Mankaryous N, et al. Nonnucleoside reverse transcriptase inhibitor resistance and the role of the second-generation agents. Ann Pharmacother. 2010;44:157–165. doi: 10.1345/aph.1M359. [DOI] [PubMed] [Google Scholar]
- 55.Lubomirov R, Rotger M, Fayet A, et al. Pharmacogenetics-based population pharmacokinetic analysis of etravirine in HIV-1 infected individuals [abstract]. Proceedings of the 18th Conference on Retroviruses and Opportunistic Infections; February 27–March 2, 2011; Boston, MA. 2011. p. 479. [Google Scholar]
- 56.Siccardi M, D'Avolio A, Baietto L, et al. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR 63396 C>T) with reduced concentrations of unboosted atazanavir. Clin Infect Dis. 2008;47(9):1222–1225. doi: 10.1086/592304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shipani A, Siccardi M, D'Avolio A, et al. Population pharmacokinetic modelling of the association between 63396 C>T pregnane X receptor polymorphism and unboosted atazanavir clearance. Antimicrob Agents Chemother. 2010;54(12):5242–5250. doi: 10.1128/AAC.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Svard J, Spiers JP, Mulcahy F, Hennessy M. Nuclear Receptor-Mediated Induction of CYP450 by Antiretrovirals: Functional Consequences of N1I2(PXR) Polymorphisms and Differential Prevalence in Whites and Sub-Saharan Africans. J Acquir Immune Defic Syndr. 2010;55(5):536–549. doi: 10.1097/QAI.0b013e3181f52f0c. [DOI] [PubMed] [Google Scholar]
- 59.Hartkoorn RC, Kwan WS, Shallcross V, et al. HIV protease inhibitors are substrates for OATP1A2, OATP1B1 and OATP1B3 and lopinavir plasma concentrations are influenced by SLCO1B1 polymorphisms. Pharmacogenet Genomics. 2010;20(2):112–120. doi: 10.1097/FPC.0b013e328335b02d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohlrausch FB, de Cassia Estrela R, Barroso PF, Suarez-Kurtz G. The impact of SLCO1B1 polymorphisms on the plasma concentrations of lopinavir and ritonavir in HIV-infected men. Br J Clin Pharmacol. 2010;69(1):95–98. doi: 10.1111/j.1365-2125.2009.03551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lubomirov R, di Iulio J, Fayet A, et al. ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir co-formulated with ritonavir. Pharmacogenet Genomics. 2010;20(4):217–230. doi: 10.1097/FPC.0b013e328336eee4. [DOI] [PubMed] [Google Scholar]
- 62.Wenning LA, Petry AS, Kost JT, et al. Pharmacokinetics of raltegravir in individuals with UGT1A1 polymorphisms. Clin Pharmacol Ther. 2009;85(6):623–627. doi: 10.1038/clpt.2009.12. [DOI] [PubMed] [Google Scholar]
- 63.Fakenheuer G, Nelson M, Lazzarin A, et al. Subgroup analysis of maraviroc in previously treated R5 HIV-1 infection. N Eng J Med. 2008;359(14):1442–1455. doi: 10.1056/NEJMoa0803154. [DOI] [PubMed] [Google Scholar]
- 64.Symonds W, Cutrell A, Edwards M, et al. Risk factor analysis of hypersensitivity reactions to abacavir. Clin Ther. 2002;24(4):565–573. doi: 10.1016/s0149-2918(02)85132-3. [DOI] [PubMed] [Google Scholar]
- 65.Peyieere H, Nicolas J, Siffert M, et al. Hypersensitivity related to abacavir in two members of a family. Ann Pharmacother. 2001;35(10):1291–1292. doi: 10.1345/aph.1A022. [DOI] [PubMed] [Google Scholar]
- 66.Hughes AR, Mosteller N, Bansal AT, et al. Association of genetic variation in HLA-B region with hypersensitivity to abacavir in some, but not all, populations. Pharmacogenomics. 2004;5(2):203–211. doi: 10.1517/phgs.5.2.203.27481. [DOI] [PubMed] [Google Scholar]
- 67.Dejesus E, Herreera G, Teofilo E, et al. Abacavir versus zidovudine combined with lamivudine and efavirenz for the treatment of antiretroviral-naive HIV-infected adults. Clin Infect Dis. 2004;39(7):1038–1046. doi: 10.1086/424009. [DOI] [PubMed] [Google Scholar]
- 68.Guilick R, Ribaudo H, Shikuma CM, et al. Three-versus four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA. 2006;296(7):769–781. doi: 10.1001/jama.296.7.769. [DOI] [PubMed] [Google Scholar]
- 69.Dart Trial Team Twenty-four week safety and tolerability of nevirapine vs abacavir in combination with zidovudine/lamivudine as first-line antiretroviral therapy: a randomized double-blind trial (NORA) Trop Med Int Health. 2008;13(1):6–16. doi: 10.1111/j.1365-3156.2007.01973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hernandez J, Cutrell A, Bonny T, et al. Diagnosis of abacavir hypersensitivity reactions among patients receiving abacavir in two blinded studies. Antivir Ther. 2003;8:L88. [Google Scholar]
- 71.Phillips EJ, Sullivan JR, Knowles R, Shear NH. Utility of patch testing in patients with hypersensitivity reaction associated with abacavir. AIDS. 2002;16(16):2223–2225. doi: 10.1097/00002030-200211080-00017. [DOI] [PubMed] [Google Scholar]
- 72.Phillips EJ, Wong GA, Kaul R, et al. Clinical and immunogenetic correlates of abacavir hypersensitivity. AIDS. 2005;19(9):979–981. doi: 10.1097/01.aids.0000171414.99409.fb. [DOI] [PubMed] [Google Scholar]
- 73.Shear NH, Milipied B, Bruynzeel DP, Phillips E. A review of drug patch testing and implications for HIV clinicians. AIDS. 2008;22(9):999–1007. doi: 10.1097/QAD.0b013e3282f7cb60. [DOI] [PubMed] [Google Scholar]
- 74.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701screening for hypersensitivity to abacavir. N Engl J Med. 2008;358(6):568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 75.Saag M, Balu R, Phillips E, et al. High sensitivity of human leukocyte antigen-B*5701 as a marker for immunogenetically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis. 2008;46(7):1111–1118. doi: 10.1086/529382. [DOI] [PubMed] [Google Scholar]
- 76.Rauch A, Nolan D, Martin A, et al. Prospective genetic screening decreases the incidence of abacavir hypersensitivity reactions in the Western Australian HIV cohort study. Clin Infect Dis. 2006;43(1):99–102. doi: 10.1086/504874. [DOI] [PubMed] [Google Scholar]
- 77.Zucman D, Truchis P, Majerholc C, et al. Prospective genetic screening for human leuckocyte antigen-B*5701 avoid abacavir hypersensitivity in the ethnically mixed French HIV population. J Acquir Immune Defic Syndr. 2007;45(1):1–3. doi: 10.1097/QAI.0b013e318046ea31. [DOI] [PubMed] [Google Scholar]
- 78.Reeves I, Churchill D, Fisher M. Screening for HLA-B*5701 reduces the frequency of abacavir hypersensitivity reactions. Antiviral Ther. 2006;11:L11. [Google Scholar]
- 79.Trottier B, Thomas R, Nguyen VK, Machouf N. How effectively HLA screening can reduce the early discontinuation of abacavir in real life [abstract]. Presented at the International AIDS Society Meeting; July 22–25, 2007; Sydney, Australia. 2007. MOPEB002. [Google Scholar]
- 80.Lalonde RG, Thomas R, Rachlis A, et al. Successful implementation of a national HLA-B*5701 genetic testing service in Canada. Tissue Antigens. 2009;75(1):12–18. doi: 10.1111/j.1399-0039.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 81.Waters LJ, Mandalia S, Gazzard B, et al. Prospective HLA-B*5701 screening and abacavir hypersensitivity: a single centre experience. AIDS. 2007;21(18):2533–2534. doi: 10.1097/QAD.0b013e328273bc07. [DOI] [PubMed] [Google Scholar]
- 82.Young B, Squires K, Patel P, et al. First large multicentre open-label study utilizing HLA-B*5701screening for abacavir hypersensitivity in North America. AIDS. 2008;22(13):1673–1675. doi: 10.1097/QAD.0b013e32830719aa. [DOI] [PubMed] [Google Scholar]
- 83.Martin AM, Nolan D, Gaudieri S, et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci U S A. 2004;101(12):4180–4185. doi: 10.1073/pnas.0307067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chessman D, Kostenko L, Lethborg T, et al. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity. 2008;28(6):822–832. doi: 10.1016/j.immuni.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 85.Martin AM, Nolan D, Mallal S. HLA-B*5701 typing by sequence-specific amplification: validation and comparison with sequence-based typing. Tissue Antigens. 2005;65(6):571–574. doi: 10.1111/j.1399-0039.2005.00401.x. [DOI] [PubMed] [Google Scholar]
- 86.Hammond E, Mamotte C, Nolan D, et al. HLA-B*5701 typing: evaluation of an allele-specific polymerase chain melting assay. Tissue Antigens. 2007;70(1):58–61. doi: 10.1111/j.1399-0039.2007.00840.x. [DOI] [PubMed] [Google Scholar]
- 87.Martin AM, Krueger R, Almeida CA, et al. A sensitive and rapid alternative to HLA typing as a genetic screening test for abacavir hypersensitivity syndrome. Pharmacogenet Genomics. 2006;16(5):353–357. doi: 10.1097/01.fpc.0000197468.16126.cd. [DOI] [PubMed] [Google Scholar]
- 88.Dello Russo C, Lisi L, Lofaro A, et al. Novel sensitive, specific and rapid pharmacogenomic test for the prediction of abacavir hypersensitivity reaction: HLA-B*5701 detection by real-time PCR. Pharmacogenomics. 2011;12(4):567–576. doi: 10.2217/pgs.10.208. [DOI] [PubMed] [Google Scholar]
- 89.Kostenko L, Kjer-Nielsen L, Nicholson I, et al. Rapid screening for the detection of HLA-B57 and HLA-B58 in prevention of drug hypersensitivity. Tissue Antigens. 2011;78(1):11–20. doi: 10.1111/j.1399-0039.2011.01649.x. [DOI] [PubMed] [Google Scholar]
- 90.Hammond E, Almeida CA, Mamotte C, et al. External quality assessment of HLA-B*5701 reporting and international multicentre survey. Antivir Ther. 2007;12(7):1027–1032. [PubMed] [Google Scholar]
- 91.Phillips E, Mallal S. Drug hypersensitivity in HIV. Curr Opin Allergy Clin Immunol. 2007;7(4):324–330. doi: 10.1097/ACI.0b013e32825ea68a. [DOI] [PubMed] [Google Scholar]
- 92.Martin AM, Nolan D, James I, et al. Predisposition to nevirapine hypersensitivity associated with HLA-DRB1*0101 and abrogated by low CD4 T cell counts. AIDS. 2005;19(1):97–99. doi: 10.1097/00002030-200501030-00014. [DOI] [PubMed] [Google Scholar]
- 93.Littera R, Carcassi C, Masala A, et al. HLA-dependent hypersensitivity to nevirapine in Sardinian HIV patients. AIDS. 2006;20(12):1621–1626. doi: 10.1097/01.aids.0000238408.82947.09. [DOI] [PubMed] [Google Scholar]
- 94.Gatanaga H, Yazaki H, Tanuma J, et al. HLA-Cw8 primarily associated with hypersensitivity to nevirapine. AIDS. 2007;21(2):264–265. doi: 10.1097/QAD.0b013e32801199d9. [DOI] [PubMed] [Google Scholar]
- 95.Chantarangsu S, Mushiroda T, Mahasirimongkol S, et al. HLA-B*3505 allele is a strong predictor for nevirapine-induced skin adverse drug reactions in HIV-infected Thai patients. Pharmacogent Genomics. 2009;19(2):139–146. doi: 10.1097/FPC.0b013e32831d0faf. [DOI] [PubMed] [Google Scholar]
- 96.Phillips E, Lucas M, Keane N, Lucas A, McKinnon E, Mallal S. HLA-B*35 is associated with nevirapine hypersensitivity in the contemporary Western Australian HIV cohort study. Eur Ann Allergy Clin Immunol. 2010;42:48. [Google Scholar]
- 97.Yuan J, Guo S, Hall D, et al. Toxicogenomics of nevirapine-associated cutaneous and hepatic adverse events among populations of African, Asian and European descent. AIDS. 2011;25(10):1271–1280. doi: 10.1097/QAD.0b013e32834779df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haas DW, Bartlett JA, Anderson JW, et al. Pharmacogenetics of nevirapine-associated hepatotoxicity: an Adult AIDS Clinical Trials Group collaboration. Clin Infect Dis. 2006;43(6):783–786. doi: 10.1086/507097. [DOI] [PubMed] [Google Scholar]
- 99.Ritchie MD, Haas DW, Motsinger AA, et al. Drug transporter and metabolizing enzyme gene variants and nonnucleoside reverse-transcriptase inhibitor hepatotoxicity. Clin Infect Dis. 2006;43(6):779–782. doi: 10.1086/507101. [DOI] [PubMed] [Google Scholar]
- 100.Ciccacci C, Borgiani P, Ceffa S, et al. Nevirapine-induced hepatotoxicity and pharmacogenetics: a retrospective study in a population from Mozambique. Pharmacogenomics. 2010;11(1):23–31. doi: 10.2217/pgs.09.142. [DOI] [PubMed] [Google Scholar]
- 101.Phillips E, Bartlett J, Sanne I, et al. Associations between HLA-DRB1*0102, HLA-B*5801 and hepatotoxicity in patients who initiated nevirapine containing regimens in South Africa [abstract]. Proceedings of the 18th Conference on Retroviruses and Opportunistic Infections; February 27–March 1, 2011; Boston, MA. 2011. p. 949. [Google Scholar]
- 102.Vitezica ZG, Milipied B, Lonjou C, et al. HLA-DRB1*01 associated with cutaneous hypersensitivity induced by nevirapine and efavirenz. AIDS. 2008;22(4):540–541. doi: 10.1097/QAD.0b013e3282f37812. [DOI] [PubMed] [Google Scholar]
- 103.Dalakas MC. Peripheral neuropathy and antiretroviral drugs. J Peripher Nerv Syst. 2001;6(1):14–20. doi: 10.1046/j.1529-8027.2001.006001014.x. [DOI] [PubMed] [Google Scholar]
- 104.Hulgan T, Haas DW, Haines JL, et al. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS. 2005;19(13):1341–1349. doi: 10.1097/01.aids.0000180786.02930.a1. [DOI] [PubMed] [Google Scholar]
- 105.Canter JA, Haas DW, Kallianpur AR, et al. The mitochondrial pharmacogenomics of haplogroup T: MTND2*LHON4917G and antiretroviral therapy-associated peripheral neuropathy. Pharmacogenomics J. 2008;8(1):71–77. doi: 10.1038/sj.tpj.6500470. [DOI] [PubMed] [Google Scholar]
- 106.Kallianpur AR, Hulgan T, Canter JA, et al. Hemochromatosis (HFE) gene mutations and peripheral neuropathy during antiretroviral therapy. AIDS. 2006;20(11):1503–1513. doi: 10.1097/01.aids.0000237366.56864.3c. [DOI] [PubMed] [Google Scholar]
- 107.Chew CS, Cherry CL, Imran D, et al. Tumour necrosis factor haplotypes associated with sensory neuropathy in Asian and Caucasian human immunodeficiency virus patients. Tissue Antigens. 2011;77(2):126–130. doi: 10.1111/j.1399-0039.2010.01570.x. [DOI] [PubMed] [Google Scholar]
- 108.Cherry CL, Rosenow A, Affandi JS, et al. Cytokine genotype suggests a role for inflammation in nucleoside analog-associated sensory neuropathy (NRTI-SN) and predicts and individuals NRTI-SN risk. AIDS Res Hum Retroviruses. 2008;24(2):117–123. doi: 10.1089/aid.2007.0168. [DOI] [PubMed] [Google Scholar]
- 109.Afandi JS, Price P, Imran D, et al. Can we predict neuropathy risk before stavudine prescription in a resource-limited setting? AIDS Res Hum Retroviruses. 2008;24(1):1281–1284. doi: 10.1089/aid.2008.0045. [DOI] [PubMed] [Google Scholar]
- 110.Yamanaka H, Gatanaga H, Kosalaraksa P, et al. Novel mutation of human DNA polymerase gamma associated with mitochondrial toxicity induced by anti-HIV treatment. J Infect Dis. 2007;195(10):1419–1425. doi: 10.1086/513872. [DOI] [PubMed] [Google Scholar]
- 111.Nolan D, Moore C, Castley A, et al. Tumour necrosis factor-alpha gene 239G/A promoter polymorphism is associated with a more rapid onset of lipodystrophy. AIDS. 2003;17(1):121–123. doi: 10.1097/00002030-200301030-00017. [DOI] [PubMed] [Google Scholar]
- 112.Maher B, Alfrevic A, Vilar FJ, et al. TNF-alpha promoter region gene polymoprhisms in HIV-positive patients with lipodystrophy. AIDS. 2003;16(15):2013–2018. doi: 10.1097/00002030-200210180-00005. [DOI] [PubMed] [Google Scholar]
- 113.Hendrickson SL, Kingsley LA, Ruiz-Pesini E, et al. Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;51(2):111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hulgan T, Tebas P, Canter JA, et al. Hemochromatosis gene polymorphisms, mitochondrial haplogroups, and peripheral lipoatrophy during antiretroviral therapy. J Infect Dis. 2008;197(6):858–866. doi: 10.1086/528697. [DOI] [PubMed] [Google Scholar]
- 115.Asensi V, Rego C, Montes AH, et al. IL-1beta (+3954C/T) polymorphism could protect human immunodeficiency virus (HIV)-infected patients on highly active antiretroviral treatment (HAART) against lipodystrophic syndrome. Gent Med. 2008;10(3):215–223. doi: 10.1097/GIM.0b013e3181632713. [DOI] [PubMed] [Google Scholar]
- 116.Ranade K, Geese WJ, Noor M, et al. Genetic analysis implicates resistin in HIV lipodystrophy. AIDS. 2008;22(13):1561–1568. doi: 10.1097/QAD.0b013e32830a9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wangsomboonsiri W, Mahasirimongkol S, Chantarangsu S, et al. Association between HLA-B*4001 and lipodystrophy among HIV-infected patients from Thailand who received a stavudine-containing antiretroviral regimen. Clin Infect Dis. 2010;50(4):597–604. doi: 10.1086/650003. [DOI] [PubMed] [Google Scholar]
- 118.Nelson M, Katlama C, Montaner JS, et al. The safety of tenofovir disoproxil fumarate for the treatment of HIV infection in adults: the first 4 years. AIDS. 2007;21(10):1273–1281. doi: 10.1097/QAD.0b013e3280b07b33. [DOI] [PubMed] [Google Scholar]
- 119.Cooper RD, Wiebe N, Smith N, Keiser P, Naicker S, Tonelli M. Systematic review and meta-analysis: renal safety of tenofovir disoproxil fumarate in HIV-infected patients. Clin Infect Dis. 2010;51(5):496–505. doi: 10.1086/655681. [DOI] [PubMed] [Google Scholar]
- 120.Goicoechea M, Liu S, Best B, et al. Greater tenofovir-associated renal function decline with protease inhibitor-based versus nonnucleoside reverse-transcriptase inhibitor based therapy. Journal of Inf Dis. 2008;197(1):102–108. doi: 10.1086/524061. [DOI] [PubMed] [Google Scholar]
- 121.Kiser JJ, Aquilante CL, Anderson PL, King TM, Carten ML, Fletcher CV. Clinical and genetic determinants of intracellular tenofovir diphosphate concentrations in HIV-infected patients. Clin Pharmacol Ther. 2008;83(2):265–272. doi: 10.1038/sj.clpt.6100269. [DOI] [PubMed] [Google Scholar]
- 122.Izzedine H, Hulot JS, Villard E, et al. Association between ABBCC2 gene haplotypes and tenofovir-induced proximal tubulopathy. J Infect Dis. 2006;194(11):1481–1491. doi: 10.1086/508546. [DOI] [PubMed] [Google Scholar]
- 123.Rodriguez-Novoa S, Labarga P, Soriano V, et al. Predictors of kidney tubular dysfunction in HIV-infected patients treated with tenofovir: a pharmacogenetic study. Clin Infect Dis. 2009;48(11):E108–E116. doi: 10.1086/598507. [DOI] [PubMed] [Google Scholar]
- 124.Pushpakom SP, Liptrott NJ, Rodrigues-Novoa S, et al. Genetic variants of ABCC10, a novel tenofovir transporter, are associated with kidney tubular dysfunction. J Infect Dis. 2011;204(1):145–153. doi: 10.1093/infdis/jir215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zucker SC, Qin X, Rouster SC. Mechanism of indinavir-induced hyperbilirubinemia. Proc Natl Acad Sci U S A. 2001;98(22):12671–12676. doi: 10.1073/pnas.231140698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Solas C, Gagnieu MC, Ravaux I, et al. Population pharmacokinetics of atazanavir in human immunodeficiency virus-infected patients. Ther Drug Monit. 2008;30(6):670–673. doi: 10.1097/FTD.0b013e3181897bff. [DOI] [PubMed] [Google Scholar]
- 127.Barrios A, Rendon AL, Gallego O, et al. Predictors of virological response to atazanavir in protease inhibitor exposed patients. HIV Clin Trials. 2004;5(4):201–205. doi: 10.1310/3HL3-HHBD-WKLR-XELL. [DOI] [PubMed] [Google Scholar]
- 128.Strassburg CP. Pharmacogenetics of Gilberts's syndrome. Pharmacogenomics. 2008;9(6):703–715. doi: 10.2217/14622416.9.6.703. [DOI] [PubMed] [Google Scholar]
- 129.Rotger M, Taffe P, Bleiber G, et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J Infect Dis. 2005;192(8):1381–1386. doi: 10.1086/466531. [DOI] [PubMed] [Google Scholar]
- 130.Rodriguez-Novoa S, Martin-Carbonero L, Barreiro P, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS. 2007;21(1):41–46. doi: 10.1097/QAD.0b013e328011d7c1. [DOI] [PubMed] [Google Scholar]
- 131.Park WB, Choe PG, Song KH, et al. Genetic factors influencing severe atazanavir-associated hyperbilirbuinemia in a population with low UDP-glucuronosyltransferase 1A1*28 allele frequency. Clin Infect Dis. 2010;51(1):101–106. doi: 10.1086/653427. [DOI] [PubMed] [Google Scholar]
- 132.Kummer O, Mossdorf E, Battegay M, et al. Treatment of an atazanavir associated grade 4 hyperbilirubinemia with efavirenz. Gut. 2007;56(10):1477–1478. doi: 10.1136/gut.2007.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodriguez-Novoa S, Barreiro P, Rendon A, et al. Plasma levels of atazanavir and the risk of hyperbilirubinemia are predicted by the 3435 C-->T polymorphism at the multidrug resistance gene 1. Clin Infect Dis. 2006;42(2):291–295. doi: 10.1086/499056. [DOI] [PubMed] [Google Scholar]
- 134.Lubomirov R, Colombo S, di Iulio J, et al. Association of pharmacogenetic markers with premature discontinuation of first-line anti HIV therapy: an observational cohort study. J Infect Dis. 2011;203(2):246–257. doi: 10.1093/infdis/jiq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arnedo M, Tarffe P, Sahli R, et al. Contribution of 20 single nucleotide polymorphisms of 13 genes to dyslipidemia associated with antiretroviral therapy. Pharmacogenet Genomics. 2007;17(9):755–764. doi: 10.1097/FPC.0b013e32814db8b7. [DOI] [PubMed] [Google Scholar]
- 136.Rotger M, Bayard C, Taffe P, et al. Contribution of genome-wide significant single-nucleotide polymorphisms and antiretroviral therapy to dyslipidemia in HIV-infected individuals: a longitudinal study. Circ Cardiovasc Genet. 2009;2(6):621–628. doi: 10.1161/CIRCGENETICS.109.874412. [DOI] [PubMed] [Google Scholar]
- 137.Chang SY, Ko WS, Kao JT, et al. Association of single-nucleotide polymorphism 3 and c.553G >T of APOA5 with hypertriglyceridemia after treatment with highly active antiretroviral therapy containing protease inhibitors in hiv-infected individuals in Taiwan. Clin Infect Dis. 2009;48(6):832–835. doi: 10.1086/597099. [DOI] [PubMed] [Google Scholar]
- 138.Grunfeld C, Kotler DP, Hamadeh R, Tierney A, Wang J, Pierson RN. Hypertriglyceridemia in the acquired immunodeficiency syndrome. Am J Med. 1989;86(1):27–31. doi: 10.1016/0002-9343(89)90225-8. [DOI] [PubMed] [Google Scholar]
- 139.Tarr PE, Taffe P, Bleiber G, et al. Modeling the influence of APOC3, APOE and TNF polymorphisms on the risk of antiretroviral therapy-associated lipid disorders. J Infect Dis. 2005;191(9):1419–1426. doi: 10.1086/429295. [DOI] [PubMed] [Google Scholar]
- 140.Tarr PE, Rotger M, Telenti A. Dyslipidemia in HIV-infected individuals: from pharmacogenetics to pharmacogenomics. Pharmacogenomics. 2010;11(4):587–594. doi: 10.2217/pgs.10.35. [DOI] [PubMed] [Google Scholar]
- 141.Cespedes MS, Aberg JA. Neuropsychiatric complications of antiretroviral therapy. Drug Saf. 2006;29(10):865–874. doi: 10.2165/00002018-200629100-00004. [DOI] [PubMed] [Google Scholar]
- 142.Van Luiri M, Bannister WP, Mocroft A, et al. Absence of a relation between efavirenz plasma concentrations and toxicity-driven efavirenz discontinuations in the EuroSIDA study. Antivir Ther. 2009;14(1):75–83. [PubMed] [Google Scholar]
- 143.Wyen C, Hendra H, Siccardi M. Cytochrome P450 2B6(CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J Antimicrob Chemother. 2011;66(9):2092–2098. doi: 10.1093/jac/dkr272. [DOI] [PubMed] [Google Scholar]
- 144.Burger D, Hugen P, Reiss P, et al. Therapeutic drug monitoring of nelfinavir and indinavir in treatment-naive HIV-1 infected individuals. AIDS. 2003;17(8):1157–1165. doi: 10.1097/00002030-200305230-00007. [DOI] [PubMed] [Google Scholar]
- 145.Demeter LM, Mukherjee AL, DiFrancesco R, et al. The design and implementation of A5146, a prospective trial assessing the utility of therapeutic drug monitoring using an inhibitory quotient in antiretroviral-experienced HIV-infected patients. HIV Clin Trials. 2008;9(1):61–72. doi: 10.1310/hct0901-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Best BM, Stek AM, Mirochnick M, et al. Lopinavir tablet pharmacokinetics with an increased dose during pregnancy. J Acquir Immune Defic Syndr. 2010;54(4):381–388. doi: 10.1097/qai.0b013e3181d6c9ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mirochnick M, Capparelli E. Pharmacokinetics of antiretrovirals in pregnant women. Clin Pharmacokinet. 2004;43(15):1071–1087. doi: 10.2165/00003088-200443150-00002. [DOI] [PubMed] [Google Scholar]
- 148.http://www.hiv-pharmacogenomics.org/pdf/ref_tbl_pharmaco/HIV_Pharmacogenomics_reference_table.pdf. Accessed October 11, 2011
- 149.Phillips EJ, Mallal SA. Pharmacogenetics of drug hypersensitivity. Pharmacogenomics. 2010;11(7):973–987. doi: 10.2217/pgs.10.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 151.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Gent. 2009;41(10):1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 152.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(1):1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 153.Bachmann H, Siffert W, Skaletz-Rorowski A, et al. Impact of IL28B polymorphisms on sustained virologic response of HCV/HIV co-infected patients [abstract]. Proceedings of the 18th Conference on Retroviruses and Opportunistic Infections; 2011 Feb 27–Mar 1; Boston, MA. 2011. p. 946. [Google Scholar]
- 154.Labarga P, Barreiro P, Mira JA, et al. Impact of IL28B polymorphisms on response to peginterferon and ribavirin in HIV-hepatitis C virus-co-infected patients with prior nonresponse or relapse. AIDS. 2011;25(8):1131–1133. doi: 10.1097/QAD.0b013e3283471d83. [DOI] [PubMed] [Google Scholar]