Abstract
Mu opioid receptor (OPRM1) gene variants, particularly the common A118G single nucleotide polymorphism (SNP), are among the most frequently studied candidate genes associated with opioid dependence. However, despite numerous case-control studies and meta-analyses, no definitive conclusion has been reached regarding the association of the A118G SNP and risk of developing opioid dependence. This study aimed to resolve this discrepancy by reinvestigating the association between A118G SNP allelic, and for the first time, genotype frequencies and opioid dependence. A meta-analysis of sixteen case-control studies of opioid dependence was performed with a total of 5169 subjects. No association between the A118G allele (P = 0.23) and genotype (P = 0.34) frequencies and opioid dependence was found. However, significant heterogeneity between studies precluded highly definitive conclusions. In addition, the possibility that other OPRM1 SNPs albeit rarer may influence the risk of opioid dependence remains to be investigated at this level. Nonetheless, despite no evidence of a direct association with risk of dependence, A118G may still influence the pharmacological response to opioids impacting on an individual’s dosage requirements.
Keywords: mu opioid receptor, opioid dependence, A118G genotype, meta-analysis
Introduction
The development of opioid tolerance and dependence, through chronic and/or inappropriate use, increases the risk of problematic adverse effects and imposes severe social costs. Unfortunately, the high fatality rate of heroin overdose and incidences of addiction to prescription opiates continue to escalate.1–3 However, the associated risk of developing opiate dependence is difficult to predict due to large individual variability in opiate sensitivity.3 This imposes major implications to the likelihood of opioid addiction, addiction treatment, and analgesic drug dosing due to the possible risk of under or overdosing.
There are at least three key interrelated factors that contribute to the individual vulnerability to developing opioid addiction once self-exposed, including psychosocial and environmental factors, neurological and pharmacological adaptions to exposure and genetic factors.4 Genetic susceptibility to drug abuse and dependence has been studied based on dependence in general and also and in relation to specific drug categories, particularly opioids,5,6 with up to 54% of the associated risk in developing opioid dependence reported as being attributable to genetic factors.7
The mu opioid receptor has a clear involvement in mediating the analgesic and rewarding effects of endogenous and exogenous opioids, and has been tightly linked to the development of addiction disorders due to its involvement in the brain reward pathway.1,8,9 The OPRM1 gene encodes for the mu opioid receptor, and sequence variants within its coding region are the most frequently studied candidates in association with opioid dependence.9 However, to date no definitive conclusion has been reached regarding the role of OPRM1 polymorphisms in determining susceptibility to opioid dependence.5,9
The most frequently studied OPRM1 variant is the single nucleotide polymorphism (SNP) A118G (recently renamed 304A/G10) in exon 1 that causes an Asn40Asp substitution2 at a putative glycosylation site in the extracellular domain and occurs at an allelic frequency of between 10% and 40% depending on ethnicity. Numerous case-control association studies have attempted to identify underlying neurobiological correlates between the A118G variant and the risk of opioid dependence, but so far have provided contradictory results. Studies have identified a significant positive correlation between A118G and heroin addiction,8 and an increased affinity for endogenous opioids, particularly beta-endorphin, and agonist-induced activation in vitro.2,4 In contrast, opposite and null findings have also been reported.11,12 Thus, an association between A118G and the risk of opioid dependence has neither been confirmed nor refuted.
The A118G variant has emerged as a prime candidate for genes associated with the risk of addiction; however, the vast discrepancy between published results needs to be addressed. This may be because the majority of the current literature is restricted to data collected from classical case-control studies, which may possibly lead to false negatives or false positives depending on the types of approach used, thus making results difficult to compare.9 Meta-analysis allows a substantially larger sample size to be analysed and therefore allows a greater statistical power. Two recent meta-analyses have attempted to elucidate the nature of the relationship between the A118G polymorphism and either opioid9 or substance5 dependence. Although neither study found an overall association, both studies only evaluated an allelic association. Furthermore, additional case-control studies have been conducted since these publications,13,14 including data from our laboratory. Therefore, capitalizing on the power of meta-analysis, the aims of the present study were to reinvestigate the association between A118G allele and genotype frequencies and the occurrence of opioid dependence.
Methods
Literature search
A systematic, computerized search of the current literature up to September 2008 was conducted using the PubMed©, Scopus, and Web of Science® online databases. Keywords used to identify articles examining the association between the A118G polymorphism and the occurrence of opioid dependence were Asn40Asp, A118G, opioid, opiate, heroin, dependence, addiction, mu-receptor polymorphism, OPRM1, and all the possible combinations thereof. The reference lists of identified studies as well as relevant reviews were also examined in an attempt to identify additional publications. Finally, the two existing meta-analyses were consulted5,9 and unpublished data from our laboratory was included.
Subjects and methods of our retrospective study from which unpublished data were obtained for inclusion in analysis are as follows. Fifty six methadone maintenance opioid-dependent and 83 healthy control subjects were included. These subjects participated in a number of clinical studies (including a pregnancy study) conducted within the discipline of pharmacology at the University of Adelaide from January 2001–2007, during which written informed consent and whole blood or tissue samples for the genotyping analysis were obtained. Approval for all studies was obtained from the Royal Adelaide Hospital Research Ethics Committee. Original study case notes were used to obtain subject demographic data. Genomic DNA was isolated from samples using a QIAamp® DNA mini kit according to the manufacturer’s instructions (QIAGEN Pty Ltd, Doncaster, VIC, Australia). Genotyping of the A118G SNP was performed using allele-specific polymerase chain reaction (PCR) as previously described.15
Inclusion criteria
Only studies investigating the association between Asn40Asp (A118G) and opioid dependence were considered for inclusion in the meta-analysis. In addition, studies had to meet the following criteria: 1. have a case-control design; 2. report Asn40Asp allele and/or genotype frequency for a group of individuals with a diagnosis of opioid dependence and also a group for a control sample; 3. present original data; and 4. have samples that were in Hardy–Weinberg equilibrium, meaning that the samples are considered representative of the populations from which they were derived. Sixteen studies met these criteria including our own unpublished data (Coller et al unpublished) and were included in the meta-analysis (for summary see Table 1), providing a total of 2324 opioid-dependent subjects and 2845 control subjects.1,2,8,11–14,16–23 It should be stated that poly-drug dependency was not an exclusion criteria per se. In reality, opioid dependents may also abuse and be dependent on other drugs (for example alcohol, nicotine, cocaine). Therefore, as long as the case subjects in the studies were dependent on opioids, regardless of the presence of other dependencies, that study was included in the meta-analysis.
Table 1.
Characteristics of the studies investigating the association between the A118G OPRM1 SNP and opioid dependence
| Study | Ethnicity | Drug dependence | Dependent cases (n) | Controls (n) |
|---|---|---|---|---|
| Bart et al8 | Caucasian (Swedish) | Heroin | 139 | 170 |
| Bond et al2 | Caucasian (European American) + Hispanic + African-American | Heroin | 111 | 39 |
| Coller et al (unpublished data) | Caucasian (Australian) | Heroin | 53 | 80 |
| Crettol et al14 | Caucasian (Swiss) | Heroin | 238 | 217 |
| Crowley et al11 | Caucasian (European American) + African-American | Opioid/cocaine/nicotine/alcohol | 225 | 200 |
| Drakenberg et al1 | Caucasian (European) | Heroin | 39 | 26 |
| Franke et al12 | Caucasian (German) | Heroin | 287 | 365 |
| Gelernter et al16 | Caucasian (European American) + Hispanic + African-American | Opioid | 79 | 228 |
| Hoehe et al17 | African-Americans | Heroin/cocaine | 137 | 35 |
| Kapur et al13 | Asian (Indian) | Opioida | 126 | 156 |
| Li et al18 | Asian (Chinese) | Heroin | 226 | 208 |
| Luo et al19 | Caucasian (European American) + African-American | Opioid/cocaine/alcohol | 131 | 234 |
| Shi et al20 | Asian (Chinese) | Heroin | 145 | 48 |
| Szeto et al21 | Asian (Chinese) | Heroin | 200 | 97 |
| Tan et al22 | Asian (Indian + Malaysian + Chinese) | Heroin | 97 | 404 |
| Zhang et al23 | Caucasian (European American) | Opioid | 91 | 338 |
| Pooled | 11 heroin, 5 opioid | 2324 | 2845 |
Note:
Either noninjectable (morphine, smack (heroin)) or injectable (pentazocine, buprenorphine) opioids.
Study characteristics
Of the 16 studies included, 11 stated that their case subjects were heroin-dependent; one stated that their case subjects were mainly (85%) dependent on morphine or smack (heroin), though some (15%) were addicted to pentazocine/buprenorphine;13 and the remaining four did not indicate to which specific opioid(s) their case subjects were addicted (Table 1). Most studies used either the Diagnostic and Statistical Manual 24-III-R14 or -IV15 criteria to diagnose opioid dependence. Exceptions to this were the studies by Bond and colleagues, which used federally-regulated criteria for admission to a methadone maintenance program;2 Crowley and colleagues which used medical records and urine screening;11 Drakenberg and colleagues, who used post-mortem opiate toxicology, history of opiate abuse, and the presence or absence of physical body-needle tracks;1 and Shi and colleagues who interviewed physicians.20 All but two studies excluded individuals with major psychiatric disorders (for example depression and schizophrenia) from their control groups.16,22 Li and colleagues excluded such patients, but these authors relied on subject’s self-reported medical history, rather than formal psychiatric evaluation, which may have failed to exclude control subjects who in fact had psychiatric illness(es).18 All studies genotyped their subjects by means of the PCR or one of its variants (such as restriction fragment length polymorphism, denaturing high performance liquid chromatography, or allele-specific PCR), except that of Hoehe and colleagues, who used a novel multiplex sequence comparison technique.17
Statistical analysis
For all 16 studies, the allele counts, that is the number of A (wild-type) and G (variant) alleles, of the case and control groups were ascertained, either directly from the article or, if not indicated there, were calculated from the genotype counts. Three studies did not report the genotype counts of their subjects (the number of subjects with A/A, A/G, and G/G genotypes), but they did report allele counts.16,17,19 For each study that reported allele counts (all 16 studies), an odds ratio (OR) was calculated to compare the frequency of the G variant allele between case (opioid-dependent) and control subjects. Similarly, for each study that reported genotype counts (13 studies), an OR (with 95% CI) was calculated to compare the frequency of the homozygous mutant genotype (G/G) between opioid-dependent and control subjects.
Pooled ORs were calculated using a random effects (meta-analytic) model according to the DerSimonian–Laird method.27 This model automatically assigns the weighting that each individual study contributes to the pooled OR depending on that study’s sample size and yielded OR. Forest plots were used to depict variant allele (G) and genotype (G/G) ORs for each individual study as well as overall OR. The degree of heterogeneity, that is the variation that cannot be explained by random chance,28 in the ORs across studies was assessed using the I-squared statistic and a χ2-test of goodness of fit.29 When high levels of heterogeneity were observed (I-squared values > 40%), meta-regression models were fitted to the data in an attempt to explain the heterogeneity.30 The following explanatory variables were included in the meta-regression models: 50% or more participants Caucasian (yes/no); 50% or more participants Asian (yes/no); mean age (continuous variable); percentage of males (continuous variable); type of dependence (opioid/heroin); and majority ethnicity (Caucasian/Asian). The potential for publication bias in the meta-analysis was assessed statistically using Egger’s test31 and visually using Begg’s funnel plot.32 The funnel plot shows the standard error (SE) of the log OR, a measure of study precision, against the log OR. In the absence of bias the plot should be symmetric and funnel-shaped, with smaller less precise studies being more widely scattered around the overall estimate of the log OR than larger studies. An asymmetric plot is typically interpreted as providing evidence of publication bias. All calculations were performed using Stata version 10 (Stata Corp., College Station, TX, USA). A P-value < 0.05 was considered statistically significant.
Results
A total of 2324 opioid-dependent (case) and 2845 control subjects from 16 studies were included in the meta-analysis. The age (mean ± SD) and percentage of males in the opioid-dependent and control groups were 33 ± 8 and 34 ± 10 years, respectively, and 78% and 56%, respectively. Table 2 summarizes the A118G allele and genotype data from each individual study.
Table 2.
A118G OPRM1 SNP allele and genotype frequency (n (%)) data in opioid-dependent cases and control groups from individual studies
| Study |
Opioid-dependent cases
|
Control subjects
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Allele frequency
|
Genotype frequency
|
Allele frequency
|
Genotype frequency
|
|||||||||
| n | A | G | A/A | A/G | G/G | n | A | G | A/A | A/G | G/G | |
| Bart et al8 | 139 | 235 (84.5) | 43 (15.4) | 98 (70.5) | 39 (28) | 2 (1.5) | 170 | 315 (92.6) | 25 (7.4) | 147 (86.4) | 21 (12.4) | 2 (1.2) |
| Bond et al2 | 111 | 202 (91) | 20 (9) | 92 (83) | 18 (16) | 1 (1) | 39 | 66 (85) | 12 (15) | 29 (74) | 8 (21) | 2 (5) |
| Coller et al (unpublished data) | 53 | 85 (80) | 21 (20) | 37 (70) | 11 (21) | 5 (9) | 80 | 136 (85) | 24 (15) | 57 (71) | 22 (28) | 1 (1) |
| Crettol et al14 | 238 | 411 (86) | 65 (14) | 177 (74) | 57 (24) | 4 (2) | 217 | 376 (87) | 58 (13) | 167 (77) | 42 (19) | 8 (4) |
| Crowley et al11 | 225 | 401 (89) | 49 (11) | 177 (79) | 47 (21) | 1 (0.4) | 200 | 359 (90) | 41 (10) | 161 (81) | 37 (19) | 2 (1) |
| Drakenberg et al1 | 39 | 66 (85) | 12 (15) | 28 (72) | 10 (26) | 1 (3) | 26 | 49 (94) | 3 (6) | 24 (92) | 1 (4) | 1 (4) |
| Franke et al12 | 287 | 498 (87) | 76 (13) | 217 (76) | 64 (22) | 6 (2) | 365 | 642 (88) | 88 (12) | 284 (78) | 74 (20) | 7 (2) |
| Gelernter et al16 | 79 | 143 (91) | 15 (9) | NR | NR | NR | 228 | 400 (88) | 56 (12) | NR | NR | NR |
| Hoehe et al17 | 137 | 263 (96) | 11 (4) | NR | NR | NR | 35 | 67 (96) | 3 (4) | NR | NR | NR |
| Kapur et al13 | 126 | 173 (69) | 79 (31) | 70 (56) | 33 (26) | 23 (18) | 156 | 276 (88) | 36 (12) | 124 (79) | 28 (18) | 4 (3) |
| Li et al18 | 226 | 297 (65) | 155 (34) | 95 (42) | 107 (47) | 24 (11) | 208 | 292 (70) | 124 (30) | 99 (48) | 94 (45) | 15 (7) |
| Luo et al19 | 131 | 240 (92) | 22 (8) | NR | NR | NR | 234 | 416 (89) | 52 (11) | NR | NR | NR |
| Shi et al20 | 145 | 217 (75) | 73 (25) | 85 (59) | 47 (32) | 13 (9) | 48 | 70 (73) | 26 (27) | 26 (54) | 18 (38) | 4 (8) |
| Szeto et al21 | 200 | 242 (61) | 158 (40) | 75 (38) | 92 (46) | 33 (17) | 97 | 137 (71) | 57 (29) | 51 (53) | 35 (36) | 11 (11) |
| Tan et al22 | 97 | 132 (68) | 62 (32) | 47 (48) | 38 (39) | 12 (12) | 404 | 464 (57) | 344 (43) | 128 (32) | 208 (51) | 68 (17) |
| Zhang et al23 | 91 | 158 (87) | 24 (13) | 69 (76) | 20 (22) | 2 (2) | 338 | 589 (87) | 87 (13) | 256 (76) | 78 (23) | 4 (1) |
| Pooled | 3763 | 885 | 1267 | 583 | 127 | 4654 | 1036 | 1553 | 666 | 129 | ||
Abbreviation: NR, not reported.
Comparison of cases and controls: Genotype frequencies
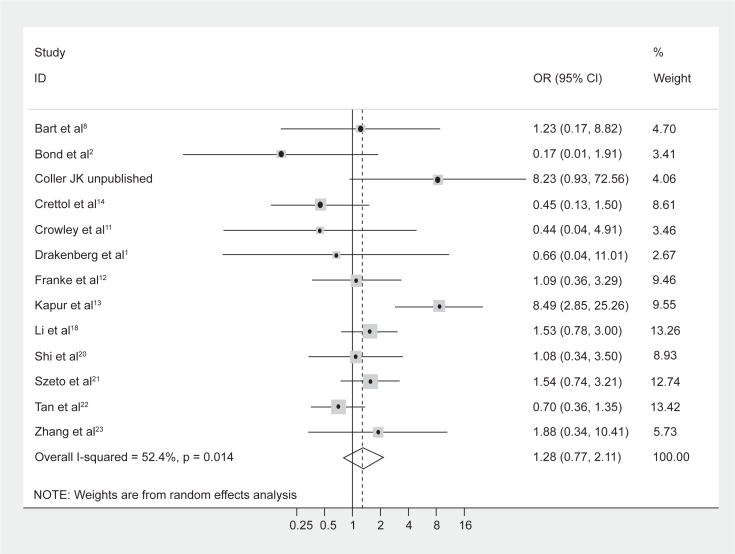
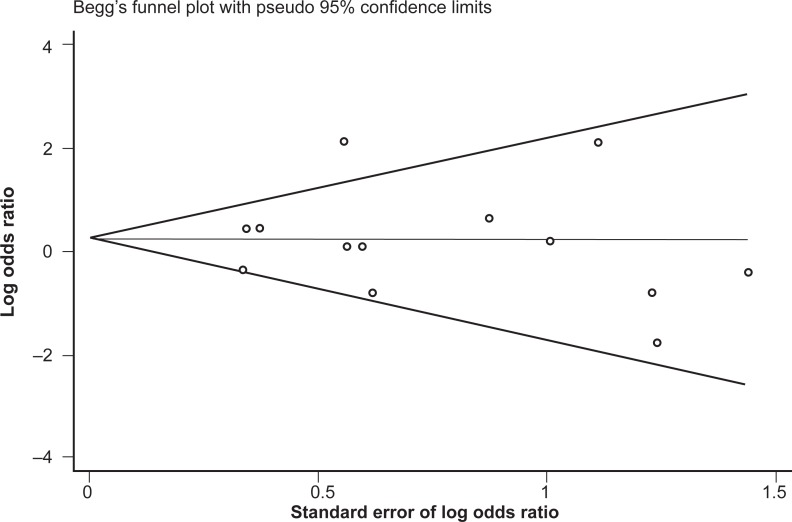
No difference in genotype frequencies between opioid-dependent and control groups was observed, with a pooled OR (95% CI) from the 13 studies of 1.28 (0.77–2.11), P = 0.34 (Figure 1). In addition, there was no evidence of publication bias as indicated by the funnel plot (Figure 2) and Egger’s test value (t = −0.12, P = 0.91). The high level of heterogeneity (I-squared = 52.4%; χ2 (12) = 25.18, P = 0.014) suggested the presence of some potential moderator variable(s). However, meta-regression models failed to identify significant moderating effects by the following potential explanatory variables: 50% or more participants being Caucasian (yes/no) P = 0.66; 50% or more participants being Asian (yes/no) P = 0.33; mean age (continuous) P = 0.51; type of drug dependence (opioid/heroin) P = 0.07; and percentage of males (continuous) P = 0.13.
Figure 1.
Forest plot depicting the odds ratios (OR) and 95% confidence interval (CI) of each of the 13 studies investigating the association between A118G OPRM1 genotype frequencies and opioid dependence. The solid line represents an OR of 1 and the stippled line represents the pooled OR.
Figure 2.
Begg’s funnel plot of the 13 studies investigating the association between A118G OPRM1 genotype frequencies and opioid dependence.
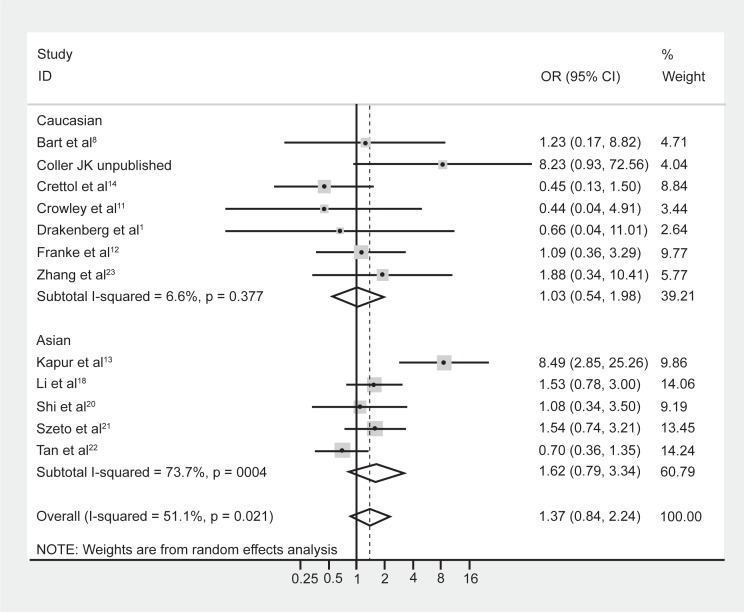
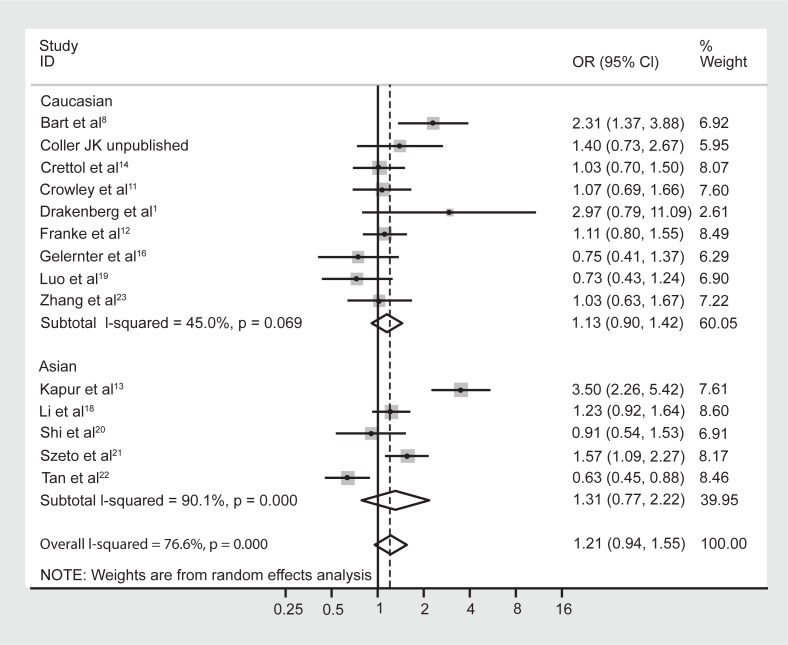
Although meta-regression failed to highlight ethnicity as a significant moderating effect of the heterogeneity, it is well known that the prevalence of the A118G polymorphism is dependent on ethnicity.5,9 Hence the data were reanalyzed following stratification by majority ethnicity, either Asian (Chinese, Indian, Malaysian) or Caucasian as these were the two prominent ethnicities studied in the literature. Results indicated that there was acceptable heterogeneity within the Caucasian studies (Figure 3, I-squared = 6.6%; χ2 (6) = 6.43, P = 0.38). In contrast however, there was still substantial heterogeneity within the Asian studies (I-squared = 73.7%; χ2 (4) = 15.22, P = 0.004). Grouping the studies with regard to Caucasian and Asian ethnicity did not alter the association between genotype frequencies and opioid dependence, with a pooled OR (95% CI) for Caucasian studies of 1.03 (0.54–1.98), P = 0.93, and for Asian studies of 1.62 (0.79–3.34), P = 0.19.
Figure 3.
Forest plot depicting the odds ratios (OR) and 95% confidence interval (CI) of each of the 13 studies investigating the association between A118G OPRM1 genotype frequencies and opioid dependence stratified as majority Caucasian and Asian ethnic groups. The solid line represents an OR of 1 and the stippled line represents the pooled OR.
Comparison of cases and controls: Allele frequencies
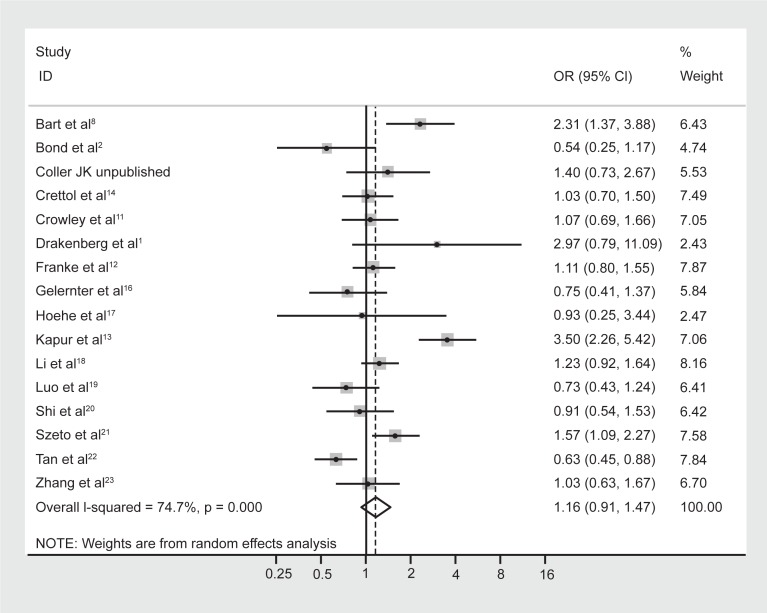
No difference in allele frequencies was observed between opioid-dependent and control groups with a pooled OR (95% CI) of the 16 studies of 1.16 (0.91–1.47), P = 0.23 (Figure 4). There was no evidence of publication bias within 16 articles included as indicated by the funnel plot (Figure 5) and Egger’s test value (t = 0.14, P = 0.89). Similar to genotype frequencies, significant heterogeneity existed between studies (I-squared = 74.7%; χ2 (15) = 59.40, P < 0.0001). Meta-regression analysis again failed to identify significant moderating effects for: 50% or more participants being Caucasian (yes/no) P = 1.0; 50% or more participants being Asian (yes/no) P = 0.50; mean age (continuous) P = 0.17; or type of dependence (opioid/heroin) P = 0.89. However, distinct from the genotype analysis, the percentage of males in the study was found to be a significant predictor of the log OR (P = 0.023), such that as the percentage of males in the study increased by 1 the OR increased by an average of 1.8%.
Figure 4.
Forest plot depicting the odds ratios (OR) and 95% confidence interval (CI) of each of the 16 studies investigating the association between A118G OPRM1 allele frequencies and opioid dependence. The solid line represents an OR of 1 and the stippled line represents the pooled OR, P = 0.000 equivalent to P < 0.0001.
Figure 5.
Begg’s funnel plot of the 16 studies investigating the association between A118G OPRM1 allele frequencies and opioid dependence.
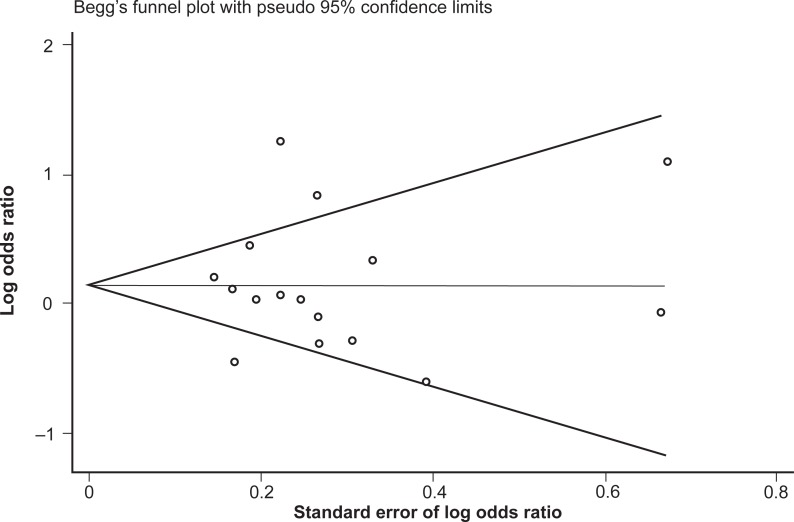
Similar to the reasoning with the genotype data, the studies were once again reanalyzed following stratification by majority ethnicity, either Asian or Caucasian (Figure 6). This, however, did not reduce the heterogeneity in either group to acceptable levels: for Asian studies the observed I-squared = 90.1%, χ2 (4) = 40.25, P < 0.0001; and for Caucasian studies the observed I-squared = 45.0%, χ2 (8) = 14.54, P = 0.07. Therefore, in this instance, stratifying by majority ethnicity was not useful in accounting for the high degree of heterogeneity between studies. Furthermore, grouping the studies with regard to Caucasian and Asian ethnicity did not alter the association between allele frequencies and opioid dependence, with a pooled OR (95% CI) for Caucasian studies of 1.13 (0.90–1.42), P = 0.29, and for Asian studies of 1.31 (0.77–2.22), P = 0.33.
Figure 6.
Forest plot depicting the odds ratios (OR) and 95% confidence interval (CI) of each of the 16 studies investigating the association between A118G OPRM1 allele frequencies and opioid dependence stratified as majority Caucasian and Asian ethnic groups. The solid line represents an OR of 1 and the stippled line represents the pooled OR, P = 0.000 equivalent to P < 0.0001.
Discussion
The present meta-analysis, which included 5169 subjects (2324 of whom were opioid dependent), showed no evidence of a relationship between the A118G OPRM1 SNP and opioid dependence in terms of either genotype or allele frequencies. These results both corroborate and extend previous meta-analyses which found no association between A118G and substance or opioid dependence.5,9 These authors, however, did not study the genotype frequency of A118G, only the allele frequency. In contrast, the current meta-analysis investigated both the allele and genotype frequency of A118G, and also included data from three additional studies conducted following the publication of these two meta-analyses. In comparison to the prior meta-analysis with opioid-dependent cases, the addition of the three new studies to the present meta-analysis resulted in a larger sample size (2324 cases and 2845 controls versus 1675 cases and 2465 controls9) and therefore potentially an increased power to detect a difference between the groups. Nonetheless, the overall conclusion regarding the association remained unchanged.
It should be emphasized that significant heterogeneity exists in the current body of case-control studies investigating the association between the A118G SNP and opioid dependence, which may have prevented the association from being reliably investigated. The heterogeneity detected in the present meta-analysis was not fully accounted for, even when studies were separated into majority Caucasian and Asian ethnicity groups. In fact this stratification only reduced heterogeneity levels to within acceptable limits for one of our four analyses, namely the genotype frequency of A118G in majority Caucasian studies, but did not alter the observation that there was no association between the SNP and opioid dependence. In the case of the majority Asian analysis, this may be explained by the inclusion of an Indian study.13 This study yielded the largest OR of A118G between opioid-dependent and control groups with regard to both allele and genotype frequency (Figures 1, 2, 4, 6). As India is not part of mainland Asia, its inhabitants have a different ancestral and hence genetic background and thus it may not be a valid inclusion. However, it should be noted that the impact of this study on the pooled Asian OR was relatively low in comparison to the other studies included (weights of 7.6% for allele data and 9.9% for genotype data), hence the inclusion in this grouping is not predicted to alter the overall ability to observe an association.
Of considerable interest with regard to modulators explaining heterogeneity was the observation that among the 16 studies with allele frequency data the percentage of males in the study was found to be a significant predictor of the log OR (P = 0.023). This was not specifically discussed in the previous meta-analyses and to the best of our knowledge there are no reports in the literature citing gender differences in the occurrence of the A118G SNP. However, research has suggested a gender difference in analgesic response to opioids33 and recently, a gender difference in the reinforcing effect of nicotine has been reported, such that the G variant allele was associated with reduced reinforcement in women with no association in men (n = 60).34 Therefore, this potential impact of gender on the association warrants further investigation.
With the exception of this gender modulator, this study was unable to determine the other cause(s) of the high degree of heterogeneity, however, it is possible to speculate on some potential sources. Firstly, four studies did not use standardized methods (DSM-III-R or -IV criteria) for diagnosing opioid dependence.1,2,11,20 For example, the dependence status of Drakenberg and colleagues’ subjects was determined postmortem by examining opiate toxicology, history of opiate abuse, and whether or not physical body-needle tracks were present. This method of assessing opioid dependence has not been formally validated and, furthermore, the authors did not state the specific cut-offs they used for each criterion. Thus, it is impossible to know how the authors decided in borderline situations and, if this may have impacted on results.
Secondly, due to the stigma attached to drug dependence, many opioid-dependent subjects may be unwilling or unable to participate in case-control studies. Thus, the case subjects may not be representative of the general opioid-dependent population. Thirdly, multiple drug dependency was not an exclusion criterion. Therefore, other drug dependencies such as alcohol and nicotine may be acting on the brain-reward pathway, making investigation of a direct association between opioid dependence and A118G difficult to observe. Finally, two of the included studies did not screen their control groups for psychiatric disorders,16,22 whilst a third relied on participants’ self-reported psychiatric history for excluding subjects.18 Consequently, in these three studies, it is possible that their respective control groups included subjects with psychiatric illness(es) which may have impacted on the phenotypes of the cases versus the controls to mask the association. It should be noted that the degree of heterogeneity observed in the present study was similarly reported by the two previous meta-analyses5,9 (χ2 = 4.35, P = 0.037 and χ2 = 41.72, P = 0.003, respectively). This heterogeneity is suggestive of potential moderating variables that may be influencing the association, including environmental differences. Consequently, it raises questions regarding the ability, at this point in time, to make reliable conclusions regarding an association, or lack thereof, between A118G OPRM1 genetics and opioid dependence.
Another potential issue that may mask the association observed in meta-analysis is the presence of publication bias. This is largely a result of studies that yield significant findings being more likely to be published, therefore, it is important to search for and include “nonsignificant” studies. Missing such studies in a literature search could possibly bias the meta-analysis towards a significant association when in fact it does not exist.26 The inclusion of relevant unpublished data can negate this possible problem. In the current study, the two Begg’s funnel plots had the expected funnel shape and, in addition, the Egger’s tests for publication bias were not significant (genotype frequency data, P = 0.91; allele frequency data, P = 0.89), indicating that there was little or no publication bias in our analyses that may have impacted on the ability to observe an association. Therefore, this is unlikely to have impacted on the overall conclusions drawn from the analysis.
In conclusion, although an association between the A118G OPRM1 SNP and opioid dependence was not supported in the current meta-analysis, further analysis of A118G is warranted which also investigates the impact of gender as a modulator. In addition, previous case studies have reported possible association of A118G SNP with opioid dependence in other ethnic groups, such as Hispanics,2,16 African-Americans,17 and Indians.13 However, these ethnicities were not represented in enough case-control studies to be included in the present meta-analysis as a separate ethnicity modulator. Furthermore, despite having no clear evidence of a direct association to the risk of developing opioid dependence, A118G may mediate or otherwise influence pharmacological and therapeutic responses to opioids, as evidenced by altered opioid dosage requirements,35 thereby possibly indirectly leading to dependence. Thus, a deeper investigation into its functionality is needed which should also consider the complex regulation of activity of the endproduct, the mu opioid receptor. Finally, the A118G polymorphism is only one of more than 100 known SNPs in OPRM1.3 Therefore, future studies should consider other candidate albeit rarer SNPs for influencing the risk of developing opioid dependence.
Acknowledgments
The authors would like to thank all the investigators of the original studies from which the subjects were recruited: Peter Athanasos, Andrea Gordon, Justin Hay, Mark Hutchinson, Erin Morton, Mario Nguyen, Sophie La Vincente, and Belinda Washbourne. JK Coller is a FTT Fricker Research Fellow at the University of Adelaide. This research was financially supported by the National Health and Medical Research Council of Australia (Grant No 299050) and by the Faculty of Health Sciences of the University of Adelaide.
References
- 1.Drakenberg N, Nikoshkov A, Horvath MC, et al. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc Natl Acad Sci U S A. 2006;103:7883–7888. doi: 10.1073/pnas.0600871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond C, LaForge KS, Tian M, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters b-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda K, Ide S, Han W, et al. How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol Sci. 2005;26:311–317. doi: 10.1016/j.tips.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Kreek MJ, LaForge KS. Stress responsivity, addiction, and a functional variant of the human mu-opioid receptor gene. Mol Intervent. 2007;7:74–78. doi: 10.1124/mi.7.2.7. [DOI] [PubMed] [Google Scholar]
- 5.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the m-opioid receptor gene with substance dependence: A meta-analysis. Drug Alcohol Depend. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 6.Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Tsuang MT, Lyons MJ, Meyer JM, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 8.Bart G, Heilig M, LaForge KS, et al. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glatt SJ, Bousman C, Wang RS, et al. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug Alcohol Depend. 2007;90:159–165. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landau R, Kern C, Columb MO, Smiley RM, Blouin J-L. Genetic variability of the mu-opioid receptor influences intrathecal fentanyl analgesia requirements in laboring women. Pain. 2008;139:5–14. doi: 10.1016/j.pain.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley JJ, Oslin DW, Patkar AA, et al. A genetic association study of the mu opioid receptor and severe opioid dependence. Psychiatr Genet. 2003;13:169–173. doi: 10.1097/00041444-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Franke P, Wang T, Nothen MM, et al. Nonreplication of association between m-opioid receptor gene (OPRM1) A118G polymorphism and substance dependence. Am J Med Genet. 2001;105:114–119. [PubMed] [Google Scholar]
- 13.Kapur S, Sharad S, Singh RA, Gupta AK. A118G polymorphism in mu opioid receptor gene (OPRM1): association with opiate addiction in subjects of Indian origin. J Integr Neurosci. 2007;6:511–522. doi: 10.1142/s0219635207001635. [DOI] [PubMed] [Google Scholar]
- 14.Crettol S, Besson J, Croquette-Krokar M, et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuro-Psychopharmacol Biol Psych. 2008;32:1722–1727. doi: 10.1016/j.pnpbp.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Jorm AF, Prior M, Sanson A, et al. Lack of association of a single-nucleotide polymorphism of the m-opioid receptor gene with anxiety-related traits: results from a cross-sectional study of adults and a longitudinal study of children. Am J Med Genet. 2002;114:659–664. doi: 10.1002/ajmg.10643. [DOI] [PubMed] [Google Scholar]
- 16.Gelernter J, Kranzler H, Cubells J. Genetics of two mu opioid receptor gene (OPRM1) exon 1 polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjects. Mol Psychiatry. 1999;4:476–483. doi: 10.1038/sj.mp.4000556. [DOI] [PubMed] [Google Scholar]
- 17.Hoehe MR, Kopke K, Wendel B, et al. Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum Mol Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- 18.Li T, Liu X, Zhu ZH, et al. Association analysis of polymorphisms in the mu opioid gene and heroin abuse in Chinese subjects. Addict Biol. 2000;5:181–186. doi: 10.1080/13556210050003775. [DOI] [PubMed] [Google Scholar]
- 19.Luo X, Kranzlet HR, Zhao H, Gelernter J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am J Med Genet. 2003;120B:97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- 20.Shi J, Hui L, Xu Y, et al. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum Mutat. 2002;19:459–460. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- 21.Szeto CY, Tang NL, Lee DT, Stadlin A. Association between mu opioid receptor gene polymorphisms and Chinese heroin addicts. Neuroreport. 2001;12:1103–1106. doi: 10.1097/00001756-200105080-00011. [DOI] [PubMed] [Google Scholar]
- 22.Tan EC, Tan CH, Karupathivan U, Yap EP. Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport. 2003;14:569–572. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Luo X, Kranzler HR, et al. Association between two m-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders (DSM-III-R) 3rd revised ed. Washington, DC: American Psychiatric Association; 1987. [Google Scholar]
- 25.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) 4th ed. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 26.Coffman BL, Kearney WR, Goldsmity S, Knosp BM, Tephly TR. Opioids bind to the amino acids 84 and 118 of UDP-glucuronosyltransferase UGT2B7. Mol Pharmacol. 2003;63:283–288. doi: 10.1124/mol.63.2.283. [DOI] [PubMed] [Google Scholar]
- 27.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 28.Livingston M, Messura J, Dellinger T, Holder R, Hyde J. Meta-analysis: an introduction into a research process. Spec Care Dentist. 2008;28:125–130. doi: 10.1111/j.1754-4505.2008.00033.x. [DOI] [PubMed] [Google Scholar]
- 29.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 30.Sharp S. sbe23: Meta-analysis regression. Stata Tech Bull. 1998;42:16–24. [Google Scholar]
- 31.Egger M, Davey Smith G, Schnider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 33.Pud D, Yarnitsky D, Sprecher E, et al. Can personality traits and gender predict the response to morphine? An experimental cold pain study. Eur J Pain. 2006;10:103–112. doi: 10.1016/j.ejpain.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Ray R, Jepson C, Patterson F, et al. Association of OPRM1 A118G variant with the relative reinforcing value of nicotine. Psychopharmacology. 2006;188:355–363. doi: 10.1007/s00213-006-0504-2. [DOI] [PubMed] [Google Scholar]
- 35.Somogyi A A, Barratt DT, Coller JK. Pharmacogenetics of opioids. Clin Pharmacol Ther. 2007;81:429–444. doi: 10.1038/sj.clpt.6100095. [DOI] [PubMed] [Google Scholar]