Abstract
To identify the proteins induced by Fe deficiency, we have compared the proteins of Fe-sufficient and Fe-deficient barley (Hordeum vulgare L.) roots by two-dimensional polyacrylamide gel electrophoresis. Peptide sequence analysis of induced proteins revealed that formate dehydrogenase (FDH), adenine phosphoribosyltransferase, and the Ids3 gene product (for Fe deficiency-specific) increased in Fe-deficient roots. FDH enzyme activity was detected in Fe-deficient roots but not in Fe-sufficient roots. A cDNA encoding FDH (Fdh) was cloned and sequenced. Fdh expression was induced by Fe deficiency. Fdh was also expressed under anaerobic stress and its expression was more rapid than that induced by Fe deficiency. Thus, the expression of Fdh observed in Fe-deficient barley roots appeared to be a secondary effect caused by oxygen deficiency in Fe-deficient plants.
In Fe-deficient calcareous soils graminaceous plants secrete mugineic acid family phytosiderophores, which are natural Fe chelators, from the roots (Takagi, 1976) to solubilize Fe required for plant growth. This Fe-acquisition mechanism in graminaceous plants is called strategy II and in nongraminaceous plants it is called strategy I (Takagi et al., 1984; Marschner et al., 1986). The pathway of the biosynthesis of mugineic acid family phytosiderophores has been established (Mori and Nishizawa, 1987, 1989; Shojima et al., 1989, 1990; Mori et al., 1990; Ma and Nomoto, 1993). Among the enzymes involved in this biosynthetic pathway, Higuchi et al. (1994, 1996) purified nicotianamine synthase and Kanazawa et al. (1994) purified nicotianamine aminotransferase. Comparison of 2D profiles of proteins in barley (Hordeum vulgare L.) roots under Fe-sufficient and Fe-deficient conditions (Suzuki et al., 1995, 1997) allowed us to identify a 36-kD protein that was specifically induced by Fe deficiency. In addition, several genes related to the Fe-deficiency response have been reported: Ids1 (Okumura et al., 1991), Ids2 (Okumura et al., 1994), and Ids3 (Nakanishi et al., 1993). In this study, we characterized several other proteins induced by Fe-deficiency stress in barley roots, one of which was identified as FDH. FDH was induced not only by Fe deficiency but also by anaerobic stress. The relationship between Fe deficiency and anaerobic stress in barley roots is discussed.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of barley (Hordeum vulgare L. cv Ehimehadaka no. 1) were germinated at room temperature on paper towels soaked with distilled water. Plants were transferred 4 d after germination to a plastic net floating on tap water at pH 5.5 in a greenhouse under natural light. On d 10, plants were transferred to a continuously aerated nutrient solution of the following composition: 0.7 mm K2SO4, 0.1 mm KCl, 0.1 mm KH2PO4, 2.0 mm Ca(NO3)2, 0.5 mm MgSO4, 10 μm H3BO3, 0.5 μm MnSO4, 0.2 μm CuSO4, 0.5 μm ZnSO4, 0.01 μm (NH4)6Mo7O24, and 0.1 mm Fe-EDTA. The pH of the culture solution was adjusted to 5.5 daily with 1 n HCl. Fe deficiency was started on d 20 using the same solution, but without Fe-EDTA. The nutrient solution was changed every 7 d. Plant roots were harvested 40 d after germination. Anaerobiosis was achieved by bubbling nitrogen gas through the nutrient solution overnight to purge oxygen gas in the solution, followed by a continuous flow of nitrogen gas throughout the anaerobic experiment.
Protein Extraction for 2D PAGE
The procedure for extraction of proteins was as described by Damerval et al. (1986) with slight modifications. The roots were homogenized in liquid nitrogen with a mortar and pestle, and the powder was resuspended in a cold solution of 10% (w/v) TCA in acetone with 0.1% (v/v) 2-ME. Proteins were allowed to precipitate for 60 min at −20°C and were then centrifuged at 16,000g for 30 min at 4°C. The supernatant solution was discarded and the pellet was rinsed with cold acetone containing 0.1% (v/v) 2-ME for 60 min at −20°C and then centrifuged at 16,000g for 30 min at 4°C. The supernatant solution was discarded and the pellet was dried under reduced pressure, dissolved (50 μL mg−1 dry weight) in sample buffer (9.5 m urea, 2% [w/v] Triton X-100, and 5% [v/v] 2-ME), and centrifuged at 16,000g for 10 min at room temperature. The supernatant solution was used for 2D PAGE. Protein concentrations were estimated by the method of Bradford (1976).
2D PAGE
2D PAGE was performed following the method of O'Farrell (1975). Gel length in the column (2.5 × 130 mm) was 100 mm. To cover the pI range from 5.0 to 8.0, the gel contained 1.6% (v/v) pH 5.0 to 8.0 ampholines and 0.4% (v/v) pH 3.0 to 10.0 ampholines. Protein extracts (200 μg) were subjected to IEF at 400 V for 15 h and at 800 V for 1 h, and the gels were equilibrated for 15 min in the SDS-PAGE sample buffer (2.3% [w/v] SDS, 10% [w/v] glycerol, 5% [v/v] 2-ME, 62.5 mm Tris-HCl, pH 6.8, and 0.1% [w/v] bromphenol blue), before loading onto slab gels for 12.5% (w/v) SDS-PAGE in the second dimension. The gel was stained with 0.25% (w/v) Coomassie brilliant blue R-250 in a mixture of 50% (v/v) methanol and 10% (v/v) acetate and destained in a solution of 50% (v/v) methanol and 10% (v/v) acetate.
Chemical and Enzymatic Digestion of Proteins and Amino Acid Sequence Analysis
Chemical or enzymatic digestion was used to determine the internal sequence of the proteins. Chemical digestion with CNBr was according to the method of Gross (1967) with the following modifications. Isolated protein spots from 50 2D PAGE gels were pooled by electroblotting onto a PVDF membrane according to the method of Towbin et al. (1979). Proteins were eluted from the membrane by soaking in a 10-fold volume of 70% (v/v) formic acid containing 1% (w/v) CNBr in a 1.5-mL microtube and incubating overnight at 4°C. The supernatant was collected, dried under reduced pressure, resuspended in the SDS-PAGE sample buffer, and incubated overnight at room temperature. Enzymatic digestion was performed according to the method of Cleveland et al. (1977) or Aebersold et al. (1987). After digestion of proteins, the peptides were separated by electrophoresis using Tricine/SDS-PAGE (Schägger and von Jagow, 1987) in 16.5% (w/v) acrylamide gels. Peptides were transferred by electroblotting onto a PVDF membrane and stained with Coomassie brilliant blue. Each band on the PVDF membrane was cut out and the amino acid sequence was determined by automated Edman degradation on a gas-phase sequencer (model 477A protein sequencer and model 120A PTH analyzer, Applied Biosystems).
FDH Assay
For the assay of FDH activity, the roots were homogenized in liquid nitrogen with a mortar and pestle, and the powder was resuspended in the following buffer: 100 mm Tris-HCl, pH 8.0, 1 mm PMSF, 1 mm EDTA, and 0.2% (w/v) Triton X-100. FDH activity on nondenaturing polyacrylamide gels was visualized according to the method of Uotila and Koivusalo (1979) as follows. A 7% (w/v) native acrylamide gel was used with the discontinuous buffer system of Laemmli (1970). One-hundred micrograms of soluble protein from roots or leaves was fractionated at 100 V for 2 h at room temperature and then incubated in darkness for 30 min at room temperature in the following solution: 100 mm sodium phosphate buffer, pH 7.0, 50 mm sodium formate, 0.8 mm NAD+, 0.03 mg mL−1 phenazine methosulfate, and 0.4 mg mL−1 nitroblue tetrazolium.
Cloning of cDNA Encoding FDH
A pYH23 cDNA library prepared from poly(A+) RNA of Fe-deficient barley roots (kindly provided by Hirotaka Yamaguchi, The University of Tokyo) was screened with an FDH PCR product corresponding to the partial amino acid sequences of FDH: GGIGTITTYTAYCARGCIGGIGARTAY and GCRTCIGCIACIGCYTGIGTRTCCAT (shaded sequences in Fig. 3). The PCR probe was labeled with a random-primer-labeling kit (version 2, Takara Biomedicals, Gennevilliers, France) in the presence of [α-32P]dATP. The cloned cDNA was sequenced according to the protocol of a sequencing kit (Dye Terminator Cycle Sequencing Ready Reaction kit, Perkin-Elmer) using a DNA sequencer (model A373, Applied Biosystems). Hybridization probes for Southern blotting were prepared by digesting the cloned cDNA corresponding to the FDH gene (Fdh) with SacI, and the smaller fragment (Fig. 3, underlined) was radiolabeled as described above. The labeled DNA was purified on a Nick column (Pharmacia) and used as a probe for both Southern and northern hybridization analyses.
Figure 3.
DNA and deduced amino acid sequence of barley FDH. Position of primers for PCR are shaded and the SacI fragment is underlined.
Genomic Southern Hybridization
Barley genomic DNA was prepared from leaves by the method of Murray and Thompson (1980) using cetyltrimethylammonium bromide. The DNA was digested with BamHI, EcoRI, or HindIII, separated on a 0.8% (w/v) agarose gel (30 μg per lane), and alkali-transferred onto a nylon membrane (Hybond-N+, Amersham). The membrane was hybridized with the labeled SacI fragment of Fdh with 5× SSPE, 4× Denhardt's solution, and 100 μg mL−1 salmon-sperm DNA at 65°C overnight. The washing conditions were three times with 2× SSPE plus 0.1% (w/v) SDS at 65°C for 30 min.
RNA Isolation and Northern Hybridization
Total RNA was isolated from roots or leaves according to the procedure of Logmann et al. (1987). RNA (10 μg per lane) was separated on 1.2% agarose gels containing 5% (v/v) formaldehyde and blotted onto nylon membranes (Hybond-N+, Amersham). The membrane was hybridized with the SacI fragment of Fdh under the same conditions described above. The washing conditions were: 6× SSPE at 65°C for 10 min and then twice with 2× SSPE plus 0.1% (w/v) SDS at 65°C for 10 min. The radioactivity was detected and quantified using an image analyzer (BAS-2000, Fuji, Tokyo, Japan).
RESULTS
2D Electrophoresis of Barley Root Proteins
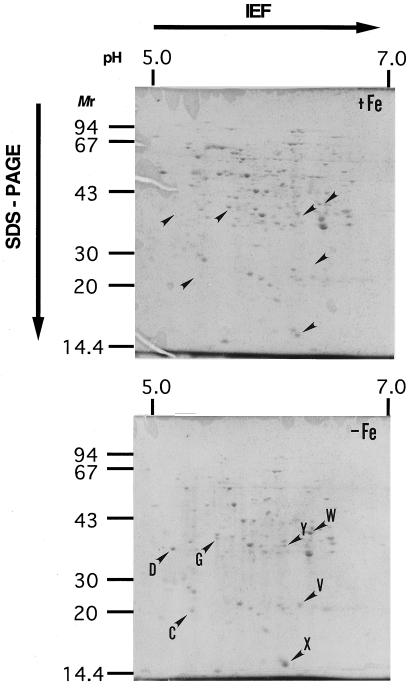
Proteins prepared from Fe-sufficient and Fe-deficient roots and analyzed on 2D PAGE gels showed different protein patterns after Coomassie brilliant blue staining (Fig. 1). In the roots of Fe-deficient plants, the protein spots named C, D, G, V, W, X, and Y were present at higher concentrations than in Fe-sufficient plants (Fig. 1). Protein spots C, D, G, and V had previously been observed to increase during Fe deficiency (Mori et al., 1988; Suzuki et al., 1995), but the W, X, and Y spots were newly identified in this study.
Figure 1.
2D PAGE gel from barley roots grown under Fe-sufficient (+Fe) and Fe-deficient (−Fe) conditions. Each gel was loaded with 200 μg of root proteins. Several spots of proteins that increased under Fe deficiency are indicated with arrowheads. C, Adenine phosphoribosyltransferase; Y, IDS3; and W, FDH. Other spots are unknown (Table I). Mr is reported in thousands.
Amino Acid Sequences of Each Protein
Among the seven proteins with increased concentrations in Fe-deficient roots, the D protein has been previously identified as a 36-kD peptide (Suzuki et al., 1995). Three proteins (C, W, and Y) were successfully sequenced. The N-terminal sequence of protein W was sequenced by Edman degradation, but the N termini of C and Y appeared to be blocked. The W protein appeared to be FDH (EC 1.2.1.2) based on homology of the sequences of internal peptides from CNBr digestion fragments (WCN-3, -4, -5, -6, and -7 in Table I) to other FDH sequences (the SwissProt and GenBank databases were used for the homology search). FDH was previously reported to be expressed in Fe-deficient roots of tomato and in the roots of the nicotianamine-free mutant chloronerva by Herbik et al. (1996). The C protein was transferred onto a PVDF membrane and digested with CNBr (CCNS-2 and CCNM-1 in Table I) or lysilendopeptidase (CLP-1 and CLP-2 in Table I). The sequences of peptides from C obtained by chemical or enzymatic digestion suggested that it is adenine phosphoribosyltransferase (EC 2.4.2.7). The sequences of CNBr digestion fragments from the Y protein (YCN-2, -3, and -4 in Table I) completely matched the translated products from the cDNA sequence of the Ids3 gene (Nakanishi et al., 1993, accession nos. D37796 and D10058). Thus, FDH, adenine phosphoribosyltransferase, and the Ids3 gene product showed increased accumulation in Fe-deficient barley roots.
Table I.
Sequences of CNBr digestion fragments
| Spot | Fragment | Sequence |
|---|---|---|
| W | N terminus | AHT?AGLKKI |
| WCN-7 | DTQAVADA?SRGHIA?YGS? | |
| WCN-6 | FVLITGPFHAPYVTHGERIK | |
| WCN-5 | AHTSAGSKKIVGVFYQAGEY | |
| WCN-4 | RILKLLRN | |
| WCN-3 | RILFKLRNFLPGYQQVMKGE | |
| Y | N terminus | Blocked |
| YCN-4 | ENILHATPAPV | |
| YCN-3 | ?EQFFHLPA?DKA?LY?E | |
| YCN-2 | GIQADYFEGD L?G?NVIL?I | |
| C | N terminus | Blocked |
| CLP-1 | GKPGEVISEEYSLEYG?DKI | |
| CLP-2 | RIPGEVIP | |
| CCNS-2 | HVGHVSPNDRDLIVDDLIHQ | |
| CCNM-1 | HDGAVAKLASRLGAKVVEIA |
FDH Assay
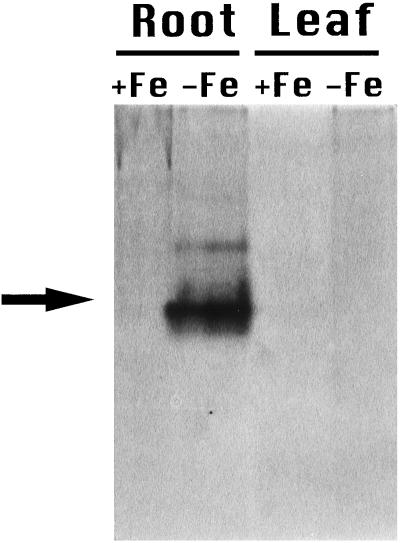
An increase in FDH activity (Fig. 2) was also detected in Fe-deficient roots, confirming the accumulation of FDH observed by 2D PAGE. FDH activity was not detected in Fe-deficient leaves, Fe-sufficient roots, or Fe-sufficient leaves at the protein concentration tested (100 μg).
Figure 2.
FDH assay on nondenaturing polyacrylamide gel. Each lane was loaded with 100 μg of soluble proteins from barley roots or leaves of Fe-sufficient (+Fe) or Fe-deficient (−Fe) plants.
Isolation of Barley cDNA Encoding FDH
A cDNA library prepared from poly(A+) RNA of Fe-deficient barley roots was screened with a PCR probe designed to encode FDH (Fig. 3). Several clones were thus identified. The cloned cDNA encoded 377 amino acids of an open reading frame corresponding to a protein with a molecular mass of 41.5 kD and a pI of 6.7. The deduced molecular mass and pI were those of W in 2D PAGE. A sequence consisting of 21 amino acids at the N terminus might be a transit peptide targeted to mitochondria, based on comparison with potato (Solanum tuberosum) tuber FDH (Colas des Francs-Small et al., 1993). Comparison of barley FDH with FDH from other plants or bacteria is shown in Figure 4. The NAD+-binding site from Lys-193 to Asn-228 (Lamzin et al., 1992) and the formate-binding site from Arg-285 are indicated. The sequences were highly conserved between barley and potato in contrast to the sequences at the N terminus, which displayed very low sequence homology.
Figure 4.
Comparison of amino acid sequences from barley FDH with FDH from other organisms. The partial amino acid sequences from the W protein are underlined. The probable NAD+-binding site is boxed, and the formate-binding site is shaded (Lamzin et al., 1992). The homology between barley FDH and other FDHs was 82.7% (S. tuberosum; Colas des Francs-Small et al., 1993), 53.0% (N. crassa; Chow and RajBhandary, 1993), 50.9% (Pseudomonas sp.; Tishkov et al., 1991), 50.0% (H. polymorpha; sequence A06214 was submitted to EMBL data bank by C.P. Hollenberg and Z. Janowicz in 1989), and 49.6% (C. methylica; Allen and Holbrook, 1995).
Southern Hybridization Analysis
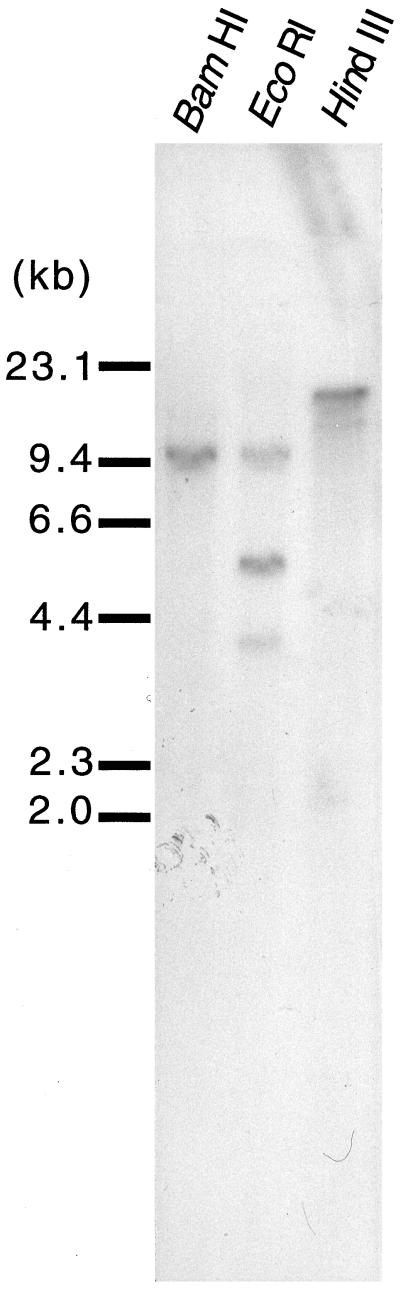
The copy number of Fdh in barley was assessed by Southern hybridization analysis (Fig. 5). One fragment was observed in the BamHI- and HindIII-digested DNA. Three fragments were detected in the EcoRI lane but the largest (10 kb) may represent an incomplete digestion product, since the sum of the molecular masses of the other two fragments (5.5 and 4.2 kb) is approximately 10 kb. Since there were no restriction enzyme sites for BamHI, EcoRI, and HindIII in the cloned Fdh cDNA, we conclude that the Fdh gene is a single copy in the barley genome and that an EcoRI site is probably present within an intron.
Figure 5.
Southern hybridization analysis of Fdh. Genomic DNA from barley was digested with BamHI, EcoRI, and HindIII and then blotted onto a nylon membrane and hybridized with a 32P-labeled SacI fragment of Fdh.
Northern Hybridization Analysis
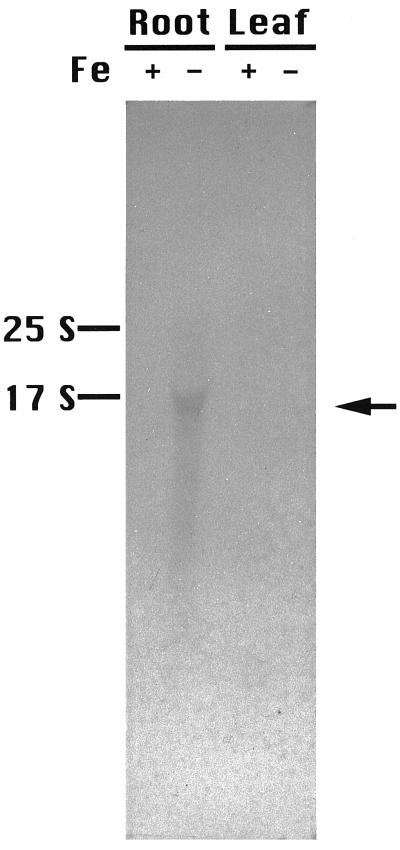
To investigate the expression of Fdh, northern hybridization analysis was performed (Fig. 6). In the control (Fe-sufficient) plants, no Fdh mRNA was detected in either the leaves or the roots (Fig. 6, compare +Fe leaf and +Fe root). In contrast, Fdh was strongly expressed in the roots of Fe-deficient plants but not in the leaves (Fig. 6, compare −Fe leaf and −Fe root).
Figure 6.
Northern hybridization analysis of Fdh. RNA was extracted from Fe-deficient (−) or Fe-sufficient (+) roots or leaves. Each lane was loaded with 10 μg of RNA. Total RNA was extracted after 14 d of Fe deficiency.
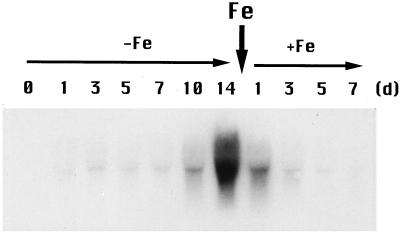
The induction of Fdh expression required 1 d of Fe deficiency, with the amount of Fdh mRNA increasing gradually day by day (Fig. 7). After 14 d, Fdh expression reached a maximum and remained constant for 28 d. When Fe was resupplied in the form of Fe-EDTA to the culture solution of Fe-deficient plants, Fdh mRNA quickly diminished and was barely detectable on d 7 after the addition of Fe.
Figure 7.
Expression of Fdh during Fe deficiency in barley roots. Each lane was loaded with 10 μg of RNA. Total RNA was isolated after 0, 1, 3, 5, 7, 10, and 14 d of Fe deficiency and 1, 3, 5, and 7 d after Fe resupply.
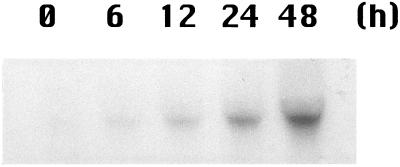
In bacteria and unicellular algae, formate is produced in large quantities under anaerobic conditions (Kreuzberg, 1984; Ferry, 1990). We therefore examined the expression of barley Fdh under anaerobic conditions. As in prokaryotes, Fdh expression increased in barley under anaerobic conditions (Fig. 8); the increase in Fdh mRNA began at 12 h compared with 1 d for Fe deficiency. The amount of Fdh mRNA present after 48 h of anaerobiosis was approximately equivalent to 10 to 14 d of Fe deficiency (quantification by the image analyzer is not shown).
Figure 8.
Expression of Fdh under anaerobic conditions. Each lane was loaded with 10 μg of RNA. Total RNA was isolated after 0, 6, 12, 24, and 48 h of anaerobic treatment.
DISCUSSION
Based on peptide sequencing of protein spot W, FDH was among the proteins induced by Fe deficiency in barley roots. FDH activity and mRNA were detected in Fe-deficient barley roots but were undetectable in Fe-deficient barley leaves (Figs. 2 and 6). In Pseudomonas sp. FDH, Arg-288 has been proposed to be a formate-binding site (Lamzin et al., 1994; Popov and Lamzin, 1994), and the His-341–Gln-317 pair is necessary for the binding of formate (Tishkov et al., 1996). These amino acids are conserved in all plant FDHs, including that of barley (Fig. 4). Yeast (Hansenula polymorpha and Candida methylica) FDH and Neurospora crassa FDH have two inserted regions that are absent in barley, potato, and Pseudomonas sp. (Fig. 3, residues 134–135 and 331–335).
Colas des Francs-Small et al. (1993) reported that FDH in potato tubers was located in the mitochondria. The N-terminal sequence of barley FDH shows little homology to potato FDH, but its hydropathy plot is similar to that of transit peptides for mitochondria targeting. Therefore, FDH activity detected in barley roots might be derived from mitochondria.
FDH mRNA was detectable after 1 d of Fe deficiency, reaching the maximal level after 2 weeks. A few days after the addition of Fe into the Fe-deficient solution, Fdh expression decreased and on d 7 was undetectable. Although this inductive response pattern to Fe deficiency is very similar to that of Ids3, the response to the Fe resupply is slower (Nakanishi et al., 1993). Ids3 is one of the clones we have isolated in barley roots by the differential hybridization method, and it supposedly encodes a putative mugineic acid synthase. We observed that the transcript of Ids3 gene is increased by Fe deficiency and we have confirmed in this experiment that Ids3 is actually translated and the Ids3 protein is accumulated in Fe-deficient barley roots (Fig. 1; Table I).
In addition to Fe-deficient conditions, the barley Fdh was also expressed under anaerobic conditions (Fig. 8). Moreover, this inductive response to anaerobic stress began 12 h after treatment and was more rapid than the response to Fe deficiency. Formate is reported to be produced in large quantities in bacteria (Ferry, 1990) and unicellular algae (Kreuzberg, 1984) under anaerobic conditions. Colas des Francs-Small et al. (1993) suggested that a major, uncharacterized metabolic pathway exists in the mitochondria of nonphotosynthetic tissues, which produces large quantities of formate. Therefore, induction of FDH by anaerobic stress in barley roots (Figs. 7 and 8) is conceivable in light of the above-mentioned results in bacteria and algae.
The more rapid response of Fdh transcript to anaerobic stress than that to Fe deficiency indicates that the expression of Fdh is primarily induced by anaerobic stress. The expression of Fdh in Fe-deficient barley roots suggests that Fe deficiency caused changes similar to the ones caused by anoxia through changes in heme biosynthesis. For example, Fe regulates the biosynthesis of δ-aminolevulinic acid (by δ-aminolevulinic acid synthase; Pushnik and Miller, 1989) and protoporphyrinogen IX (by co-proporphyrinogen III oxidase). These are the common precursors of protoporphyrin IX, from which chlorophyll a in the chloroplasts and heme in the mitochondria are synthesized. Moreover, Fe regulates the biosynthesis of divinyl protochlorophyllide (by Mg-protoporphyrin IX monomethylester cyclase), which is the precursor of chlorophyll in the chloroplasts (von Wettstein et al., 1995).
Fe is also incorporated into protoporphyrin IX to become heme in the mitochondria. Therefore, Fe deficiency not only lowers the amount of chlorophyll in the chloroplasts of shoots but also the amount of heme in the mitochondria of both shoots and roots (Marschner, 1995). Since large amounts of heme are needed for energy production by the respiratory chain, the inhibition of energy production by decreasing respiration in mitochondria could be caused by Fe deficiency. In addition, we previously reported that Fe deficiency in rice roots caused morphological malformation of mitochondria and a decrease in the energy charge from 0.748 to 0.520 (Mori et al., 1991). Therefore, anaerobiosis-like changes may be produced in mitochondria of Fe-deficient barley roots.
Anaerobic stress causes acute damage to the plant by reducing available energy, which may result in its turning to formate metabolism to produce NADH by FDH. On the other hand, Fe deficiency may cause anoxia by the depletion of Fe from heme and, secondarily, by the depletion of heme protein as the result of inhibition of heme biosynthesis. If the metabolism of formate in plant roots is similar to that of bacteria, as was suggested by Colas des Francs-Small et al. (1993), the formate pathway would be induced either by the decrease of heme (Fe deficiency) or by reduced electron transport in the respiratory chain of mitochondria (anaerobiosis). In conclusion, FDH induction might be caused by the anoxia induced by Fe deficiency in spite of the presence of oxygen.
ACKNOWLEDGMENT
We thank Professor Elizabeth C. Theil of North Carolina State University (Raleigh) for editing English usage.
Abbreviations:
- CNBr
cyanogen bromide
- FDH
formate dehydrogenase
- 2D
two-dimensional
- 2-ME
2-mercaptoethanol
Footnotes
This work has been supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation (Tsukuba, Japan).
LITERATURE CITED
- Aebersold RH, Leavitt J, Saavedra RA, Hood LE, Kent SBH. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ digestion on nitrocellulose. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SJ, Holbrook JJ. Isolation, sequence and overexpression of the gene encoding NAD-dependent formate dehydrogenase from the methylotrophic yeast Candida methylica. Gene. 1995;162:99–104. doi: 10.1016/0378-1119(95)00347-9. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chow CM, RajBhandary UL. Developmental regulation of the gene for formate dehydrogenase in Neurospora crassa. J Bacteriol. 1993;175:3703–3739. doi: 10.1128/jb.175.12.3703-3709.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Fischer SG, Kirshner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Rémy R. Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol. 1993;102:1171–1177. doi: 10.1104/pp.102.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, de Vienne D, Zivy M, Thiellement H. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis. 1986;7:52–54. [Google Scholar]
- Ferry JG. Formate dehydrogenase. FEMS Microbiol Rev. 1990;87:377–382. doi: 10.1111/j.1574-6968.1990.tb04940.x. [DOI] [PubMed] [Google Scholar]
- Gross E. The cyanogen bromide reaction. Methods Enzymol. 1976;11:238–255. [Google Scholar]
- Herbik A, Giritch A, Horstmann C, Becker R, Blazer HJ, Bäumlein H, Stephan UW. Iron and copper nutrition-dependent changes in protein expression in a tomato wild type and the nicotianamine-free mutant chloronerva. Plant Physiol. 1996;111:533–540. doi: 10.1104/pp.111.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Chino M, Mori S. Purification and characterization of nicotianamine synthase from Fe-deficient barley roots. Plant Soil. 1994;165:173–179. [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Mori S. The role of nicotianamine synthase in response to Fe nutrition status in Gramineae. Plant Soil. 1996;178:171–177. [Google Scholar]
- Kanazawa K, Higuchi K, Nishizawa NK, Fushiya S, Chino M, Mori S. Nicotianamine aminotransferase activities are correlated to the phytosiderophore secretions under Fe-deficient conditions in Gramineae. J Exp Bot. 1994;45:1903–1906. [Google Scholar]
- Kreuzberg K. Starch fermentation via formate producing pathway Chlamydomonas reinhardii, Chlorogonium elongatum, and Chlorella fusca. Physiol Plant. 1984;61:87–94. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamzin VS, Aleshin AE, Srtokopytov BV, Yukhnevich MG, Popov VO, Harutyunyan EH, Wilson KS. Crystal structure of NAD-dependent formate dehydrogenase. Eur J Biochem. 1992;206:441–452. doi: 10.1111/j.1432-1033.1992.tb16945.x. [DOI] [PubMed] [Google Scholar]
- Lamzin VS, Dauter Z, Popov VO, Harutyunyan EH, Wilson KS. Structure of holo and high resolution apo formate dehydrogenase. J Mol Biol. 1994;236:759–785. doi: 10.1006/jmbi.1994.1188. [DOI] [PubMed] [Google Scholar]
- Logmann J, Schelland J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Ma JF, Nomoto K. Two related biosynthetic pathway of mugineic acid in graminaceous plants. Plant Physiol. 1993;102:373–378. doi: 10.1104/pp.102.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Functions of mineral nutritions: micronutrients. In: Marschner R, editor. Mineral Nutrition of Higher Plants, Ed 2. London: Academic Press; 1995. pp. 313–404. [Google Scholar]
- Marschner H, Römheld V, Kissel M. Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr. 1986;9:695–713. [Google Scholar]
- Mori S, Hachisuka M, Kawai S, Takagi S, Nishizawa NK. Peptides related to phytosiderophore secretion by Fe-deficient barley roots. J Plant Nutr. 1988;11:653–662. [Google Scholar]
- Mori S, Nishizawa N. Methionine as a dominant precursor of phytosiderophores in Gramineae plants. Plant Cell Physiol. 1987;28:1081–1092. [Google Scholar]
- Mori S, Nishizawa N. Identification of barley chromosome no. 4, possible encoder of genes of mugineic acid synthesis from 2′-deoxymugineic acid using wheat-barley addition lines. Plant Cell Physiol. 1989;30:1057–1060. [Google Scholar]
- Mori S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T. Identification of rye chromosome 5R as a carrier of the genes for mugineic acid synthase and hydroxymugineic acid synthase using wheat-rye addition lines. Jpn J Genet. 1990;65:343–352. [Google Scholar]
- Mori S, Nishizawa NK, Hayashi H, Chino M, Yoshimura E, Ishihara J. Why are young rice plants highly susceptible to iron deficiency? Plant Soil. 1991;130:143–156. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Okumura N, Umehara Y, Nishizawa NK, Chino M, Mori S. Expression of a gene specific for iron deficiency (Ids3) in the roots of Hordeum vulgare. Plant Cell Physiol. 1993;34:401–410. [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Okumura N, Nishizawa NK, Umehara Y, Mori S. An iron deficiency specific cDNA (Ids1) with two homologous cysteine rich domains. Plant Mol Biol. 1991;17:531–535. doi: 10.1007/BF00040651. [DOI] [PubMed] [Google Scholar]
- Okumura N, Nishizawa NK, Umehara Y, Ohata T, Nakanishi H, Yamaguchi T, Chino M, Mori S. A dioxygenase (Ids2) expressed under iron deficiency conditions in the roots of Hordeum vulgare. Plant Mol Biol. 1994;25:705–719. doi: 10.1007/BF00029608. [DOI] [PubMed] [Google Scholar]
- Popov VO, Lamzin VS. NAD+-dependent formate dehydrogenase. Biochem J. 1994;301:625–643. doi: 10.1042/bj3010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushnik JC, Miller GW. Iron regulation of chloroplast photosynthetic function: mediation of PSI development. J Plant Nutr. 1989;12:407–421. [Google Scholar]
- Schägger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S. Biosynthesis of phytosiderophores: in vitro biosynthesis of 2′-deoxymugineic acid from l-methionine and nicotianamine. Plant Physiol. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Mori S. Establishment of a cell free system for the biosynthesis of nicotianamine. Plant Cell Physiol. 1989;30:673–677. [Google Scholar]
- Suzuki K, Hirano H, Yamaguchi H, Irifune T, Nishizawa NK, Chino M, Mori S. Partial amino acid sequences of a peptide induced by Fe deficiency in barley roots. In: Abadía J, editor. Iron Nutrition in Soils and Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 363–369. [Google Scholar]
- Suzuki K, Kanazawa K, Higuchi K, Nishizawa NK, Mori S. Immunological characterization of a 36 kDa Fe-deficiency specific peptide in barley roots. Biometals. 1997;10:77–84. [Google Scholar]
- Takagi S. Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Sci Plant Nutr. 1976;22:423–433. [Google Scholar]
- Takagi S, Nomoto K, Takemoto S. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J Plant Nutr. 1984;7:469–477. [Google Scholar]
- Tishkov VI, Galkin AG, Egorov AM. NAD-dependent formate dehydrogenase from the methylotrophic bacterium Pseudomonas sp. 101: cloning, expression and the study of gene structure. Dokl Acad Nauk USSR. 1991;317:745–748. [PubMed] [Google Scholar]
- Tishkov VI, Matorin AD, Rojkova AM, Fedorchuk VV, Savitsky PA, Dementieva LA, Lamzin VS, Mezentzev AV, Popov VO. Site-directed mutagenesis of the formate dehydrogenase active center: role of the His332-Gln313 pair in enzyme catalysis. FEBS Lett. 1996;390:104–108. doi: 10.1016/0014-5793(96)00641-2. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uotila L, Koivusalo M. Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys. 1979;196:33–45. doi: 10.1016/0003-9861(79)90548-4. [DOI] [PubMed] [Google Scholar]
- von Wettstein D, Gough S, Kannangara G. Chlorophyll biosynthesis. Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]