Abstract
Advanced stage non-small cell lung cancer and head and neck squamous cell carcinoma are both treated with DNA damaging agents including platinum-based compounds and radiation therapy. However, at least one quarter of all tumors are resistant or refractory to these genotoxic agents. Yet the agents are extremely toxic, leading to undesirable side effects with potentially no benefit. Alternative therapies exist, but currently there are no tools to predict whether the first-line genotoxic agents will work in any given patient. To maximize therapeutic success and limit unnecessary toxicity, emerging clinical trials aim to inform personalized treatments tailored to the biology of individual tumors. Worldwide, significant resources have been invested in identifying biomarkers for guiding the treatment of lung and head and neck cancer. DNA repair proteins of the nucleotide excision repair pathway (ERCC1) and of the base excision repair pathway (XRCC1), which are instrumental in clearing DNA damage caused by platinum drugs and radiation, have been extensively studied as potential biomarkers of clinical outcomes in lung and head and neck cancers. The results are complex and contradictory. Here we summarize the current status of single nucleotide polymorphisms, mRNA, and protein expression of ERCC1 and XRCC1 in relation to cancer risk and patient outcomes.
Keywords: nucleotide excision repair, base excision repair, DNA damage, DNA repair, chemotherapy, NSCLC, HNSCC, single nucleotide polymorphism
Introduction
Lung cancer is the second most common cancer in the USA and is the leading cause of cancer-related death.1 Based on the predicted response to treatment and known risk factors, lung cancers are categorized in two groups: small cell and non-small cell lung cancers (NSCLC). NSCLC are more frequent, and smoking is a risk factor. Histologically, NSCLC are composed mainly of adenocarcinoma and, to a lesser degree, of squamous cell carcinoma (SCC) and large cell carcinoma. Treatment varies based on clinical stage. Early stage NSCLC is treated with surgery, while loco-regionally advanced and metastatic cancers are treated with multidrug systemic chemotherapy, which often includes a platinum compound.2
Head and neck cancers are similar to NSCLC in many respects, although they are less common, representing the eighth most frequent type of cancer in the USA.1 Smoking is a recognized risk factor for head and neck cancers, like for NSCLC. Pathologically, cancers of the aerodigestive tract are mostly head and neck squamous cell carcinoma (HNSCC). As for NSCLC, early stage HNSCC is successfully treated with surgery, while treatment of loco-regionally advanced tumors includes systemic therapy.2–4 Frequently, concomitant radiotherapy and chemotherapy with a platinum-based DNA damaging agent (cisplatin or carboplatin) is used, either as primary treatment or as adjuvant post-operative therapy. Alternative systemic treatments that do not rely upon DNA damage, such as taxanes, base analogs, and anti-metabolites can also be used.4 However, currently we do not have the tools to predict which patients will respond best to the various possible therapies.
To maximize treatment success of NSCLC and HNSCC, and to reduce unnecessary toxicity, there is great demand for identifying biomarkers that predict clinical outcomes prospectively. The goal is to measure validated biomarker(s) in individual tumors to probe the biology of each tumor and predict whether it is likely to be vulnerable to genotoxic agents such radiation and platinum drugs. This would enable identification of patients likely to be resistant to these modalities, allowing use of alternative therapies, preventing unnecessary toxic side-effects, and improving clinical outcomes.
Choosing a biomarker
Biomarkers in DNA repair pathways
DNA repair proteins are obvious candidate biomarkers for predicting how tumors will respond to genotoxic stress. The prediction is that overexpression of DNA repair proteins in tumors could mediate resistance to genotoxic therapies and therefore poor outcomes. In turn, persons with inherited defects in DNA repair mechanisms are frequently exquisitely hypersensitive to radiation and/or genotoxic agents. This is true of patients with ataxia telangiectasia (AT), ataxia telangiectasia-like disorder, severe combined immunodeficiency, Ligase IV syndrome, Rothmund–Thompson syndrome, Seckel syndrome, Werner syndrome, Nijmegen breakage syndrome, all due to defective repair of double-strand breaks (DSBs)5 or stalled replication forks.6 It is also true of patients with Fanconi anemia caused by defective repair of DNA interstrand crosslinks (ICLs) and patients with xeroderma pigmentosum due to a defect in nucleotide excision repair (NER) of helix-distorting DNA adducts.7,8 Since NSCLC and HNSCC are treated with cisplatin and radiation therapy, it is logical to predict that patients with reduced DSB repair, single-strand break (SSB) repair, ICL repair, or NER due to polymorphisms affecting the expression or function of DNA repair proteins might be most responsive to DNA damaging agents.
ERCC1-XPF repair endonuclease
ERCC1 is an attractive candidate biomarker. ERCC1 partners with XPF to form a bi-partite nuclease that is essential for NER and ICL repair, and participates in DSB repair (Figure 1).9–12 Platinum-based chemotherapy drugs react with DNA to induce adducts that affect one strand of DNA (monoadducts and intrastrand crosslinks), which are repaired by NER, as well as adducts that affect both strands (ICLs), which are repaired by a distinct DNA repair mechanism: ICL repair.13–15 Because ERCC1-XPF is unique in being required for both NER and ICL repair pathways, it is the only enzyme required for removal of all types of DNA lesions caused by cisplatin and carboplatin. In addition, it facilitates the repair of DNA lesions caused by radiation therapy (bulky oxidative lesions and DSBs).10 Hence, it has been proposed that decreased expression of ERCC1-XPF might mediate increased susceptibility to chemoradiation and improved clinical outcome. It is therefore not surprising that ERCC1 has been extensively evaluated as a biomarker in NSCLC and HNSCC, with over 90 peer-reviewed reports published on the subject. However, it is important to emphasize that the expression level of ERCC1-XPF has not been established as rate limiting for NER, ICL, or DSB repair, therefore the influence of ERCC1-XPF protein levels on the DNA repair capacity of cells or tumors is not known.
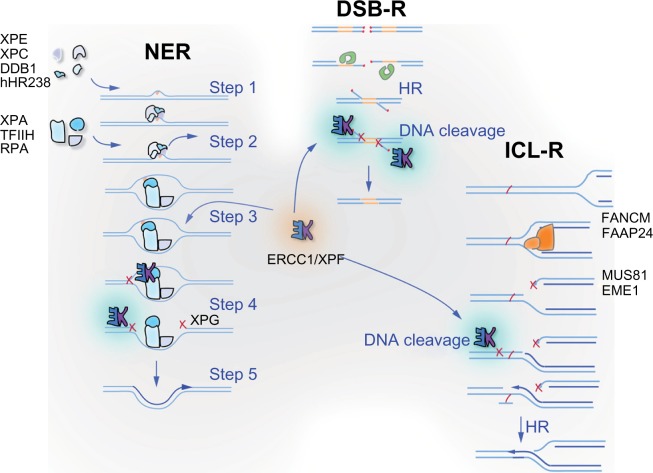
Figure 1.
ERCC1 and its obligate binding partner XPF are involved in multiple DNA repair pathways. ERCC1-XPF heterodimer is an endonuclease that cuts one strand of DNA at a double-strand:single-strand junction. It is critical for nucleotide excision repair (NER) of bulky chemical DNA adducts like cisplatin intrastrand crosslinks, the repair of double-strand breaks that cannot be directly ligated back together like those induced by ionizing radiation, and the repair of interstrand crosslinks (ICLs). In NER (represented on the left), adducts that cause distortion of the DNA double helix are detected by XPC-hHR23B, in some cases with the assistance of XPE-DDB1 (Step 1). These complexes recruit of TFIIH, which unwinds the DNA around the adduct and XPA and RPA, which stabilize the open complex (Step 2). XPA recruits ERCC1-XPF to cut the damaged strand 5′ to the adduct (Step 3), while TFIIH recruits a second endonuclease XPG to cut 3′ of the lesion (Step 4). The damaged base is removed as part of a single-stranded oligonucleotide. The replication machinery uses the 3′-OH created by ERCC1-XPF incision to prime DNA synthesis to fill the gap (Step 5). After ligation, the integrity of the DNA is fully restored. In double-strand breaks (DSB) repair (represented in the middle), two broken ends can be spliced together if they have long patches of sequence homology via homologous recombination (labeled HR) or if they have small patches of homology, known as microhomology, very close to the broken ends via alternative end-joining. In both cases, ERCC1-XPF is needed to remove 3′ single-stranded flaps of non-homologous sequence at the ends of the breaks (labeled DNA cleavage) to allow sealing of the spliced ends by a DNA ligase. ICLs (represented on the right) are predominantly repaired during S phase of the cell cycle. ICLs are an absolute block to replication and when encountered by the replication machinery lead to the collapse of the replication fork and creation of a DSB. This DSB cannot be repaired until ERCC1-XPF cuts near the ICL to release it from one strand (DNA cleavage), allowing bypass of the adduct by a translesion polymerase such as REV1/Polζ.
XRCC1 scaffold protein
XRCC1 is an equally promising candidate biomarker involved in the repair of oxidative DNA damage and single-strand breaks (SSBs) (Figure 2), two types of DNA damage abundantly produced by ionizing radiation. XRCC1 does not have enzymatic activity, but it is a critical scaffold protein for base excision repair (BER) and SSB repair (reviewed in Kennedy and D’Andrea,8 Hoeijmakers,16 Ladiges,17 and Almeida and Sobol).18 XRCC1 interacts strongly with PARP1, which recognizes SSBs, and LIGIII that seals SSBs and BER intermediates.17,19 Cells lacking XRCC1 are hypersensitive to ionizing radiation, oxidative stress and alkylating agents (reviewed by Caldecott).19 It is therefore plausible that reduced expression of XRCC1 in cancer patients may lead to increased susceptibility to chemoradiation and improved patient survival. However, like ERCC1-XPF, XRCC1 has not been established as rate limiting for DNA repair. Thus, the impact of low expression of XRCC1 on a cell’s capacity for BER and SSB is not known.
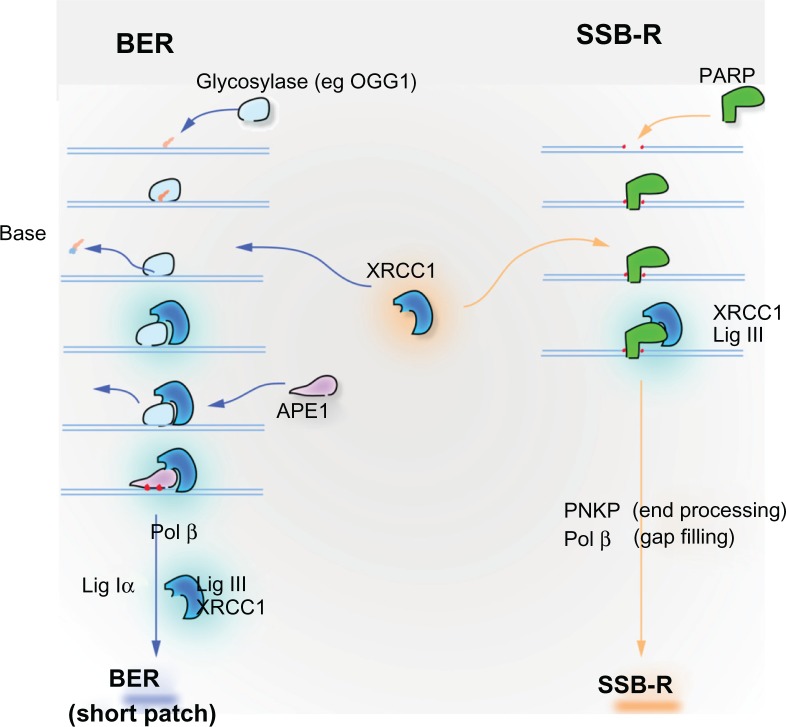
Figure 2.
XRCC1 is instrumental in base excision repair (BER) of small oxidative lesions and a related mechanism for the repair of single-strand breaks (SSB-R), both caused by ionizing radiation. Oxidative damage and alkylation leads to small alterations of bases that are principally repaired through BER pathway. Damaged bases are recognized and excised by glycosylases, such as OGG1, which removes the abundant oxidative lesion 8-oxodeoxyguanosine. Excision of the damaged base leaves an abasic (AP) site. The DNA backbone adjacent to the AP site is incised by APE1 endonuclease to create a single-strand break (SSB). XRCC1 has no enzymatic activity, but is critical as a scaffolding protein in BER. It is recruited to the site of damage by the glycosylase or by PARP1, which binds the newly created SSB. XRCC1 forms a tight complex with LIG3, the ligase that seals the SSB repair intermediate to complete BER. Primary SSBs, a common consequence of ionizing radiation, are directly recognized by PARP1, which recruits XRCC1-LIG3 to repair the broken strand. PNKP removes 3′ phosphate groups that block DNA ligation by LIG3. Polβ may be required to replace missing nucleotides at the site of the break.
Methods to assess biomarkers and clinical endpoints
Available methods to interrogate DNA repair
Directly measuring NER, DSB repair, ICL repair, or BER would be the ideal method for predicting an individual’s DNA repair capacity. However measuring DNA repair requires viable, and for some pathways, replicating cells. Thus, currently it is not possible to rapidly measure DNA repair in clinical samples because it first requires establishing a cell line from peripheral blood mononuclear cells, dermal fibroblasts, or tumors. Hence measuring DNA repair protein expression is used as a surrogate. Multiple techniques are available to measure ERCC1 and XRCC1 expression including immunohistochemistry or immunofluorescence of fixed tissue sections, quantification of mRNA expression by qRT-PCR, or quantification of protein expression by immunoblot if frozen specimens are available. It must be strongly emphasized, however, that it is not established that ERCC1 is rate limiting for NER or ICL repair, or that XRCC1 is rate limiting for BER or SSB repair. ERCC1 and XRCC1 can also be investigated by sequencing DNA to detect functional single nucleotide polymorphisms (SNP) affecting protein function or expression level.
Measuring protein expression
Immunohistochemistry (IHC) and immunofluorescence are semi-quantitative methods that permit estimation of protein expression level in clinical samples. The intensity of the histochemical reaction or fluorescent signal varies with the expression level of the protein of interest and can be scored as positive versus negative or on a graded scale. These methods are advantageous since they employ paraffin embedded tissue specimens, which are readily available. However, several caveats must be considered while interpreting data from immunohistochemical methods. Protein expression within a given tumor may vary from one area to another.20,21 Therefore expression measured on a biopsy specimen or in a tissue core in an array, which represent only a small fraction of a tumor, may not reflect overall expression. In one patient cohort, however, it was established that ERCC1 expression in biopsies correlated with expression measured in tumor sections.22 Another important technical consideration is the fact that tissue collection method, handling, storage, fixation, processing, and analysis influence the biomarker readout, and causes inter-study variability.23 This has led to the publication of guidelines for evaluation of biomarkers, in an attempt to unify methods of biomarker analysis.24
Equally important, immunodetection methods are by definition indirect measures of protein expression, dependent upon the sensitivity and specificity of the antibody used. The specificity of the commercially available antibodies is rarely rigorously tested. ERCC1 protein expression was erroneously quantified in virtually all oncology studies prior to 2010 due to the implementation of an antibody raised against ERCC1 that lacks specificity.25 Finally, methods for quantifying and scoring biomarker expression vary from study to study, and are somewhat subjective. For instance, biomarker positivity can be defined as the presence of any staining detected by a pathologist, calculated as an H-score based on the staining intensity and number of positive cells, or quantified by an automated system to minimize subjectivity. Thus, while immunohistochemical methods are potentially useful for quantifying biomarker protein expression, multiple factors can introduce intra- or inter-study variability.
Measuring mRNA expression
mRNA expression is often used as a surrogate marker for protein expression. Typically this is done by quantitative RT-PCR, using primers specific for the target biomarker. The advantages of quantifying mRNA are that the method is very sensitive, highly specific, and can be applied to fixed specimens. However, quantitative methods to measure mRNA levels are not readily available outside of biomedical research facilities. Importantly, mRNA and protein expression do not always correlate.26,27 Translational regulation, post-translational modification and protein stability alter protein levels independently of mRNA.28 So while mRNA levels can be a useful biomarker to predict clinical outcomes, mRNA levels do not necessarily reflect protein levels. Therefore, changes in mRNA levels should not be used to infer changes in biological activity in the absence of experimental evidence.
Genomic approaches
Base changes in a gene can lead to reduced expression of the encoded protein if they affect the promoter, 5′ or 3′ untranslated sequence, regulatory miRNA binding sites, splice sites, or the coding sequence if the change leads to protein misfolding or destabilization, or utilization of a less abundant tRNA during translation. Missense mutations in the coding sequence can also alter protein function by affecting protein:protein interactions or catalytic activity. Single nucleotide polymorphisms (SNPs) are defined as single base changes that occur in more than 1% of the population. They occur every 360 bases in the human genome, and, thus, affect all genes (reviewed by Kim and Misra).29 The National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/projects/SNP reports 246 SNPs in ERCC1, and 550 SNPs in XRCC1. In silico, in vitro, or epidemiological studies can be used to identify SNPs with the highest likelihood of being a useful biomarker. This includes SNPs with a known impact on mRNA level or protein expression, or activity. Fourteen SNPs in ERCC1 and eleven for XRCC1 have been investigated in NSCLC and/or HNSCC. The advantages of analyzing SNPs as biomarkers are that multiple SNPs can be evaluated in one sample using an array and DNA hybridization method and require only DNA extracted from a simple blood draw.29,30 However, it is important to remember that the genotype of a tumor may differ from the germline genotype found in the rest of the body, as tumors are inherently genomically unstable and accumulate DNA mutations. Therefore SNPs identified in a patient’s blood sample may not reflect a patient’s tumor’s genotype.31 Furthermore, because SNPs are much more abundant than recombination events in the human genome, they are inherited in clusters, referred to as haplotypes. Thus, a SNP in ERCC1 or XRCC1 could be a useful biomarker for predicting outcomes in cancer without having any impact on DNA repair.
Clinical endpoints
In oncology, clinical outcomes for which it would be desirable to have biomarkers include: (1) risk of cancer, (2) prognosis in untreated patients, (3) tumor response to therapy, (4) severity of treatment-related toxicities, (5) progression-free survival, and (6) overall survival. DNA repair-related endpoints could logically contribute to any of these endpoints, in particular when genotoxic chemotherapeutics or radiation is the therapy of choice.
One of the most widely recognized risk factors for NSCLC and HNSCC is smoking. The pathogenesis of these tumors involves tobacco-related DNA damage. It is rational to hypothesize that persons with low expression of ERCC1 or XRCC1 may have impaired ability to remove tobacco-induced DNA damage and therefore are more likely to develop smoking-related cancers. The best way to test this hypothesis is with well-powered prospective risk analysis. But these types of studies are difficult to conduct because they necessitate large cohorts and long follow-up times. For instance, >520,000 patients would have to be followed for 10 years to find 116 lung cancer and 82 HNSCC.32 Thus, most published studies evaluating cancer risk associated with ERCC1 and XRCC1 are retrospective case-control studies, which have their inherent limitations.
Since DNA repair-related biomarkers could have value for multiple clinical endpoints, they could potentially have prognostic or predictive value. Prognostic biomarkers estimate progression-free or overall survival in an untreated patient population. It gives information on the natural course of the disease.33 In contrast, predictive biomarkers estimate how likely a given treatment is expected to work (efficacy). Predictive value is determined in prospective randomized trial settings with treatment and control arms. Both prognostic and predictive biomarkers are useful but they require different study designs. Once identifying a bio-marker of interest, validation is essential and ultimately the greatest barrier to implementation of the biomarker in clinic practice.34 Validation includes establishing that a biomarker of interest (expression, genotype) consistently predicts a particular clinical outcome (response rate, progression free survival, overall survival). Thus, validation requires multiple clinical studies conducted by multiple independent groups. With these considerations in mind, we now critically review the literature on ERCC1 and XRCC1 SNPs as biomarkers in NSCLC and HNSCC.
ERCC1 as biomarker for NSCLC and HNSCC
ERCC1 as a biomarker for cancer risk
Two SNPs, Asn118Asn and C8092A, have been described as potentially affecting ERCC1 expression. Asn118Asn involves a synonymous polymorphism at codon 118, where AAC is changed to AAT. While the amino acid sequence does not change, the variant (T) allele is associated with lower mRNA and protein levels in ovarian cancer cells.35,36 C8092 is in the 3′-UTR of ERCC1. The 3′-UTR is implicated in translational repression of ERCC1 mRNA.28 However the impact of the polymorphism on ERCC1 protein expression has not been critically evaluated to date. In patients, the C8092A polymorphism correlates neither with mRNA,37 nor with protein levels.38 Numerous other SNPs in ERCC1 have been studied, but like C8092, their functional impact on ERCC1 expression or activity has not been clearly established.
Studies evaluating ERCC1 as a potential biomarker to predict the risk of developing NSCLC or HNSCC rest principally on SNP analysis. There are ten studies examining ERCC1 SNPs in relation to NSCLC.32,39–47 In these studies, only 14 of 246 reported SNPs in ERCC1 were evaluated, with just six SNPs analyzed in greater than one study (Table 1). Most report retrospective case-controlled studies focused on Asn118, C8092, and IVS3. While case-control studies are important for identifying new biomarkers, they have inherent biases that can limit the generalization of the results. For instance, if the biomarker is not robust, confounding factors in the cohort may lead to erroneous conclusions. In most of the retrospective studies, SNPs in ERCC1 were not significantly associated with susceptibility of developing NSCLC.32,39–42,46–48 However, there was not good concordance between studies.42–45 To clarify the role of SNPs in ERCC1 as risk factor for NSCLC, meta-analyses were done. When patients from the diverse studies were combined into large data pools, none of the four SNPs in ERCC1 meeting study inclusion criteria reached statistical significance as a risk factor for NSCLC.48–50 Furthermore, mRNA levels in blood samples were not identified as a risk factor for lung cancer.51 In summary, our review of the literature suggests that neither SNPs in ERCC1 studied to date by more than one group, nor peripheral mRNA levels, constitute a risk factor for NSCLC.
Table 1.
Association between SNPs in ERCC1 and cancer risk
| Cancer | rs | SNPs | Alternate names | Reference | n (case-control) | Riska |
|---|---|---|---|---|---|---|
| NSCLC | rs11615 | Asn118 Asn | C118T; 354 C > T; T19007C; C19007T; 3525 C > T | Zhou et al39 | 1752–1358 | 0 |
| Matullo et al32,# | 116–> 520,000 | 0 | ||||
| Yin et al40 | 151–143 | 0 | ||||
| Hung et al41 | 4460–5217 | 0 | ||||
| Yu et al42 | 988–986 | 0 | ||||
| Deng et al43 | 315–315 | 1 | ||||
| Zienolddiny et al44 | 343–413 | 1 | ||||
| rs3212986 | C8092A | 14443 C > A | Zhou et al39 | 1752–1358 | 0; 1 in heavy smokers | |
| Zienolddiny et al44 | 343–413 | 0 | ||||
| Yu et al42 | 988–986 | 0 | ||||
| Hung et al41 | 4688–4546 | 0 | ||||
| rs3212948 | 19716 C > G | IVS3 174G > C | Shen et al46 | 122–122 | 0 | |
| Jones et al167 | 452–790 | 0 | ||||
| Zienolddiny et al44 | 343–413 | 0 | ||||
| Ma et al45 | 1010–1011 | 2 | ||||
| rs3212930 | (−)433 T > C | Ma et al45 | 1010–1011 | 0 | ||
| Yu et al42 | 988–986 | 1 | ||||
| rs3212961 | 4855 C > T | IVS5 + 33 C > A; 17677 C > A | Shen et al46 | 122–122 | 0 | |
| Yu et al42 | 1000–1000 | 0 | ||||
| Zienolddiny et al44 | 343–413 | 0 | ||||
| rs3212955 | Ma et al45 | 1010–1011 | 0 | |||
| Jones et al167 | 452–790 | 0 | ||||
| rs3212981 | Ma et al45 | 1010–1011 | 0 | |||
| rs16979802 | 15310 C > G | Zienolddiny et al44 | 343–413 | 1 | ||
| rs3212951 | Ma et al45 | 1010–1011 | 0 | |||
| rs1007616 | Ma et al45 | 1010–1011 | 2 | |||
| rs1319052 | Jones et al167 | 452–790 | 0 | |||
| rs735482 | Jones et al167 | 452–790 | 0 | |||
| rs2298881 | 262 G > T | Yu et al42 | 988–986 | 0; (1) in smokers | ||
| unnamed | Ma et al45 | 1010–1011 | 0 | |||
| HNSCC | rs11615 | Asn118 Asn | 354 T > C; 19007 T > C; 3525 C > T | Abbasi et al53 | 257–769 | 0 |
| Canova et al54 | 1511–1457 | 0 | ||||
| Matullo et al32 | 82–> 520,000 | 0 | ||||
| rs3212986 | C8092A | 14443 C > A | Abbasi et al53 | 257–769 | 0 | |
| Sugimura et al52 | 122–244 | (1); 1 in smoker | ||||
| Sturgis et al55 | 313–313 | (2) | ||||
| rs3212948 | 19716 C > G | IVS3 + 74C > G | Canova et al54 | 1511–1457 | 0 | |
| Jones et al167 | 175–790 | 0 | ||||
| rs3212961 | 4855 C > T | IVS5 + 33C > A | Abbasi et al53 | 257–769 | 0 | |
| Canova et al54 | 1511–1457 | 2 | ||||
| rs1319052 | Jones et al167 | 175–790 | 0 | |||
| rs735482 | Jones et al167 | 175–790 | 0 | |||
| rs3212955 | Jones et al167 | 175–790 | 0 |
Notes:
Risk for variable allele, 0 = non significant, (1) = trend to increased, 1 = increased, (2) = trend to protective, 2 = protective;
retrospective analysis of prospective study.
Abbreviations: HNSCC, head and neck squamous cell carcinoma; NSCLC, non-small cell lung cancers; rs, reference SNP; SNPs, single nucleotide polymorphisms.
Head and neck cancers are less common than lung cancer. Hence clinical studies to identify biomarkers that predict the risk of developing HNSCC are less frequent and smaller. We identified six studies evaluating whether polymorphisms in ERCC1 are a risk factor for HNSCC (Table 1).32,47,52–55 Only four SNPs were assessed more than once: (Asn118Asn), (C8092A), 119216 C > G, and 4855 C > T. None showed statistically significant association with risk of HNSCC, with the exception of one large case control study in which 4855 C > T appeared to be protective.54 One small retrospective case-controlled study suggested that low ERCC1 mRNA in peripheral blood might be a risk factor for HNSCC,56 but the findings could not be confirmed by others after multivariate analysis.37 Therefore, we conclude that none of the SNPs in ERCC1 tested thus far, nor peripheral ERCC1 mRNA levels are definitive risk factors for HNSCC. However, 4855 C > T deserves close attention in future studies. Further, we cannot exclude the possibility that these or other ERCC1 SNPs may be useful biomarkers in selected subpopulations for predicting cancer risk.
ERCC1 SNPs as biomarkers for clinical outcome
Polymorphisms in ERCC1 could affect tumor sensitivity to treatment, and hence influence patient outcomes. Patients with a polymorphic variant of ERCC1, which results in impaired NER and/or ICL repair capacity, may be exquisitely sensitive to chemotherapy with genotoxic agents or radiation. This could mean their tumors respond better to chemoradiation therapy and outcomes are improved. Alternatively, the host may be hypersensitive to genotoxic stress leading to exaggerated side effects of therapy and poor outcomes.
In NSCLC, we identified sixteen studies testing whether ERCC1 polymorphisms influence clinical outcome,38,57–71 including five prospective studies (Table 2).58,62,69,70 The only two SNPs tested were Asn118 and C8092. The results are inconsistent, weakening the generalizability of the conclusions. When more than 500 patients from multiple studies were pooled into a single meta-analysis, Asn118 Asn was predictive of tumor response to chemotherapy.72 As expected, the variant allele (C→T), which presumably causes lower ERCC1 expression, correlated with a higher response rate.72 However, this meta-analysis excluded one important report, a large phase Phase III study (n = 526) in which Asn118 did not predict clinical outcome, including response to treatment.58 These conflicting results, derived from equally large studies, suggest that this ERCC1 SNP is not a robust predictive biomarker in an unselected population. To our knowledge, C8092 has not been evaluated in a large prospective study or in a meta-analysis as a predictor of clinical outcomes in NSCLC. In retrospective cohorts, C8092 showed mixed results as predictive biomarker. The general tendency was slightly weighed toward the variant allele (C→A) predicting worse outcomes.38,59,63,73 In summary, none of the SNPs in ERCC1 tested have been identified as strongly predictive biomarkers for outcomes in NSCLC, but C8092 emerges as a potentially promising candidate.
Table 2.
Association between SNPs in ERCC1 and clinical outcome
| Cancer | rs | SNPs | Alternate names | Reference | n | Outcomea |
|---|---|---|---|---|---|---|
| NSCLC | rs11615 | Asn118 Asn | C118T; 354 T > C; 19007 T > C; 3525 C > T | Zhou et al63 | 128 | 0 |
| Gandara et al (2005)b | 526 | 0 | ||||
| Suk et al59 | 214 | 0 (toxicity) | ||||
| De Las Penas et al71,b | 135 | 0 | ||||
| Tibaldi et al61 | 65 | 0 | ||||
| Takenaka et al73 | 122 | 0 | ||||
| Vinolas et al62,b | 94 | 0 | ||||
| Park et al64 | 178 | (1); 1 for stage III | ||||
| Ryu et al65 | 109 | 1 | ||||
| Isla et al68 | 62 | 1 | ||||
| Su et al66 | 230 | 1 | ||||
| Kalikaki et al57 | 119 | 1 | ||||
| Okuda et al38 | 90 | 1 | ||||
| Yin et al67 | 257 | 1 | ||||
| Li et al70,b | 115 | 2 | ||||
| Zhou et al69,b | 130 | 2 | ||||
| rs3212986 | C8092A | 14443 C > A | Zhou et al63 | 128 | 1 | |
| Suk et al59 | 214 | 1 (toxicity) | ||||
| Park et al64 | 178 | 0 | ||||
| Okuda et al38 | 90 | 1 | ||||
| Takenaka et al73 | 122 | 1 | ||||
| Kalikaki et al57 | 119 | 2 | ||||
| Li et al70,b | 115 | 2 | ||||
| HNSCC | rs3212986 | C8092A | 14443 C > A | Quintela-Fandino et al74 | 103 | −1 |
| rs735482 | Lys259Thr | 1264 A > C | Grau et al75,b | 47 | 0 | |
| Carles et al76 | 108 | 1 (but only 4% of carrier) |
Notes:
Outcome for variable allele, 0 = non significant, (1) = trend to worse, 1 = worse, (2) = trend to better, 2 = better;
prospective study.
Abbreviations: HNSCC, head and neck squamous cell carcinoma; NSCLC, non-small cell lung cancers; rs, reference SNP; SNPs, single nucleotide polymorphisms.
In HNSCC, we identified only three studies evaluating the predictive value of SNPs in ERCC1 (Table 2).74–76 Like NSCLC, in HNSCC, there was a trend towards an association between the variant allele of C8092 (C→A) with poor response to chemoradiation, and no correlation with survival.74 A new SNP (rs735482) located in the 3′UTR of ERCC1 was evaluated for predictive value of clinical outcome in two separate cohorts, but results were mixed.75,76 Therefore, we conclude that there is currently no strong evidence that SNPs in ERCC1 can predict clinical outcome in HNSCC.
ERCC1 protein expression as a biomarker of patient outcomes in NSCLC
While SNPs are often used as a crude estimate of ERCC1 expression or activity, immunodetection approaches permit a more direct quantification of ERCC1 protein level in tumor samples. We identified 17 studies addressing whether quantification of ERCC1 expression in NSCLC tumors by immunohistochemistry has prognostic or predictive value (Table 3).27,38,60,73,77–91 In a seminal retrospective analysis of a phase III trial, more than 780 patients with fully resected early stage NSCLC were randomized to observation versus multidrug chemotherapy.81 The results suggested that tumoral ERCC1 protein expression was a biomarker with a complex profile. High ERCC1 levels correlated with good prognosis for untreated cases. But patients with low ERCC1 levels did significantly better when treated with multidrug chemotherapy. These results are consistent with the prediction that decreased expression of ERCC1 could promote sensitivity to genotoxic chemotherapy. Most studies agree that low ERCC1 protein expression is a marker for better clinical outcome after genotoxic therapy in NSCLC. Thirteen of 17 studies reported that low ERCC1 correlated with better clinical outcome (total n = 1815),77–85,87,91,92 or had a statistical trend towards better outcome (total n = 218).38 Two studies showed no correlation between ERCC1 level and outcome (n = 218),89,90 while two studies showed a significantly worse outcome (total n = 269)27,88 in patients with tumors expressing low levels of ERCC1. A recent meta-analysis evaluated NSCLC patients treated with platinum compounds.93 Low expression of ERCC1 in tumors quantified by immunohistochemistry was associated with a better clinical response to cisplatin, which translated into better survival.93 Despite some variability between individual studies, ERCC1 appears to emerge as a good candidate biomarker predictive of clinical outcome in NSCLC. An important point, however, is that in all 18 of the studies the monoclonal antibody, 8F1 was used to measure ERCC1 expression, and this antibody is not specific for ERCC1.25 Therefore, the claim that low ERCC1 expression correlates with better outcome is inaccurate. The more precise conclusion is that low 8F1 signal correlates with better outcome. More recent studies comparing 8F1 and another antibody specific for ERCC1 reveal that they have different predictive capacities with relation to clinical outcomes in cervical cancer.94
Table 3.
Association between ERCC1 protein expression and clinical outcome
| Cancer | Reference | n | Outcomea |
|---|---|---|---|
| NSCLC | Planchard et al90 | 188 | 0 |
| Koh et al89 | 130 | 0 | |
| Zheng et al27 | 187 | 1 | |
| Kang et al88 | 82 | 1 | |
| Okuda et al38 | 55 | (2) | |
| Okuda et al91 | 90 | 2 | |
| Olaussen et al81 | 783 | 2 | |
| Azuma et al84 | 67 | 2 | |
| Fujii et al83 | 35 | 2 | |
| Lee et al87 | 130 | 2 | |
| Holm et al86 | 163 | 2; men P = 0.005, women P = 0.7 | |
| Azuma et al85 | 34 | 2 | |
| Lee et al82 | 50 | 2 | |
| Ota et al80 | 156 | 2 | |
| Reynolds et al79,b | 69 | 2 | |
| Vilmar et al78,b | 264 | 2 | |
| Wang et al77 | 214 | 2 | |
| Taillade et al22 | 34 | Biopsy vs tumor correlation | |
| Gomez-Roca et al (2009) | 49 | Primary vs metastasis | |
| Kang et al164 | 82 | Primary vs metastasis | |
| Papay et al (2009) | 17 | Change after chemotherapy | |
| Besse et al (2010)c | 761 | Brain metastasis | |
| HNSCC | Fountzilas et al31 | 37 | 0 |
| Koh et al89 | 80 | 0 | |
| Handra-Luca et al97 | 96 | 2 | |
| Jun et al98 | 45 | 2 | |
| Fountzilas et al31,b | 26 | 2 |
Notes:
Outcome for low ERCC1 expression, 0 = non significant changes, (1) = trend to worse, 1 = worse, (2) = trend to better, 2 = better;
prospective study;
retrospective analysis of prospective study.
Abbreviations: HNSCC, head and neck squamous cell carcinoma; NSCLC, non-small cell lung cancers.
In HNSCC, only five studies (total n = 285) evaluated whether ERCC1 protein expression in tumors correlated with clinical outcome (Table 3).31,95–98 The 8F1 antibody was used in all of the studies. Low 8F1 signal was associated with better outcome in three studies (total n = 168),95,97,98 while no significant association was found in the other two (n = 117).31,96
ERCC1 transcript levels as a biomarker in NSCLC and HNSCC
As a surrogate marker of ERCC1 expression, ERCC1 mRNA was measured in NSCLCs in cell lines,99 and in six retrospective68,100–104 and six prospective studies.105–110 The results were mixed, but most studies showed an association between low ERCC1 mRNA and better clinical outcome, either significantly (seven studies)100,102–105,108,109 or with a statistical trend (three studies).68,105,110 In a meta-analysis, both low tumoral mRNA and protein levels correlated with a better response rate to chemoradiation and overall patient survival.93 While assays used to measure mRNA levels in tumors are not yet readily available for clinical use in all cancer centers, ERCC1 mRNA may prove to be a reasonable predictive biomarker of outcome in NSCLC patients treated with platinum-based chemotherapy.93 Interestingly, ERCC1 mRNA and protein levels were found to be not correlated in NSCLC27 and inversely correlated in ovarian cancer.111 Furthermore, mRNA levels were not correlated with chemosensitivity in NSCLC cell lines99 nor with response to chemotherapy in HNSCC.31 Thus, the relationship between ERCC1 mRNA and DNA repair capacity is not direct and remains to be clarified.
XRCC1 as biomarker for NSCLC and HNSCC
XRCC1 as a biomarker for cancer risk
Similar studies have sought to establish whether XRCC1 is linked with cancer risk, prognosis, or treatment outcome. SNPs in XRCC1 have been extensively studied in NSCLC, although only 9 SNPs out of 550 possible have been evaluated in published reports. The majority of trials focus on Arg194Trp, Arg280His, and Arg399Gln, three nonsynonymous SNPs in XRCC1 (reviewed by Schneider et al).112 Four studies, including two large ones, also analyzed a SNP in the XRCC1 promoter (−77T→C).113–116 The variant allele −77T→C alters a binding site for the zinc finger transcription factor SP1, leading to reduced transcription of XRCC1.113 The variant allele at position 399 (Gln) correlates with lower DNA repair capacity and increased genomic instability in multiple studies.117–121 These functional SNPs in XRCC1 are attractive candidate biomarkers in cancer.
XRCC1 SNPs as biomarkers for cancer risk
The assessment of SNPs in XRCC1 as risk factors for developing NSCLC has focused mainly on XRCC1 Arg194-Trp, Arg280His and Arg399Gln, and to a lesser degree on −77T→C (Table 4).32,41,44,67,112–116,122–143 Studies failed to identify significant association between Arg194Trp, Arg280His, and Arg399Gln genotypes and NSCLC risk. However, −77T→C did emerge as a significant risk factor in two large studies.113,114 This is consistent with the notion that low XRCC1 expression leads to impaired BER and SSB repair, greater mutational load and therefore increased cancer risk. A well conducted meta-analysis pooling more than 10,000 patients for the analysis of Arg194Trp, Arg280His, and Arg399Gln, and more than 1,000 patients for the analysis of Pro206Pro and −77T→C found that, in NSCLC, −77T→C was associated with cancer risk (P < 0.0001), while none of the other four SNPs analyzed in XRCC1 showed association.50 Furthermore, this meta-analysis reviewed a total of 241 associations in 16 genes, and XRCC1 −77T→C was one of the only two associations that maintained a significant association through the most stringent analysis. Thus, there is strong epidemiological and biological credibility supporting XRCC1–77T→C as a risk factor for NSCLC.
Table 4.
Association between SNPs in XRCC1 and cancer risk
| Cancer | rs | SNPs | Alternate names | Reference | n (case-control) | Riska |
|---|---|---|---|---|---|---|
| NSCLC | rs1799782 | Arg194Trp | 194 C > T; 194 R > W; 194 Arg > Trp; C26304T |
Butkiewicz et al124 | 96–96 | 0 |
| Hu et al114 | 710–710 | 0 | ||||
| Shen et al46 | 122–122 | 0 | ||||
| Matullo et al32 | 116–> 520,000 | 0 | ||||
| Hao et al113 | 1024–1118 | 0 | ||||
| Zienolddiny et al44 | 343–413 | 0 | ||||
| Yin et al131 | 247–253 | 0 | ||||
| Hung et al41,b | 6463–6603 | 0 | ||||
| Improta et al126 | 940–121 | 0 | ||||
| Tanaka et al130 | 50–50 | 0 | ||||
| Ratnasinghe et al128 | 108 | 0; 2 in drinkers | ||||
| David-Beabes132 | 332–704 | 0; 2 in African-Americans | ||||
| Schneider et al112 | 446–622 | 0; 2 in heavy smokers | ||||
| Hung et al127,b | 2188–2198 | 0; 2 in heavy smokers | ||||
| Chen et al56 | 109–109 | (1) | ||||
| Pachouri et al133 | 103–122 | (1) | ||||
| De Ruyck et al116 | 110–110 | 2 | ||||
| Yin et al67 | 55–74 | 2 | ||||
| rs25489 | Arg280His | 280 G > A; 280 R > H; 280 Arg > His |
Butkiewicz et al124 | 96–96 | 0 | |
| Misra et al122,b | 305–305 | 0 | ||||
| Vogel et al124 | 265–272 | 0 | ||||
| Schneider et al112 | 446–622 | 0 | ||||
| Shen et al46 | 122–122 | 0 | ||||
| Hao et al113 | 1024–1118 | 0 | ||||
| Zienolddiny et al44 | 343–413 | 0 | ||||
| Hung et al41 | 6463–6603 | 0 | ||||
| Yin et al67 | 55–74 | 0 | ||||
| Yin et al131 | 247–253 | 0; 2 in non-smokers | ||||
| Hung et al127,b | 2188–2198 | 0; 2 in heavy smokers | ||||
| Ratnasinghe et al128 | 108 | 1 | ||||
| De Ruyck et al116 | 110–110 | 2 | ||||
| rs25487 | Arg399Gln | G28152A; 399 G > A; 399 R > Q; 399 Arg > Gln |
Butkiewicz et al124 | 96–96 | 0 | |
| David-Beabes132 | 332–704 | 0 | ||||
| Ratnasinghe et al128 | 108 | 0 | ||||
| Chen et al56 | 109–109 | 0 | ||||
| Ito et al135 | 178–449 | 0 | ||||
| Popanda et al137 | 463–460 | 0 | ||||
| Vogel et al134 | 265–272 | 0 | ||||
| Zhang et al139 | 1000–1000 | 0 | ||||
| Hu et al114 | 710–710 | 0 | ||||
| Hung et al127,b | 2188–2198 | 0 | ||||
| Zienolddiny et al44 | 343–413 | 0 | ||||
| Hao et al113 | 1024–1118 | 0 | ||||
| Yin et al131 | 247–253 | 0 | ||||
| Lopez-Cima et al136 | 516–533 | 0 | ||||
| Hung et al41,b | 6463–6603 | 0 | ||||
| Improta et al126 | 940–121 | 0 | ||||
| Yin et al67 | 55–74 | 0 | ||||
| De Ruyck et al116 | 110–110 | 0; 1 in light smokers, 2; in heavy smokers | ||||
| Misra et al122,b | 305–305 | 0; (2) in heavy smokers | ||||
| Schneider et al112 | 446–622 | 0; 2 in heavy smokers | ||||
| Ryk et al138 | 177–153 | 0; 2 in non-smokers | ||||
| Park et al140,b | 192–135 | (1) for SCC | ||||
| Zhou et al141 | 1091–1240 | (1) | ||||
| Sreeja et al142 | 171–211 | 1 | ||||
| Divine et al143 | 172–143 | 1 in Caucasian but not Hispanic | ||||
| Shen et al46 | 122–122 | (2) | ||||
| Matullo et al32 | 116–> 520,000 | 2 (by stepwise regression) | ||||
| Pachouri et al133 | 103–122 | 2 | ||||
| rs3213245 | −(77) T > C | De Ruyck et al116 | 110–110 | 0 | ||
| Hsieh et al115 | 294–288 | 0 | ||||
| Hao et al113 | 1024–1118 | 1 | ||||
| Hu et al114 | 710–710 | 1 | ||||
| rs915927 | Pro206Pro | 206 A > G; 206 pro = pro |
Matullo et al32 | 116–> 520,000 | 0 | |
| Yin et al131 | 247–253 | 1 | ||||
| Yin et al67 | 55–74 | 1 | ||||
| rs17852150 | Gln632Gln | 632 G > A; 632 Gln = Gln |
Yin et al131 | 247–253 | 0 | |
| Yin et al67 | 55–74 | 0 | ||||
| rs2307191 | Pro161Leu | 161 Pro > Leu | Tanaka et al130 | 50 | 0 | |
| rs2307177 | Tyr576Ser | 576 Tyr > Ser | Tanaka et al130 | 50 | 0 | |
| n/a | Arg59Cys | Zienolddiny et al44 | 343–413 | ND | ||
| HNSCC | rs1799782 | Arg194Trp | 194 C > T; 194 R > W; 194 Arg > Trp; C26304T |
Sturgis et al151 | 203–424 | 0; 2 for oral and pharyngeal cancer |
| Olshan et al148 | 182–202 | 0 | ||||
| Varzim et al168 | 88–178 | 0 | ||||
| Matullo et al32 | 82–> 520,000 | 0 | ||||
| Harth et al146 | 312–300 | 0 | ||||
| Applebaum et al144 | 722–815 | 0 | ||||
| Csejtei et al145 | 108–102 | 0 | ||||
| Kowalski et al149 | 92–124 | (1) | ||||
| Tae et al150 | 147–168 | 1 | ||||
| rs25489 | Arg280His | 280 G > A; 280 R > H; 280 Arg > His |
Tae et al150 | 147–168 | 0 | |
| Harth et al146 | 312–300 | 0 | ||||
| Applebaum et al144 | 722–815 | 0 | ||||
| Sturgis et al151 | 203–424 | 0 | ||||
| Cho et al152 | 334–283 | 2 | ||||
| rs25487 | Arg399Gln | G28152A; 399 G > A; 399 R > Q; 399 Arg > Gln |
Varzim et al168 | 88–178 | 0 | |
| Cho et al152 | 334–283 | 0 | ||||
| Tae et al150 | 147–168 | 0 | ||||
| Huang et al154 | 555–792 | 0; 2 in Caucasian | ||||
| Harth et al146 | 312–300 | 0 | ||||
| Canova et al54 | 1478–1424 | 0 | ||||
| Applebaum et al144 | 722–815 | 0; (1) in p16 neg smokers | ||||
| Csejtei et al145 | 108–102 | 0 | ||||
| Kowalski et al149 | 92–124 | 0 | ||||
| Sturgis et al151 | 203–424 | (1) | ||||
| Olshan et al148 | 182–202 | 2 | ||||
| Gal et al153 | 279 | 2; for overall survival only | ||||
| rs915927 | Pro206Pro | Matullo et al32 | 82–> 520,000 | 0 | ||
| Canova et al54 | 1495–1436 | 0 | ||||
| rs762507 | Canova et al54 | 1447–1397 | 0 |
Notes:
Risk for variable allele, 0 = non significant, (1) = trend to increased, 1 = increased, (2) = trend to protective, 2 = protective; ND = not done;
retrospective analysis of prospective study.
Abbreviations: HNSCC, head and neck squamous cell carcinoma; NSCLC, non-small cell lung cancers; rs, reference SNP; SCC, squamous cell carcinoma; SNPs, single nucleotide polymorphisms.
In HNSCC, only five SNPs have been evaluated as cancer risk factors.32,54,144–154 Four of them have been evaluated more than once: Arg194Trp, Arg280His, Arg399Gln, and Pro206Pro (Table 4). The results were mixed for all four SNPs, but primarily showed no significant association with cancer risk, except for a tendency for the homozygous variant 399Gln-Gln to be protective in Caucasians in one large pooled study.154 Interestingly, when patients from individual studies were pooled for a meta-analysis, Arg194Trp emerged as a significant risk factor for HNSCC, as well as for other solid cancers (skin, esophageal, and stomach).50 It will be interesting to follow whether future studies can validate this SNP as a biomarker for risk stratification in HNSCC.
XRCC1 SNPs as biomarkers for clinical outcome
Biologically, genetic polymorphisms in XRCC1 could potentially predict clinical outcome, because reduced XRCC1 expression in animal models confers sensitivity to ionizing radiation. We identified eleven studies57,67,71,115,155–161 looking at XRCC1 SNPs (Arg194Trp, Arg280His, Arg399Gln, and −77T→C) including five prospective studies,71,155,157,159,160 totaling more than 1700 patients (Table 5). Results were mixed for Arg194Trp: three studies showed no association (total n = 382),155–157 one showed a worse prognosis for the allelic variant (n = 229),158 and one showed a better prognosis (n = 82).159 Results for Arg399Gln were also mixed, with significantly worse overall survival or toxicity for the allelic variant in three studies (total n = 515),57,67,156 while a better prognosis was found in two studies (n = 238)71,160 and no association was found in other studies (total n = 559).155,157–159,161 Finally, Arg280His showed no significant association with any outcome (2 studies; total n = 428). A meta-analysis and additional studies to examine −77T→C are needed to determine if SNPs in XRCC1 have any value for predicting clinical outcomes in patients with NSCLC treated with chemoradiation.
Table 5.
Association between SNPs in XRCC1 and clinical outcome
| Cancer | rs | SNPs | Alternate names | Reference | n | Outcomea |
|---|---|---|---|---|---|---|
| NSCLC | rs1799782 | Arg194Trp | 194 C > T; 194 R > W; 194 Arg > Trp; C26304T | Petty et al155,b | 49 | 0 |
| Wang et al156 | 139 | 0 | ||||
| Yuan et al157,b | 199 | 0 | ||||
| Yoon et al158 | 229 | 1 | ||||
| Sun et al159,b | 82 | 2 | ||||
| rs25489 | Arg280His | 280 G > A; 280 R > H; 280 Arg > His | Yoon et al158 | 229 | 0 | |
| Yuan et al157,b | 199 | (2) | ||||
| rs25487 | Arg399Gln | G28152 A; 399 G > A; 399 R > Q; 399 Arg > Gln | Yoon et al158 | 229 | 0 | |
| Petty et al155,b | 49 | 0 | ||||
| Sun et al159,b | 82 | 0 | ||||
| Yuan et al157 | 199 | 0 | ||||
| Gurubhagavatula et al161,c | 103 | (1) | ||||
| Kalikaki et al57 | 119 | 1 | ||||
| Yin et al67 | 257 | 1 | ||||
| Wang et al156 | 139 | 1 (toxicity) | ||||
| Giachino et al160,b | 203 | 2 (toxicity) | ||||
| De las Penas et al71,b | 135 | 2 | ||||
| rs3213245 | −(77) T > C | Hsieh et al115 | 294 | 0 | ||
| rs1799782 | Arg194Trp | 194 C > T; 194 R > W; C26304T | Geisler et al162 | 190 | 0 | |
| Csejtei et al145 | 108 | 1 | ||||
| rs25487 | Arg399Gln | G28152A; 399 G > A; 399 R > Q | Carles et al76 | 108 | 0 | |
| Csejtei et al145 | 108 | 0 | ||||
| Geisler et al162 | 190 | 2 | ||||
| Quintela-Fandino et al74 | 103 | 2 |
Notes:
Outcome for variable allele, 0 = non significant, (1) = trend to worse, 1 = worse, (2) = trend to better, 2 = better;
prospective study;
retrospective analysis of prospective study.
Abbreviations: NSCLC, non-small cell lung cancers; rs, reference SNP; SNPs, single nucleotide polymorphisms.
In HNSCC, XRCC1 has not been extensively studied. We identified only four reports assessing the predictive value of SNPs in XRCC1, focusing predominantly on Arg399Gln,74,76,145,162 and to a lesser extent Arg194Trp145,162 (Table 5). Results for Arg399Gln were mixed; two out of the four studies (total n = 293) showed a better outcome for the allelic variant.74,162 Interestingly, Arg194Trp, which was previously identified as a significant risk factor for HNSCC, did not influence treatment outcome.162 As with NSCLC, more studies and larger prospective studies are needed to evaluate whether SNPs in XRCC1 influence response to treatment in HNSCC.
XRCC1 expression as a biomarker of patient outcomes in cancer
There is very little data on XRCC1 expression in tumors, despite the fact that at least in NSCLC cell lines increased XRCC1 mRNA is significantly associated with cisplatin resistance.163 There are two studies (both using the same patient cohort) reporting XRCC1 expression in NSCLC, as measured by immunohistochemistry.88,164 XRCC1 protein expression did not correlate with either response to treatment or survival. Interestingly, more than half of the metastases had a stronger immunohistochemical signal than their matched primary tumor, suggesting that the level of XRCC1 may increase during cancer progression. This could have therapeutic implications if elevated expression of XRCC1 renders cells more resistant to treatment.
Only one study evaluated XRCC1 protein expression and clinical outcome in HNSCC.165 High XRCC1 expression was correlated with resistance to radiotherapy. There is also a paucity of studies on the predictive value of either peripheral or tumor XRCC1 mRNA in cancer. In contrast to the protein data, XRCC1 mRNA appears to be lower in early stage lung cancer compared with more advanced cancer.166
Conclusion
In summary, for the past decade the biomedical community has evaluated DNA repair genes as potential biomarkers to predict cancer risk and prognosis of cancer patients treated with genotoxic agents. There has been considerable investment toward this endeavor, yet none of the candidate biomarkers, other than BRCA1 and BRCA2, have yet to be translated to clinic use. ERCC1 and XRCC1 are two good candidate biomarkers, with robust experimental evidence demonstrating that reduced expression or activity of either protein results in increased genomic instability and sensitivity to DNA damaging agents.7,9–11,19 To date, investigations as to whether ERCC1 and XRCC1 alter cancer risk or outcomes are primarily modest-sized retrospective case controlled studies, which have yielded conflicting results. The strongest associations to date are that a CC genotype at SNP −77 of XRCC1, which causes reduced XRCC1 mRNA, predicts increased risk of NSCLC. For ERCC1, there are numerous studies indicating that low mRNA or protein expression is associated with a better prognosis in HNSCC and NSCLC, respectively. However, it is not established that ERCC1 expression is regulated at the transcriptional level. Furthermore, in the studies measuring protein level, a nonspecific antibody was used. Therefore these studies, while validating the utility of these biomarkers (ERCC1 mRNA levels or 8F1 immunohistochemical signal) for predicting clinical outcomes, do not directly demonstrate that DNA repair levels are altered in tumors.
Acknowledgments
We would like to give special thanks to Dr Laura Alonso for her careful review of the manuscript and her suggestions. AV is supported by a T32 National Institutes of Health training grant (T32 CA060397 to JR Grandis). LJN and CHF are supported by the National Institute of Environmental Health Sciences (RO1 ES016114 and −03S2, respectively).
Footnotes
Disclosure
The authors report no conflicts of interest in relation to this paper.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Seiwert TY, Salama JK, Vokes EE. The concurrent chemoradiation paradigm – general principles. Nat Clin Pract Oncol. 2007;4(2):86–100. doi: 10.1038/ncponc0714. [DOI] [PubMed] [Google Scholar]
- 3.Murdoch D. Standard, and novel cytotoxic and molecular-targeted, therapies for HNSCC: an evidence-based review. Curr Opin Oncol. 2007;19(3):216–221. doi: 10.1097/01.cco.0000264952.98166.99. [DOI] [PubMed] [Google Scholar]
- 4.Seiwert TY, Salama JK, Vokes EE. The chemoradiation paradigm in head and neck cancer. Nat Clin Pract Oncol. 2007;4(3):156–171. doi: 10.1038/ncponc0750. [DOI] [PubMed] [Google Scholar]
- 5.O’Driscoll M, Jeggo PA. The role of double-strand break repair – insights from human genetics. Nat Rev Genet. 2006;7(1):45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 6.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 7.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross) linked to DNA repair. Cell. 2005;123(7):1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24(23):3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 9.Niedernhofer LJ, Garinis GA, Raams A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad A, Robinson AR, Duensing A, et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28(16):5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhagwat N, Olsen AL, Wang AT, et al. XPF-ERCC1 participates in the fanconi anemia pathway of cross-link repair. Mol Cell Biol. 2009;29(24):6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaspers NG, Raams A, Silengo MC, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80(3):457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwelling LA, Anderson T, Kohn KW. DNA-protein and DNA inter-strand cross-linking by cis- and trans-platinum(II) diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity. Cancer Res. 1979;39(2 Pt 1):365–369. [PubMed] [Google Scholar]
- 14.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2(8):483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 15.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14(5):1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 16.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 17.Ladiges WC. Mouse models of XRCC1 DNA repair polymorphisms and cancer. Oncogene. 2006;25(11):1612–1619. doi: 10.1038/sj.onc.1209370. [DOI] [PubMed] [Google Scholar]
- 18.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6(6):695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2(9):955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Hatem J, Wang J, Quinn A, Hicks D, Tang P. Tissue microarray-based immunohistochemical study can significantly underestimate the expression of HER2 and progesterone receptor in ductal carcinoma in situ of the breast. Biotech Histochem. 2010 Aug 12; doi: 10.3109/10520295.2010.502845. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Tamaki K, Sasano H, Ishida T, et al. Comparison of core needle biopsy (CNB) and surgical specimens for accurate preoperative evaluation of ER, PgR and HER2 status of breast cancer patients. Cancer Sci. 2010;101(9):2074–2079. doi: 10.1111/j.1349-7006.2010.01630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taillade L, Penault-Llorca F, Boulet T, et al. Immunohistochemichal expression of biomarkers: a comparative study between diagnostic bronchial biopsies and surgical specimens of non-small-cell lung cancer. Ann Oncol. 2007;18(6):1043–1050. doi: 10.1093/annonc/mdm072. [DOI] [PubMed] [Google Scholar]
- 23.Babic A, Loftin IR, Stanislaw S, et al. The impact of pre-analytical processing on staining quality for H&E, dual hapten, dual color in situ hybridization and fluorescent in situ hybridization assays. Methods. 2010;52(4):287–300. doi: 10.1016/j.ymeth.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 24.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 25.Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. N Engl J Med. 2007;356(24):2538–2540. 2540–2531. doi: 10.1056/NEJMc070742. author reply. [DOI] [PubMed] [Google Scholar]
- 26.Britten RA, Liu D, Tessier A, Hutchison MJ, Murray D. ERCC1 expression as a molecular marker of cisplatin resistance in human cervical tumor cells. Int J Cancer. 2000;89(5):453–457. [PubMed] [Google Scholar]
- 27.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356(8):800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 28.McGurk CJ, Cummings M, Koberle B, Hartley JA, Oliver RT, Masters JR. Regulation of DNA repair gene expression in human cancer cell lines. J Cell Biochem. 2006;97(5):1121–1136. doi: 10.1002/jcb.20711. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- 30.Schneider J, Classen V, Philipp M, Helmig S. Rapid analysis of XRCC1 polymorphisms using real-time polymerase chain reaction. Mol Cell Probes. 2006;20(3–4):259–262. doi: 10.1016/j.mcp.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Fountzilas G, Kalogera-Fountzila A, Lambaki S, et al. MMP9 but not EGFR, MET, ERCC1, P16, and P-53 is associated with response to concomitant radiotherapy, cetuximab, and weekly cisplatin in patients with locally advanced head and neck cancer. J Oncol. 2009;2009:305908. doi: 10.1155/2009/305908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matullo G, Dunning AM, Guarrera S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27(5):997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 33.Ferte C, Andre F, Soria JC. Molecular circuits of solid tumors: prognostic and predictive tools for bedside use. Nat Rev Clin Oncol. 2010;7(7):367–380. doi: 10.1038/nrclinonc.2010.84. [DOI] [PubMed] [Google Scholar]
- 34.Srivastava S, Gray JW, Reid BJ, Grad O, Greenwood A, Hawk ET. Translational Research Working Group developmental pathway for biospecimen-based assessment modalities. Clin Cancer Res. 2008;14(18):5672–5677. doi: 10.1158/1078-0432.CCR-08-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ford BN, Ruttan CC, Kyle VL, Brackley ME, Glickman BW. Identification of single nucleotide polymorphisms in human DNA repair genes. Carcinogenesis. 2000;21(11):1977–1981. doi: 10.1093/carcin/21.11.1977. [DOI] [PubMed] [Google Scholar]
- 36.Yu JJ, Mu C, Lee KB, et al. A nucleotide polymorphism in ERCC1 in human ovarian cancer cell lines and tumor tissues. Mutat Res. 1997;382(1–2):13–20. doi: 10.1016/s1383-5726(97)00004-6. [DOI] [PubMed] [Google Scholar]
- 37.Yang M, Kim WH, Choi Y, et al. Effects of ERCC1 expression in peripheral blood on the risk of head and neck cancer. Eur J Cancer Prev. 2006;15(3):269–273. doi: 10.1097/01.cej.0000195709.79696.0c. [DOI] [PubMed] [Google Scholar]
- 38.Okuda K, Sasaki H, Hikosaka Y, et al. Excision repair cross complementation group 1 polymorphisms predict overall survival after platinum-based chemotherapy for completely resected non-small-cell lung cancer. J Surg Res. 2009 Sep 26; doi: 10.1016/j.jss.2009.09.006. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 39.Zhou W, Liu G, Park S, et al. Gene-smoking interaction associations for the ERCC1 polymorphisms in the risk of lung cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(2):491–496. doi: 10.1158/1055-9965.EPI-04-0612. [DOI] [PubMed] [Google Scholar]
- 40.Yin J, Vogel U, Guo L, Ma Y, Wang H. Lack of association between DNA repair gene ERCC1 polymorphism and risk of lung cancer in a Chinese population. Cancer Genet Cytogenet. 2006;164(1):66–70. doi: 10.1016/j.cancergencyto.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Hung RJ, Christiani DC, Risch A, et al. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu D, Zhang X, Liu J, et al. Characterization of functional excision repair cross-complementation group 1 variants and their association with lung cancer risk and prognosis. Clin Cancer Res. 2008;14(9):2878–2886. doi: 10.1158/1078-0432.CCR-07-1612. [DOI] [PubMed] [Google Scholar]
- 43.Deng Q, Sheng L, Su D, et al. Genetic polymorphisms in ATM, ERCC1, APE1 and iASPP genes and lung cancer risk in a population of southeast China. Med Oncol. 2010 Mar 31; doi: 10.1007/s12032-010-9507-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 44.Zienolddiny S, Campa D, Lind H, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27(3):560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 45.Ma H, Xu L, Yuan J, et al. Tagging single nucleotide polymorphisms in excision repair cross-complementing group 1 (ERCC1) and risk of primary lung cancer in a Chinese population. Pharmacogenet Genomics. 2007;17(6):417–423. doi: 10.1097/01.fpc.0000239975.77088.17. [DOI] [PubMed] [Google Scholar]
- 46.Shen M, Berndt SI, Rothman N, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005;116(5):768–773. doi: 10.1002/ijc.21117. [DOI] [PubMed] [Google Scholar]
- 47.Jones NR, Spratt TE, Berg AS, Muscat JE, Lazarus P, Gallagher CJ. Association studies of excision repair cross-complementation group 1 (ERCC1) haplotypes with lung and head and neck cancer risk in a Caucasian population. Cancer Epidemiol. 2011;35(2):175–181. doi: 10.1016/j.canep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Gu S, Wu Q, et al. No association of ERCC1 C8092A and T19007C polymorphisms to cancer risk: a meta-analysis. Eur J Hum Genet. 2007;15(9):967–973. doi: 10.1038/sj.ejhg.5201855. [DOI] [PubMed] [Google Scholar]
- 49.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci. 2007;4(2):59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vineis P, Manuguerra M, Kavvoura FK, et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101(1):24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 51.Cheng L, Spitz MR, Hong WK, Wei Q. Reduced expression levels of nucleotide excision repair genes in lung cancer: a case-control analysis. Carcinogenesis. 2000;21(8):1527–1530. [PubMed] [Google Scholar]
- 52.Sugimura T, Kumimoto H, Tohnai I, et al. Gene-environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med. 2006;35(1):11–18. doi: 10.1111/j.1600-0714.2005.00364.x. [DOI] [PubMed] [Google Scholar]
- 53.Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, Popanda O. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125(6):1431–1439. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- 54.Canova C, Hashibe M, Simonato L, et al. Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 European countries: the ARCAGE project. Cancer Res. 2009;69(7):2956–2965. doi: 10.1158/0008-5472.CAN-08-2604. [DOI] [PubMed] [Google Scholar]
- 55.Sturgis EM, Dahlstrom KR, Spitz MR, Wei Q. DNA repair gene ERCC1 and ERCC2/XPD polymorphisms and risk of squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2002;128(9):1084–1088. doi: 10.1001/archotol.128.9.1084. [DOI] [PubMed] [Google Scholar]
- 56.Cheng L, Sturgis EM, Eicher SA, Spitz MR, Wei Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer. 2002;94(2):393–397. doi: 10.1002/cncr.10231. [DOI] [PubMed] [Google Scholar]
- 57.Kalikaki A, Kanaki M, Vassalou H, et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10(2):118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 58.Gandara DR, Kawaguchi T, Crowley J, et al. Japanese-US common-arm analysis of paclitaxel plus carboplatin in advanced non-small-cell lung cancer: a model for assessing population-related pharmacogenomics. J Clin Oncol. 2009;27(21):3540–3546. doi: 10.1200/JCO.2008.20.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suk R, Gurubhagavatula S, Park S, et al. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. 2005;11(4):1534–1538. doi: 10.1158/1078-0432.CCR-04-1953. [DOI] [PubMed] [Google Scholar]
- 60.Takenaka T, Yoshino I, Kouso H, et al. Combined evaluation of Rad51 and ERCC1 expressions for sensitivity to platinum agents in non-small cell lung cancer. Int J Cancer. 2007;121(4):895–900. doi: 10.1002/ijc.22738. [DOI] [PubMed] [Google Scholar]
- 61.Tibaldi C, Giovannetti E, Vasile E, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14(6):1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 62.Vinolas N, Provencio M, Reguart N, et al. Single nucleotide polymorphisms in MDR1 gen correlates with outcome in advanced non-small-cell lung cancer patients treated with cisplatin plus vinorelbine. Lung Cancer. 2011;71(2):191–198. doi: 10.1016/j.lungcan.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 63.Zhou W, Gurubhagavatula S, Liu G, et al. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2004;10(15):4939–4943. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 64.Park SY, Hong YC, Kim JH, et al. Effect of ERCC1 polymorphisms and the modification by smoking on the survival of non-small cell lung cancer patients. Med Oncol. 2006;23(4):489–498. doi: 10.1385/MO:23:4:489. [DOI] [PubMed] [Google Scholar]
- 65.Ryu JS, Hong YC, Han HS, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44(3):311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 66.Su D, Ma S, Liu P, et al. Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer. 2007;56(2):281–288. doi: 10.1016/j.lungcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Yin Z, Zhou B, He Q, et al. Association between polymorphisms in DNA repair genes and survival of non-smoking female patients with lung adenocarcinoma. BMC Cancer. 2009;9:439. doi: 10.1186/1471-2407-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15(8):1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 69.Zhou C, Ren S, Zhou S, et al. Predictive effects of ERCC1 and XRCC3 SNP on efficacy of platinum-based chemotherapy in advanced NSCLC patients. Jpn J Clin Oncol. 2010;40(10):954–960. doi: 10.1093/jjco/hyq071. [DOI] [PubMed] [Google Scholar]
- 70.Li F, Sun X, Sun N, et al. Association between polymorphisms of ERCC1 and XPD and clinical response to platinum-based chemotherapy in advanced non-small cell lung cancer. Am J Clin Oncol. 2010;33(5):489–494. doi: 10.1097/COC.0b013e3181b9cedc. [DOI] [PubMed] [Google Scholar]
- 71.De las Penas R, Sanchez-Ronco M, Alberola V, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17(4):668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 72.Wei SZ, Zhan P, Shi MQ, et al. Predictive value of ERCC1 and XPD polymorphism in patients with advanced non-small cell lung cancer receiving platinum-based chemotherapy: a systematic review and meta-analysis. Med Oncol. 2011;28(1):315–321. doi: 10.1007/s12032-010-9443-1. [DOI] [PubMed] [Google Scholar]
- 73.Takenaka T, Yano T, Kiyohara C, et al. Effects of excision repair cross-complementation group 1 (ERCC1) single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients. Lung Cancer. 2010;67(1):101–107. doi: 10.1016/j.lungcan.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 74.Quintela-Fandino M, Hitt R, Medina PP, et al. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24(26):4333–4339. doi: 10.1200/JCO.2006.05.8768. [DOI] [PubMed] [Google Scholar]
- 75.Grau JJ, Caballero M, Campayo M, et al. Gene single nucleotide polymorphism accumulation improves survival in advanced head and neck cancer patients treated with weekly paclitaxel. Laryngoscope. 2009;119(8):1484–1490. doi: 10.1002/lary.20254. [DOI] [PubMed] [Google Scholar]
- 76.Carles J, Monzo M, Amat M, et al. Single-nucleotide polymorphisms in base excision repair, nucleotide excision repair, and double strand break genes as markers for response to radiotherapy in patients with Stage I to II head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2006;66(4):1022–1030. doi: 10.1016/j.ijrobp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, Zhao J, Yang L, et al. Positive expression of ERCC1 predicts a poorer platinum-based treatment outcome in Chinese patients with advanced non-small-cell lung cancer. Med Oncol. 2010;27(2):484–490. doi: 10.1007/s12032-009-9239-3. [DOI] [PubMed] [Google Scholar]
- 78.Vilmar A, Santoni-Rugiu E, Sorensen JB. ERCC1, toxicity and quality of life in advanced NSCLC patients randomized in a large multicentre phase III trial. Eur J Cancer. 2010;46(9):1554–1562. doi: 10.1016/j.ejca.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 79.Reynolds C, Obasaju C, Schell MJ, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol. 2009;27(34):5808–5815. doi: 10.1200/JCO.2009.21.9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ota S, Ishii G, Goto K, et al. Immunohistochemical expression of BCRP and ERCC1 in biopsy specimen predicts survival in advanced non-small-cell lung cancer treated with cisplatin-based chemotherapy. Lung Cancer. 2009;64(1):98–104. doi: 10.1016/j.lungcan.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 81.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355(10):983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 82.Lee HW, Choi YW, Han JH, et al. Expression of excision repair cross-complementation group 1 protein predicts poor outcome in advanced non-small cell lung cancer patients treated with platinum-based doublet chemotherapy. Lung Cancer. 2009;65(3):377–382. doi: 10.1016/j.lungcan.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 83.Fujii T, Toyooka S, Ichimura K, et al. ERCC1 protein expression predicts the response of cisplatin-based neoadjuvant chemotherapy in non-small-cell lung cancer. Lung Cancer. 2008;59(3):377–384. doi: 10.1016/j.lungcan.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 84.Azuma K, Komohara Y, Sasada T, et al. Excision repair cross-complementation group 1 predicts progression-free and overall survival in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Sci. 2007;98(9):1336–1343. doi: 10.1111/j.1349-7006.2007.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Azuma K, Sasada T, Kawahara A, et al. Expression of ERCC1 and class III beta-tubulin in non-small cell lung cancer patients treated with a combination of cisplatin/docetaxel and concurrent thoracic irradiation. Cancer Chemother Pharmacol. 2009;64(3):565–573. doi: 10.1007/s00280-008-0907-3. [DOI] [PubMed] [Google Scholar]
- 86.Holm B, Mellemgaard A, Skov T, Skov BG. Different impact of excision repair cross-complementation group 1 on survival in male and female patients with inoperable non-small-cell lung cancer treated with carboplatin and gemcitabine. J Clin Oncol. 2009;27(26):4254–4259. doi: 10.1200/JCO.2008.18.8631. [DOI] [PubMed] [Google Scholar]
- 87.Lee KH, Min HS, Han SW, et al. ERCC1 expression by immunohistochemistry and EGFR mutations in resected non-small cell lung cancer. Lung Cancer. 2008;60(3):401–407. doi: 10.1016/j.lungcan.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 88.Kang CH, Jang BG, Kim DW, et al. The prognostic significance of ERCC1, BRCA1, XRCC1, and betaIII-tubulin expression in patients with non-small cell lung cancer treated by platinum- and taxane-based neoadjuvant chemotherapy and surgical resection. Lung Cancer. 2010;68(3):478–483. doi: 10.1016/j.lungcan.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 89.Koh Y, Jang B, Han SW, et al. Expression of class III beta-tubulin correlates with unfavorable survival outcome in patients with resected non-small cell lung cancer. J Thorac Oncol. 2010;5(3):320–325. doi: 10.1097/JTO.0b013e3181ce684f. [DOI] [PubMed] [Google Scholar]
- 90.Planchard D, Domont J, Taranchon E, et al. The NER proteins are differentially expressed in ever smokers and in never smokers with lung adenocarcinoma. Ann Oncol. 2009;20(7):1257–1263. doi: 10.1093/annonc/mdn785. [DOI] [PubMed] [Google Scholar]
- 91.Okuda K, Sasaki H, Dumontet C, et al. Expression of excision repair cross-complementation group 1 and class III beta-tubulin predict survival after chemotherapy for completely resected non-small cell lung cancer. Lung Cancer. 2008;62(1):105–112. doi: 10.1016/j.lungcan.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 92.Vilmar AC, Santoni-Rugiu E, Sorensen JB. ERCC1 and histopathology in advanced NSCLC patients randomized in a large multicenter phase III trial. Ann Oncol. 2010;21(9):1817–1824. doi: 10.1093/annonc/mdq053. [DOI] [PubMed] [Google Scholar]
- 93.Chen S, Zhang J, Wang R, Luo X, Chen H. The platinum-based treatments for advanced non-small cell lung cancer, is low/negative ERCC1 expression better than high/positive ERCC1 expression? A meta-analysis. Lung Cancer. 2010;70(1):63–70. doi: 10.1016/j.lungcan.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 94.Doll CM, Prystajecky M, Eliasziw M, et al. Low ERCC1 mRNA and protein expression are associated with worse survival in cervical cancer patients treated with radiation alone. Radiother Oncol. 2010;97(2):352–359. doi: 10.1016/j.radonc.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 95.Fountzilas G, Bamias A, Kalogera-Fountzila A, et al. Induction chemotherapy with docetaxel and cisplatin followed by concomitant chemoradiotherapy in patients with inoperable non-nasopharyngeal carcinoma of the head and neck. Anticancer Res. 2009;29(2):529–538. [PubMed] [Google Scholar]
- 96.Koh Y, Kim TM, Jeon YK, et al. Class III beta-tubulin, but not ERCC1, is a strong predictive and prognostic marker in locally advanced head and neck squamous cell carcinoma. Ann Oncol. 2009;20(8):1414–1419. doi: 10.1093/annonc/mdp002. [DOI] [PubMed] [Google Scholar]
- 97.Handra-Luca A, Hernandez J, Mountzios G, et al. Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13(13):3855–3859. doi: 10.1158/1078-0432.CCR-07-0252. [DOI] [PubMed] [Google Scholar]
- 98.Jun HJ, Ahn MJ, Kim HS, et al. ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. Br J Cancer. 2008;99(1):167–172. doi: 10.1038/sj.bjc.6604464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shimizu J, Horio Y, Osada H, et al. mRNA expression of RRM1, ERCC1 and ERCC2 is not associated with chemosensitivity to cis-platin, carboplatin and gemcitabine in human lung cancer cell lines. Respirology. 2008;13(4):510–517. doi: 10.1111/j.1440-1843.2008.01302.x. [DOI] [PubMed] [Google Scholar]
- 100.Rosell R, Felip E, Taron M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res. 2004;10(12 Pt 2):4215s–4219s. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- 101.Simon GR, Sharma S, Cantor A, Smith P, Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127(3):978–983. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 102.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17(12):1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 103.Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8(7):2286–2291. [PubMed] [Google Scholar]
- 104.Ceppi P, Longo M, Volante M, et al. Excision repair cross complementing-1 and topoisomerase IIalpha gene expression in small-cell lung cancer patients treated with platinum and etoposide: a retrospective study. J Thorac Oncol. 2008;3(6):583–589. doi: 10.1097/JTO.0b013e3181734f24. [DOI] [PubMed] [Google Scholar]
- 105.Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25(19):2747–2754. doi: 10.1200/JCO.2006.09.7915. [DOI] [PubMed] [Google Scholar]
- 106.Booton R, Ward T, Ashcroft L, Morris J, Heighway J, Thatcher N. ERCC1 mRNA expression is not associated with response and survival after platinum-based chemotherapy regimens in advanced non-small cell lung cancer. J Thorac Oncol. 2007;2(10):902–906. doi: 10.1097/JTO.0b013e318155a637. [DOI] [PubMed] [Google Scholar]
- 107.Simon G, Sharma A, Li X, et al. Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2007;25(19):2741–2746. doi: 10.1200/JCO.2006.08.2099. [DOI] [PubMed] [Google Scholar]
- 108.Su C, Zhou S, Zhang L, et al. ERCC1, RRM1 and BRCA1 mRNA expression levels and clinical outcome of advanced non-small cell lung cancer. Med Oncol. 2010 May 14; doi: 10.1007/s12032-010-9553-9. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 109.Ren S, Zhou S, Zhang L, et al. High-level mRNA of excision repair cross-complementation group 1 gene is associated with poor outcome of platinum-based doublet chemotherapy of advanced nonsmall cell lung cancer patients. Cancer Invest. 2010;28(10):1078–1083. doi: 10.3109/07357901003735659. [DOI] [PubMed] [Google Scholar]
- 110.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24(29):4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 111.Baghwat Neha, Niedernhofer Laura J. Pers comm; 13th Annual Midwest DNA Repair Meeting; Toledo, Ohio, USA. 2011. [Google Scholar]
- 112.Schneider J, Classen V, Bernges U, Philipp M. XRCC1 polymorphism and lung cancer risk in relation to tobacco smoking. Int J Mol Med. 2005;16(4):709–716. [PubMed] [Google Scholar]
- 113.Hao B, Miao X, Li Y, et al. A novel T-77C polymorphism in DNA repair gene XRCC1 contributes to diminished promoter activity and increased risk of non-small cell lung cancer. Oncogene. 2006;25(25):3613–3620. doi: 10.1038/sj.onc.1209355. [DOI] [PubMed] [Google Scholar]
- 114.Hu Z, Ma H, Lu D, et al. A promoter polymorphism (−77T > C) of DNA repair gene XRCC1 is associated with risk of lung cancer in relation to tobacco smoking. Pharmacogenet Genomics. 2005;15(7):457–463. doi: 10.1097/01.fpc.0000167329.85163.0d. [DOI] [PubMed] [Google Scholar]
- 115.Hsieh WC, Cheng YW, Lin CJ, Chou MC, Chen CY, Lee H. Prognostic significance of X-ray cross-complementing group 1 T-77C polymorphism in resected non-small cell lung cancer. Jpn J Clin Oncol. 2009;39(2):81–85. doi: 10.1093/jjco/hyn130. [DOI] [PubMed] [Google Scholar]
- 116.De Ruyck K, Szaumkessel M, De Rudder I, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631(2):101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 117.Ladiges W, Wiley J, MacAuley A. Polymorphisms in the DNA repair gene XRCC1 and age-related disease. Mech Ageing Dev. 2003;124(1):27–32. doi: 10.1016/s0047-6374(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 118.Cheng J, Leng S, Li H, et al. Suboptimal DNA repair capacity predisposes coke-oven workers to accumulate more chromosomal damages in peripheral lymphocytes. Cancer Epidemiol Biomarkers Prev. 2009;18(3):987–993. doi: 10.1158/1055-9965.EPI-08-0763. [DOI] [PubMed] [Google Scholar]
- 119.Abdel-Rahman SZ, El-Zein RA. The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett. 2000;159(1):63–71. doi: 10.1016/s0304-3835(00)00532-2. [DOI] [PubMed] [Google Scholar]
- 120.Relton CL, Daniel CP, Fisher A, Chase DS, Burn J, Tawn EJ. Polymorphisms of the DNA repair gene XRCC1 and the frequency of somatic mutations at the glycophorin A locus in newborns. Mutat Res. 2002;502(1–2):61–68. doi: 10.1016/s0027-5107(02)00030-1. [DOI] [PubMed] [Google Scholar]
- 121.Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59(11):2557–2561. [PubMed] [Google Scholar]
- 122.Misra RR, Ratnasinghe D, Tangrea JA, et al. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett. 2003;191(2):171–178. doi: 10.1016/s0304-3835(02)00638-9. [DOI] [PubMed] [Google Scholar]
- 123.Yin J, Vogel U, Ma Y, Qi R, Wang H. Association of DNA repair gene XRCC1 and lung cancer susceptibility among nonsmoking Chinese women. Cancer Genet Cytogenet. 2009;188(1):26–31. doi: 10.1016/j.cancergencyto.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 124.Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis. 2001;22(4):593–597. doi: 10.1093/carcin/22.4.593. [DOI] [PubMed] [Google Scholar]
- 125.Chen S, Tang D, Xue K, et al. DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis. 2002;23(8):1321–1325. doi: 10.1093/carcin/23.8.1321. [DOI] [PubMed] [Google Scholar]
- 126.Improta G, Sgambato A, Bianchino G, et al. Polymorphisms of the DNA repair genes XRCC1 and XRCC3 and risk of lung and colorectal cancer: a case-control study in a Southern Italian population. Anticancer Res. 2008;28(5B):2941–2946. [PubMed] [Google Scholar]
- 127.Hung RJ, Brennan P, Canzian F, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst. 2005;97(8):567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 128.Ratnasinghe D, Yao SX, Tangrea JA, et al. Polymorphisms of the DNA repair gene XRCC1 and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10(2):119–123. [PubMed] [Google Scholar]
- 129.Shen M, Berndt SI, Rothman N, et al. Polymorphisms in the DNA base excision repair genes APEX1 and XRCC1 and lung cancer risk in Xuan Wei, China. Anticancer Res. 2005;25(1B):537–542. [PubMed] [Google Scholar]
- 130.Tanaka Y, Maniwa Y, Bermudez VP, et al. Nonsynonymous single nucleotide polymorphisms in DNA damage repair pathways and lung cancer risk. Cancer. 2010;116(4):896–902. doi: 10.1002/cncr.24850. [DOI] [PubMed] [Google Scholar]
- 131.Yin J, Vogel U, Ma Y, Qi R, Sun Z, Wang H. The DNA repair gene XRCC1 and genetic susceptibility of lung cancer in a northeastern Chinese population. Lung Cancer. 2007;56(2):153–160. doi: 10.1016/j.lungcan.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 132.David-Beabes GL, London SJ. Genetic polymorphism of XRCC1 and lung cancer risk among African-Americans and Caucasians. Lung Cancer. 2001;34(3):333–339. doi: 10.1016/s0169-5002(01)00256-2. [DOI] [PubMed] [Google Scholar]
- 133.Pachouri SS, Sobti RC, Kaur P, Singh J. Contrasting impact of DNA repair gene XRCC1 polymorphisms Arg399Gln and Arg194Trp on the risk of lung cancer in the north-Indian population. DNA Cell Biol. 2007;26(3):186–191. doi: 10.1089/dna.2006.9999. [DOI] [PubMed] [Google Scholar]
- 134.Vogel U, Nexo BA, Wallin H, Overvad K, Tjonneland A, Raaschou-Nielsen O. No association between base excision repair gene polymorphisms and risk of lung cancer. Biochem Genet. 2004;42(11–12):453–460. doi: 10.1023/b:bigi.0000043957.03420.7e. [DOI] [PubMed] [Google Scholar]
- 135.Ito H, Matsuo K, Hamajima N, et al. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25(8):1395–1401. doi: 10.1093/carcin/bgh153. [DOI] [PubMed] [Google Scholar]
- 136.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Popanda O, Schattenberg T, Phong CT, et al. Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis. 2004;25(12):2433–2441. doi: 10.1093/carcin/bgh264. [DOI] [PubMed] [Google Scholar]
- 138.Ryk C, Kumar R, Thirumaran RK, Hou SM. Polymorphisms in the DNA repair genes XRCC1, APEX1, XRCC3 and NBS1, and the risk for lung cancer in never- and ever-smokers. Lung Cancer. 2006;54(3):285–292. doi: 10.1016/j.lungcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 139.Zhang X, Miao X, Liang G, et al. Polymorphisms in DNA base excision repair genes ADPRT and XRCC1 and risk of lung cancer. Cancer Res. 2005;65(3):722–726. [PubMed] [Google Scholar]
- 140.Park JY, Lee SY, Jeon HS, et al. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11(1):23–27. [PubMed] [Google Scholar]
- 141.Zhou W, Liu G, Miller DP, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12(4):359–365. [PubMed] [Google Scholar]
- 142.Sreeja L, Syamala VS, Syamala V, et al. Prognostic importance of DNA repair gene polymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln in lung cancer patients from India. J Cancer Res Clin Oncol. 2008;134(6):645–652. doi: 10.1007/s00432-007-0328-4. [DOI] [PubMed] [Google Scholar]
- 143.Divine KK, Gilliland FD, Crowell RE, et al. The XRCC1 399 glutamine allele is a risk factor for adenocarcinoma of the lung. Mutat Res. 2001;461(4):273–278. doi: 10.1016/s0921-8777(00)00059-8. [DOI] [PubMed] [Google Scholar]
- 144.Applebaum KM, McClean MD, Nelson HH, Marsit CJ, Christensen BC, Kelsey KT. Smoking modifies the relationship between XRCC1 haplotypes and HPV16-negative head and neck squamous cell carcinoma. Int J Cancer. 2009;124(11):2690–2696. doi: 10.1002/ijc.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Csejtei A, Tibold A, Koltai K, et al. Association between XRCC1 polymorphisms and head and neck cancer in a Hungarian population. Anticancer Res. 2009;29(10):4169–4173. [PubMed] [Google Scholar]
- 146.Harth V, Schafer M, Abel J, et al. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A. 2008;71(13–14):887–897. doi: 10.1080/15287390801988160. [DOI] [PubMed] [Google Scholar]
- 147.Shen H, Sturgis EM, Khan SG, et al. An intronic poly (AT) polymorphism of the DNA repair gene XPC and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Res. 2001;61(8):3321–3325. [PubMed] [Google Scholar]
- 148.Olshan AF, Watson MA, Weissler MC, Bell DA. XRCC1 polymorphisms and head and neck cancer. Cancer Lett. 2002;178(2):181–186. doi: 10.1016/s0304-3835(01)00822-9. [DOI] [PubMed] [Google Scholar]
- 149.Kowalski M, Przybylowska K, Rusin P, et al. Genetic polymorphisms in DNA base excision repair gene XRCC1 and the risk of squamous cell carcinoma of the head and neck. J Exp Clin Cancer Res. 2009;28:37. doi: 10.1186/1756-9966-28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tae K, Lee HS, Park BJ, et al. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer. 2004;111(5):805–808. doi: 10.1002/ijc.20338. [DOI] [PubMed] [Google Scholar]
- 151.Sturgis EM, Castillo EJ, Li L, et al. Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis. 1999;20(11):2125–2129. doi: 10.1093/carcin/20.11.2125. [DOI] [PubMed] [Google Scholar]
- 152.Cho EY, Hildesheim A, Chen CJ, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol Biomarkers Prev. 2003;12(10):1100–1104. [PubMed] [Google Scholar]
- 153.Gal TJ, Huang WY, Chen C, Hayes RB, Schwartz SM. DNA repair gene polymorphisms and risk of second primary neoplasms and mortality in oral cancer patients. Laryngoscope. 2005;115(12):2221–2231. doi: 10.1097/01.mlg.0000183736.96004.f7. [DOI] [PubMed] [Google Scholar]
- 154.Huang WY, Olshan AF, Schwartz SM, et al. Selected genetic polymorphisms in MGMT, XRCC1, XPD, and XRCC3 and risk of head and neck cancer: a pooled analysis. Cancer Epidemiol Biomarkers Prev. 2005;14(7):1747–1753. doi: 10.1158/1055-9965.EPI-05-0162. [DOI] [PubMed] [Google Scholar]
- 155.Petty WJ, Knight SN, Mosley L, et al. A pharmacogenomic study of docetaxel and gemcitabine for the initial treatment of advanced non-small cell lung cancer. J Thorac Oncol. 2007;2(3):197–202. doi: 10.1097/JTO.0b013e318031cd89. [DOI] [PubMed] [Google Scholar]
- 156.Wang Z, Xu B, Lin D, et al. XRCC1 polymorphisms and severe toxicity in lung cancer patients treated with cisplatin-based chemotherapy in Chinese population. Lung Cancer. 2008;62(1):99–104. doi: 10.1016/j.lungcan.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 157.Yuan P, Liu L, Wu C, et al. No association between XRCC1 polymorphis and survival in non-small-cell lung cancer patients treated with platinum-based chemotherapy. Cancer Biol Ther. 2010;10(9) doi: 10.4161/cbt.10.9.13238. [DOI] [PubMed] [Google Scholar]
- 158.Yoon SM, Hong YC, Park HJ, et al. The polymorphism and haplotypes of XRCC1 and survival of non-small-cell lung cancer after radiotherapy. Int J Radiat Oncol Biol Phys. 2005;63(3):885–891. doi: 10.1016/j.ijrobp.2005.07.951. [DOI] [PubMed] [Google Scholar]
- 159.Sun X, Li F, Sun N, et al. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65(2):230–236. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 160.Giachino DF, Ghio P, Regazzoni S, et al. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13(10):2876–2881. doi: 10.1158/1078-0432.CCR-06-2543. [DOI] [PubMed] [Google Scholar]
- 161.Gurubhagavatula S, Liu G, Park S, et al. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22(13):2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 162.Geisler SA, Olshan AF, Cai J, Weissler M, Smith J, Bell D. Glutathione S-transferase polymorphisms and survival from head and neck cancer. Head Neck. 2005;27(3):232–242. doi: 10.1002/hed.20141. [DOI] [PubMed] [Google Scholar]
- 163.Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer. 2005;4(1):18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kang CH, Jang BG, Kim DW, et al. Differences in the expression profiles of excision repair crosscomplementation group 1, x-ray repair crosscomplementation group 1, and betaIII-tubulin between primary non-small cell lung cancer and metastatic lymph nodes and the significance in mid-term survival. J Thorac Oncol. 2009;4(11):1307–1312. doi: 10.1097/JTO.0b013e3181b9f236. [DOI] [PubMed] [Google Scholar]
- 165.Nix P, Greenman J, Stafford N, Cawkwell L. Expression of XRCC1 and ERCC1 proteins in radioresistant and radiosensitive laryngeal cancer. Cancer Therapy. 2004;(2):47–53. [Google Scholar]
- 166.Campioni M, Ambrogi V, Pompeo E, et al. Identification of genes down-regulated during lung cancer progression: a cDNA array study. J Exp Clin Cancer Res. 2008;27:38. doi: 10.1186/1756-9966-27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jones NR, Spratt TE, Berg AS, Muscat JE, Lazarus P, Gallagher CJ. Association studies of excision repair cross-complementation group 1 (ERCC1) haplotypes with lung and head and neck cancer risk in a Caucasian population. Cancer Epidemiol. 2010;35(2):175–81. doi: 10.1016/j.canep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Varzim G, Monteiro E, Silva RA, Fernandes J, Lopes C. CYP1A1 and XRCC1 gene polymorphisms in SCC of the larynx. Eur J Cancer Prev. 2003;12(6):495–499. doi: 10.1097/00008469-200312000-00008. [DOI] [PubMed] [Google Scholar]