Abstract
Background:
Genetic variability in ABCB1, encoding the P-glycoprotein efflux transporter, has been linked to altered methadone maintenance treatment dose requirements. However, subsequent studies have indicated that additional environmental or genetic factors may confound ABCB1 pharmacogenetics in different methadone maintenance treatment settings. There is evidence that genetic variability in OPRM1, encoding the mu opioid receptor, and ABCB1 may interact to affect morphine response in opposite ways. This study aimed to examine whether a similar gene-gene interaction occurs for methadone in methadone maintenance treatment.
Methods:
Opioid-dependent subjects (n = 119) maintained on methadone (15–300 mg/day) were genotyped for five single nucleotide polymorphisms of ABCB1 (61A > G; 1199G > A; 1236C > T; 2677G > T; 3435C > T), as well as for the OPRM1 118A > G single nucleotide polymorphism. Subjects’ methadone doses and trough plasma (R)-methadone concentrations (Ctrough) were compared between ABCB1 haplotypes (with and without controlling for OPRM1 genotype), and between OPRM1 genotypes (with and without controlling for ABCB1 haplotype).
Results:
Among wild-type OPRM1 subjects, an ABCB1 variant haplotype group (subjects with a wild-type and 61A:1199G:1236C:2677T:3435T haplotype combination, or homozygous for the 61A:1199G:1236C:2677T:3435T haplotype) had significantly lower doses (median ± standard deviation 35 ± 5 versus 180 ± 65 mg/day, P < 0.01) and Ctrough (78 ± 22 versus 177 ± 97 ng/mL, P < 0.05) than ABCB1 wild-type subjects. Among subjects with the most common ABCB1 haplotype combination (wild-type with 61A:1199G:1236T:2677T:3435T), the OPRM1 118 A/G genotype was associated with a significantly higher Ctrough than 118 A/A (250 ± 126 versus 108 ± 36 ng/mL, P = 0.016). No ABCB1 haplotype group or OPRM1 genotype was associated with dose or Ctrough without taking into account confounding genetic variability at the other locus. Therefore, two interacting pharmacogenetic determinants of methadone maintenance treatment response were identified, ie, ABCB1, where variants are associated with lower methadone requirements, and OPRM1, where the variant is associated with higher methadone requirements.
Conclusion:
These opposing pharmacogenetic effects therefore need to be considered in combination when assessing methadone maintenance treatment pharmacogenetics.
Keywords: methadone, opiate substitution treatment, ABCB1, P-glycoprotein, OPRM1, receptors, opioid, mu
Introduction
Long-term opioid maintenance remains the most cost-effective approach for managing opioid dependence.1 However, the safe and effective use of substitution opioids, such as methadone, relies heavily on optimal dosing that minimizes both withdrawal and adverse opioid side effects, whilst reducing heroin use. This is made difficult by the narrow therapeutic index and large interindividual variability observed in the dose-response and plasma concentration-response relationship for methadone.2–5 Despite application of individualized treatment strategies that titrate dose against patient symptoms, attrition rates in Australian methadone maintenance therapy programs remain high (62%–74% at 12 months).6,7 Therefore, a better understanding of the factors underlying individual responses to methadone is required in order to improve treatment individualization and enhance clinical efficacy.
In addition to pathology and environmental factors such as diet, smoking, drug-drug, and food-drug interactions, there is increasing recognition that genetic factors may play a role in shaping the pharmacokinetic and pharmacodynamic aspects of the individual response to opioids.8 Thus, patient genetics could be used to guide dose individualization further, helping to reduce the incidence of withdrawal and adverse effects, particularly during the induction phase of treatment, and improve treatment retention.
Some investigations into the pharmacogenetics of methadone metabolism have suggested a role for CYP2D6 and CYP2B6 genetic variability in influencing systemic methadone exposure,2,8–11 whilst others have not been able to confirm some of these findings.12 Even when variability in plasma methadone pharmacokinetics is accounted for, there remains a substantial (five-fold) range of methadone concentrations required to suppress opioid withdrawal.3 Therefore, methadone is clearly influenced by additional factors affecting central nervous system distribution and pharmacodynamic response.
One possible pharmacogenetic determinant of methadone central nervous system distribution is the ABCB1 (MDR1) gene encoding the P-glycoprotein efflux transporter, for which methadone is a substrate.13,14 P-glycoprotein has a wide tissue distribution, but its most important role relating to opioid drugs is likely at the blood–brain barrier, where it can directly influence central nervous system exposure to substrates.15–17 We have previously investigated the relationship between ABCB1 haplotypes [consisting of the 61A > G (rs9282564); 1199G > A (rs2229109); 1236C > T (rs1128503); 2677G > T (rs2032582); 3435C > T (rs1045642) single nucleotide polymorphisms] and methadone maintenance therapy doses (15–110 mg/day) in a South Australian treatment center, demonstrating a link between variant haplotypes and significantly lower (approximately 50%) methadone doses.18 Specifically, subjects carrying the variant AGCTT (61A; 1199G; 1236C; 2677T; 3435T) haplotype (underlined nucleotides here and throughout remainder of paper are variant) required only 62% of the dose of other subjects, presumably due to decreased P-glycoprotein function at the blood–brain barrier and thus higher brain methadone concentrations (hence effect) relative to dose.18 However, a later study in Swiss subjects receiving a much larger range of methadone doses (3–430 mg/day) failed to replicate this association between ABCB1 haplotypes and dose.19 Alternatively, a study in Israeli subjects receiving 30–280 mg/day found that the frequency of homozygous 1236TT; 2677TT; 3435TT variant haplotypes was significantly higher in high-dose (>150 mg/day) methadone maintenance therapy subjects than those designated as low-dose (<150 mg/day),20 a finding seemingly contrary to the majority of ABCB1 pharmacogenetic studies in opioids.8
Therefore, there is conflicting evidence regarding the role of variant ABCB1 haplotypes in determining the methadone maintenance therapy dose, depending on the clinical context (eg, treatment population, clinical policy, philosophy, and/or dosing practice). One notable difference between studies has been the range of methadone doses that subjects received. Not only were wider ranges of doses encompassed by the Swiss and Israeli studies when compared with the Australian study (3–430 and 30–280 versus 15–110 mg/day, respectively), on average the doses in these patient populations were also higher (107 and 160 versus 59 mg/day, respectively). Therefore, it seems plausible that the dose range investigated may impact on whether ABCB1 genetic variability significantly influences the response to methadone maintenance therapy. Indeed, we have previously proposed that as doses of methadone increase, the potential impact of genetic variability of ABCB1 haplotypes on requirements may decrease, and that the impact of pharmacogenetics at other loci, eg, genes encoding for target receptors and signal transduction or reward pathways, will become more significant factors in determining methadone response.21 However, prior to this study, the impact of ABCB1 genetic variability in higher dose ranges (>150 mg/day) in Australian methadone maintenance therapy settings was unknown.
One such genetic locus potentially altering opioid pharmacodynamics is the 118A > G (Asn40 Asp, rs1799971 recently renamed c.335A > G, Asn102 Asp, http://www.ncbi.nlm.nih.gov/SNP/) polymorphism of the mu opioid receptor gene, OPRM1.8,22,23 Clinical studies have provided strong evidence that the G variant is associated with reduced analgesic response and side effects to opioids, and have been reviewed elsewhere.24 Regarding methadone, whilst the 118A > G variant has been associated with decreased miotic potency in healthy controls,25 it has not been shown to relate to methadone dose or response in methadone maintenance therapy22 when analyzed in isolation. However, there is evidence that variants of OPRM1 and ABCB1 interact to affect morphine pain relief,26 and it is possible that a similar gene-gene interaction may occur in methadone maintenance therapy.
Therefore, the aims of this study were to investigate the impact of ABCB1 and OPRM1 genetic variability on dose requirements and pharmacokinetics in an opioid-dependent population maintained on a wide range of methadone doses, and the potential gene-gene interaction between ABCB1 and OPRM1 in this context.
It was hypothesized that, in this cohort, methadone requirements would significantly differ between ABCB1 haplotypes and OPRM1 genotypes when taking into account confounding genetic variability (from OPRM1 and ABCB1, respectively), but not when these genetic factors were analyzed in isolation.
Materials and methods
Subjects
In total, 119 subjects were included in this retrospective study. All were Caucasian and not pregnant. They consisted of 38 methadone maintenance therapy patients from our previously published ABCB1 pharmacogenetic study18 (having excluded all pregnant and non-Caucasian subjects from that study) and a further 45 methadone maintenance therapy patients who had taken part in clinical studies conducted by the Discipline of Pharmacology at The University of Adelaide since 2006 (all approved by the Royal Adelaide Hospital research ethics committee). All of these 83 subjects were originally recruited from South Australian clinics and private prescribers operating under the guidelines of Drug and Alcohol Services South Australia.27 A further 36 methadone maintenance therapy subjects, originally part of a clinical study from the Byrne Surgery, a specialist drug and alcohol medical practice in Redfern, New South Wales (approved by the South East Sydney Area Health Service ethics committee), were also included. Treated under New South Wales Department of Health clinical guidelines that allow dosing up to and above (with state approval) 200 mg/day,28 these 36 subjects represent a cohort of subjects who require relatively high (>150 mg/day) doses to reduce withdrawal symptoms and heroin use. Written informed consent for genotyping analysis was obtained for all subjects.
Genotyping
Genomic DNA was isolated from whole blood samples using a QIAamp® DNA mini kit according to the manufacturer’s instructions (Qiagen Pty Ltd, Doncaster, Australia).
ABCB1
61A > G, 1199G > A, 1236C > T and 3435C > T genotyping was performed using polymerase chain reaction (PCR) followed by restriction fragment length polymorphism (RFLP) as described previously.18 2677G > T genotyping differed from previously because it was also detected by PCR-RFLP based on the method of Cascorbi et al.29 Briefly, PCR primers were: forward – 5′-TGCAGGCTATAGGTTCCAGG-3′; reverse – 5′-TTTAGTTTGACTCACCTTCCC-3′. PCR reactions were performed in 30 μL total volume containing 100 ng DNA, 50 μM dNTPs, 0.1 μM each primer, 2.5 U Taq DNA polymerase and 1 × reaction buffer (New England Biolabs Inc, distributed by Genesearch Pty Ltd, Arundel, Australia). PCR cycling conditions were: 5 minutes at 94°C; 45 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 1.5 minutes at 72°C; and 5 minutes at 72°C. Twenty microliters of PCR product was digested with 4 U of BanI (New England Biolabs Inc) for 16 hours at 37°C, then 20 minutes of heat inactivation at 65°C. Digested PCR products were visualized by agarose gel electrophoresis as described previously.18 Restriction fragments were: wild-type (digested), 198 and 26 base pairs (bp); variant (undigested), 224 bp. An example of this 2677G > T PCR-RFLP assay is provided in Supplementary Material A.
OPRM1
118A > G genotyping was performed using allele-specific PCR as previously described.30
For all genotyping assays, genotypes of random samples were confirmed via sequencing using BigDye Terminator v3.0 chemistry and analysis on an ABI Prism 3700 DNA Sequencer (Applied Biosystems, Australia). All assays contained both negative (no DNA template) and positive (DNA of known genotype confirmed by sequencing) controls to ensure accuracy.
Demographic, dose, and pharmacokinetic data
Demographic, dose, and pharmacokinetic data were obtained from original study case notes and are summarized in Table 1. Because dose as a measure of methadone requirements can be confounded by significant variability in absorption, distribution, metabolism, and elimination unrelated to P-glycoprotein, trough plasma (R)-methadone concentrations (Ctrough, ng/mL), were also investigated. Used here as an indicator of plasma concentrations required to suppress withdrawal, Ctrough should not be confused with dose-adjusted Ctrough (Ctrough/dose), which is used here as an indicator of the potential influence of P-glycoprotein on the intestinal absorption and elimination of methadone. Eighty-four subjects had Ctrough data available and were included in pharmacokinetic analyses (the demographic, dose, and pharmacokinetic data for this subset of 84 subjects are also summarized in Table 1).
Table 1.
Subject demographic data
| All subjects (n = 119) | Subjects with PK data (n = 84) | |
|---|---|---|
| Male:female | 100:19 | 69:15 |
| Age, median ± SD (range) | 35 ± 8 (20–54) | 36 ± 8 (23–54) |
| Body weight (kg), median ± SD (range) | 75 ± 14 (48–127) | 76 ± 15 (48–127) |
| Time in treatment (months), median ± SD (range) | 12 ± 58 (1–276) | 13 ± 63 (1–276) |
| Methadone dose (mg/day), median ± SD (range) | 80 ± 72 (15–300) | 90 ± 80 (15–300) |
| Ctrough (ng/mL), median ± SD (range) | – | 153 ± 139 (18–669) |
| Ctrough/dose ((ng/mL)/mg), median ± SD (range) | – | 1.7 ± 0.7 (0.5–3.5) |
Abbreviations: PK, pharmacokinetic; Ctrough, trough plasma (R)-methadone concentration; Ctrough/dose, dose-adjusted trough plasma (R)-methadone concentration; SD, standard deviation.
Data analysis
Chi-square tests were used to test for genotype deviations from Hardy-Weinberg equilibrium. ABCB1 haplotypes were inferred from genotype data using PHASE (version 2.1) software.31,32 Subjects whose haplotypes were inferred with low confidence (PHASE call confidence probability less than 0.75) were excluded from haplotype analyses (n = 5). Dose, Ctrough and Ctrough/dose data are presented as the median ± standard deviation. For differences between ABCB1 haplotypes, four haplotype combination groups were compared based on observed frequency and our previous significant findings,18 ie, homozygous wild-type (“AGCGC/AGCGC”); combination of wild-type with variant at 1236C > T, 2677G > T and 3435C > T only (“AGCGC/AGTTT”); homozygous variant at 1236C > T, 2677G > T, and 3435C > T only (“AGTTT/AGTTT”); and combination of wild-type with variant at 2677G > T and 3435C > T only, or homozygous variant at 2677G > T and 3435C > T only (“AGCTT”). These groups were compared simultaneously using Kruskal-Wallis tests (with Dunn’s post hoc comparing variant groups with AGCGC/AGCGC). This was first done for all subjects, regardless of OPRM1 genotype, and then only within subjects who were OPRM1 118A > G wild-type (A/A) genotype (for dose and Ctrough only).
Differences in daily dose and Ctrough between OPRM1 genotypes were first investigated in all subjects, regardless of ABCB1 haplotype, using Kruskal-Wallis tests (with Dunn’s multiple comparisons post hoc). Then, within subjects with the most common ABCB1 haplotype combination (AGCGC/AGTTT), OPRM1 118A > G A/G and G/G (n = 1 and 0 for dose and Ctrough, respectively) genotype subjects were combined and compared with the A/A genotype using the Mann-Whitney U test.
The influence of demographic covariates on the relationship between genetic variability and methadone dose, Ctrough and Ctrough/dose was also examined by multiple linear regression analysis (see Supplementary Material B). P < 0.05 was considered statistically significant for all analyses.
Results
Genetic variability
OPRM1 (see Supplementary Material C, Table SC1) and ABCB1 genotype frequencies were in Hardy-Weinberg equilibrium (P ≥ 0.40). ABCB1 haplotypes could be confidently determined for 114 subjects, with 12 different haplotypes observed (see Supplementary Material C, Table SC1).
Methadone requirements and pharmacokinetics
Not taking into account OPRM1 genetic variability, there were no significant differences between ABCB1 haplotype groups for either dose (P = 0.51), Ctrough (P = 0.97), or Ctrough/dose (P = 0.60, Table 2). Not taking into account ABCB1 genetic variability, there were no significant differences between OPRM1 genotypes for either dose (P = 0.40) or Ctrough (P = 0.064, Table 2).
Table 2.
Comparison of methadone dose, (R)-methadone Ctrough and dose-adjusted (R)-methadone Ctrough between common ABCB1 haplotype combinations and OPRM1 genotypes of methadone maintenance therapy patients
|
Median ± SD (n)
|
||||||
|---|---|---|---|---|---|---|
| Dose (mg/day) | Ctrough (ng/mL) | Ctrough/dose ((ng/mL)/mg) | ||||
| ABCB1 haplotype group | ||||||
| AGCGC/AGCGC | 180 ± 60 (9) | 181 ± 113 (6) | 1.5 ± 0.5 (6) | |||
| AGCGC/AGTTT | 78 ± 80 (24) | 201 ± 144 (13) | 1.6 ± 0.7 (13) | |||
| AGTTT/AGTTT | 105 ± 67 (9) | 173 ± 143 (7) | 1.8 ± 0.8 (7) | |||
| AGCTT | 40 ± 99 (7) | 220 ± 193 (6) | 1.9 ± 0.3 (6) | |||
| P valuea | 0.51 | 0.97 | 0.60 | |||
| OPRM1 118A > G genotype | ||||||
| A/A | 80 ± 63 (77) | 124 ± 115 (52) | ||||
| A/G | 73 ± 82 (34) | 214 ± 135 (26) | ||||
| G/G | 93 ± 101 (8) | 262 ± 259 (6) | ||||
| P valuea | 0.40 | 0.064 | ||||
Notes:
P value of Kruskal-Wallis test for differences between ABCB1 haplotype groups or OPRM1 118A > G genotypes.
AGCGC/AGCGC: Subjects homozygous wild-type for ABCB1 single nucleotide polymorphisms (61A > G; 1199G > A; 1236C > T; 2677G > T; and 3435C > T. AGCGC/AGTTT: Subjects with one wild-type haplotype and one 61A:1199G:1236T:2677T:3435T haplotype. AGTTT/AGTTT: Subjects homozygous for 61A:1199G:1236T:2677T:3435T haplotype. AGCTT: Subjects with one wild-type haplotype and one 61A:1199G:1236C:2677T:3435T haplotype, or homozygous for 61A:1199G:1236C:2677T:3435T haplotype.
Abbreviations: Ctrough, Trough plasma (R)-methadone concentration; Ctrough/dose, Dose adjusted trough plasma (R)-methadone concentrations.
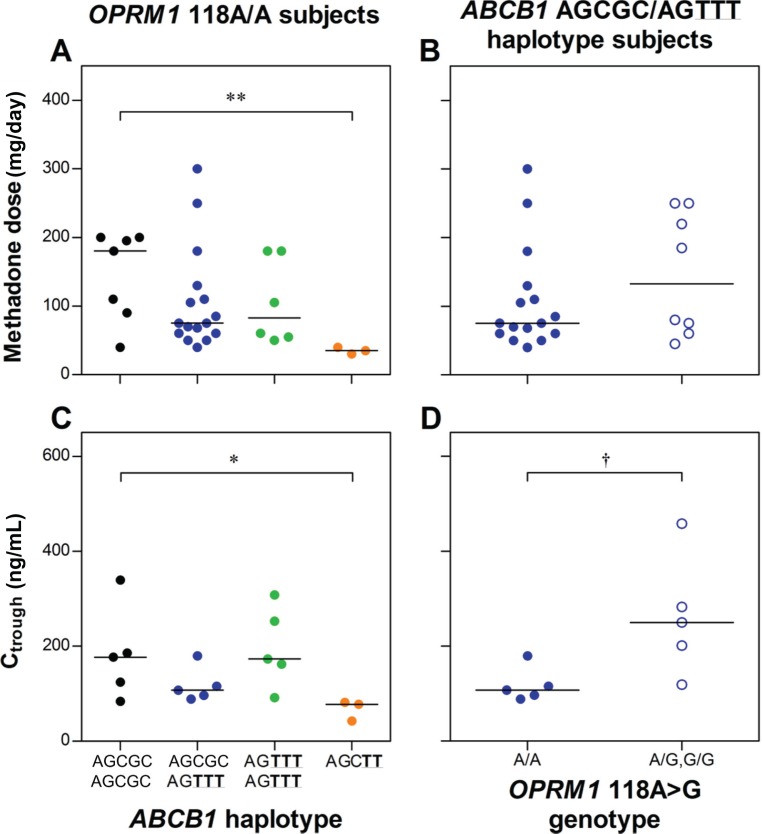
Among OPRM1 118A/A subjects, there was a significant difference in dose and Ctrough between ABCB1 haplotype groups, with the AGCTT variant haplotype group associated with a significantly lower dose (median ± standard deviation 35 ± 5 versus 180 ± 65 mg/day, P < 0.01) and Ctrough (78 ± 22 versus 177 ± 97 ng/mL, P < 0.05) than the wild-type ABCB1 subjects (Figure 1). Among subjects with the ABCB1 AGCGC/AGTTT haplotype combination, the OPRM1 118A/G genotype was associated with significantly higher Ctrough than the 118A/A genotype (250 ± 126 versus 108 ± 36 ng/mL, respectively, P = 0.016), but with no significant difference for dose (133 ± 89 versus 75 ± 75 mg/day, respectively, P = 0.37). Results of multiple linear regression analyses are provided in Supplementary Material B.
Figure 1.
Influence of ABCB1 haplotype group on (A) methadone dose and (C) trough plasma (R)-methadone concentrations (Ctrough) among OPRM1 wild-type (118A/A) subjects; and OPRM1 118A > G genotype on (B) methadone dose and (D) Ctrough among ABCB1 AGCGC/AGTTT haplotype combination subjects.
Notes: Lines are medians. *P < 0.05, **P < 0.01 versus ABCB1 AGCGC/AGCGC haplotype group, †P < 0.05 OPRM1118A/A versus A/G,G/G genotype in ABCB1 AGCGC/AGTTT haplotype group.
Discussion
Conflicting findings of previous pharmacogenetic studies of methadone maintenance therapy suggest the relative impact of ABCB1 genetic variability on methadone dose may be dependent on the clinical context.18,19 More specifically, we have previously published an association between the ABCB1 variant AGCTT (61A; 1199G; 1236C; 2677T; 3435T) haplotype and significantly lower methadone doses in a methadone maintenance therapy population receiving doses up to approximately 150 mg/day.18 However, a subsequent larger study by Crettol et al19 in patients receiving a much higher range of doses (up to 430 mg/day) failed to replicate these findings.
To investigate further the basis of these discordant findings, this study first set out to refine (by excluding all pregnant and non-Caucasian subjects) and expand upon (adding a further 81 subjects) our existing methadone maintenance therapy study population, to re-examine ABCB1 pharmacogenetics over a dose range more akin to other studies in methadone maintenance therapy.19,20
In agreement with the findings of Crettol et al,19 no significant association between ABCB1 haplotypes and methadone dose was observed when ABCB1 genetic variability was investigated in isolation. Similarly, and in line with a separate study by Crettol et al,22 the OPRM1 118A > G polymorphism had no significant influence on methadone dose, despite strong clinical evidence that it can influence the pharmacodynamics of other opioids.24
Given the complex nature of opioid dependence and the heterogeneity within the methadone maintenance therapy population, particularly over such wide ranges of doses, it is not surprising that single genetic factors cannot individually explain variability in dose requirements. However, we have been able to show that by taking into account at least part of the confounding genetic variability, the potential influence of ABCB1 and OPRM1 on methadone response can be revealed.
For ABCB1, the AGCTT haplotype group was associated with a significantly lower median dose (<20% of wild-type) when controlling for OPRM1. Alternatively, when controlling for ABCB1 variability, OPRM1 118G variant allele carriers had a nearly 1.8-fold higher median dose than 118A/A genotype subjects, although this difference was not significant, with doses within each genotype group still varying greatly (5–8-fold).
The degree of variability in dose within the OPRM1 genotype groups is not entirely surprising, given that methadone dose requirements can also be significantly influenced by additional variability in intestinal absorption, peripheral distribution, metabolism, and elimination (unrelated to P-glycoprotein activity).
Downstream of variability in pharmacokinetics affecting the dose-plasma concentration relationship, Ctrough was examined as a potentially more direct indicator of variability in pharmacodynamics due to the OPRM1 118A > G polymorphism, as well as the impact of ABCB1 haplotype on the distribution of methadone in the central nervous system. For ABCB1, as with dose, the AGCTT haplotype group was associated with a significantly lower median Ctrough (44% of wild-type) when controlling for OPRM1. In addition, variant OPRM1 118G allele carriers had significantly higher (2.3-fold) median Ctrough than 118A/A subjects, when controlling for ABCB1.
These findings suggest that when methadone potency at the mu opioid receptor is not compromised, ABCB1 haplotype (influencing exposure to methadone in the central nervous system) can have a significant effect on methadone requirements, but has less impact when higher doses/concentrations are required to overcome deficiencies in mu opioid receptor activation. Conversely, when variability in peripheral pharmacokinetics and ABCB1 genetics are controlled for, an association between the OPRM1 118G variant, and higher than normal plasma methadone concentrations to suppress withdrawal, is revealed. This effect is most likely as a result of decreased methadone potency at the mu opioid receptor, a hypothesis supported by previous studies in healthy subjects.25 Such an interaction between ABCB1 and OPRM1 genetic variability is mechanistically expected, and corresponds with that reported for morphine analgesia,26 but requires confirmation within a larger cohort of subjects.
It is interesting that, as in our previous study,18 the AGCTT, but not the AGTTT, haplotype was associated with a lower methadone dose. Certainly, in vitro evidence would suggest that these variant haplotypes should have similar phenotypes (either decreased33,34 or no effect35–37 on P-glycoprotein expression and/or function). However, findings from Levran et al20 in methadone maintenance therapy, linking the homozygous 1236TT; 2677TT; 3435TT variant haplotype to high-dose requirements (>150 mg/day), indicate that the situation in vivo may be quite different. These studies highlight the importance of examining ABCB1 haplotypes, rather than individual single nucleotide polymorphisms, at least for methadone maintenance therapy. This, along with the confounding effect of OPRM1 genetics, may explain why a significant association between individual ABCB1 single nucleotide polymorphisms and methadone requirements has not been observed in previous studies.2,11,18
Regarding the potential influence of ABCB1 genetic variability on methadone pharmacokinetics altering the dose-plasma concentration relationship, Crettol et al2 have previously reported significant differences in dose-adjusted trough (R)-, (S)- and (R,S)-methadone concentrations between ABCB1 61A > G and 3435C > T genotypes. However, in this study, ABCB1 haplotypes had no significant influence on Ctrough/dose. This is not surprising, given that for drugs such as methadone, P-glycoprotein is expected to have a relatively minor impact on intestinal absorption and elimination when compared with its influence on central nervous system distribution, especially at high doses.15,16
Multiple linear regression analyses incorporating demographic covariates did not add significantly to the interpretation of the influence of ABCB1 and OPRM1 genetic factors on methadone dose, Ctrough, or Ctrough/dose (see Supplementary Material B).
Frequencies of the common ABCB1 haplotypes and the OPRM1 118G allele in subjects on methadone maintenance therapy were similar to those reported in non-opioid-dependent Caucasians (AGCGC: 30% versus 28%; AGCGT: 10% versus 8%; AGCTT: 6% versus 3%; AGTTT: 32% versus 35%; GGTTT: 8% versus 6%; OPRM1 118G: 21% versus 20%), in line with previous findings that these genes are not associated with risk of opioid dependence.18,30
Some limitations to this study, largely brought about by the number of subjects, need to be discussed. Firstly, only a small subset of ABCB1 haplotypes were able to be analyzed, therefore it is possible that further, less common, haplotype combinations may also influence methadone distribution, and could not be analyzed here. Secondly, only the most basic of comparisons were able to be performed regarding gene-gene interactions, which encompassed only a third of the study population. Given sufficient numbers, a two-way analysis of variance or regression analysis, with inclusion of other clinical covariates, would be ideal. Thirdly, this study was limited to examination of only two genes in what is becoming increasingly recognized as a polygenetic trait. In addition to ABCB1 and OPRM1, there is evidence that genetic variants in the dopamine D2 receptor/ankyrin repeat and kinase domain containing 1 (DRD2/ANKK1),22,38 nerve growth factor β polypeptide (NGFB),39 myocardin (MYOCD),40 metabotropic glutamate receptor (GRM6),40 beta-Arrestin2 (ARRB2),41 and potassium inwardly rectifying channel (KCNJ6),42 genes may also contribute to interindividual variability in methadone maintenance therapy response in Caucasians. A recent study in Han Chinese subjects on methadone maintenance therapy has also demonstrated that ABCB1, CYP2B6, OPRM1, and ANKK1-DRD2 genetic polymorphisms jointly correlate with methadone dose.43 Therefore, consideration of multiple genetic and environmental factors will be central to the enhanced individualization, and ultimately improvement, of the treatment of opioid dependence.
Finally, no adjustments were made for multiple testing. Therefore, whilst an attempt was made to keep the number of cross-analyses to a minimum, there is a risk of type 1 error in these findings, and they require replication. A prospective, large-scale, multicenter study is currently being conducted that will be able to address the issues discussed above.
In conclusion, neither ABCB1 nor OPRM1 genetic variants independently predicted methadone requirements in methadone maintenance therapy. However, a significant association between an ABCB1 variant and lower methadone requirements was revealed when controlling for OPRM1 genetic variability. Likewise, a significant association between an OPRM1 variant and higher methadone requirements was revealed when controlling for ABCB1 genetic variability. Therefore, these two opposing pharmacogenetic effects need to be considered in combination when assessing their impact on methadone maintenance therapy.
Acknowledgments
DTB was supported by a National Health and Medical Research Council of Australia Post-graduate scholarship; JKC is a FTT Fricker Research Fellow at the University of Adelaide (Medical Endowment Funds). This study was supported by the Faculty of Health Sciences, University of Adelaide, and National Health and Medical Research Council of Australia project grants (229050, 565387).
Footnotes
Disclosure
The authors have no conflicts of interest to report in this work.
Supplementary material A
ABCB1 2677G > T genotype determination by restriction fragment length (Figure SA1).
BanI restriction fragment patterns for 2677G > T genotyping.
Notes: bp: size of marker bands in base pairs; M: pUC19/HpaII DNA molecular weight marker; 1, negative control; 2, homozygous variant genotype (T/T) with 224 bp undigested polymerase chain reaction product; 3, heterozygous genotype (G/T) with 224 bp undigested + 198 bp digested fragment; and 4, homozygous wild-type genotype (G/G) with 198 bp digested fragment. 26 bp digested fragments not visible on gel.
Supplementary material B
Regression analysis
Methods
Multiple linear regression analysis (IBM SPSS Statistics 19) was used to investigate the effects of demographic covariates on methadone dose, Ctrough, and Ctrough/dose. For these analyses, Ctrough and Ctrough/dose data were normalized by log-transformation (dose data could not be normalized by any standard transformation). Because the contributions of ABCB1 haplotype groups AGCGC/AGTTT and AGTTT/AGTTT were unclear from this study, only the AGCGC/AGCGC and AGCTT groups were included in regression analysis, and coded 0 and 1, respectively. OPRM1 118A/A, 118A/G, and 118G/G genotypes were coded 0, 1, and 2, respectively.
Firstly, gender, age, and weight (log-transformed) were submitted to stepwise inclusion (F probability < 0.05) regression for each dependent variable. Based on these results, any significant covariate was then automatically entered for all regression analyses of relevant dependent variable (dose, Ctrough, or Ctrough/dose) prior to stepwise inclusion (F probability < 0.05) of: OPRM1 genotype then ABCB1 haplotype; ABCB1 haplotype in OPRM1 118 A/A subjects; or OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects. A subsequent conditional removal (F probability > 0.1) step for weight was also included for comparison.
Results
Of the demographic variables examined, weight was the only significant predictor of methadone dose by regression analysis (positive correlation, model P = 0.013). Subsequent regression analyses identified ABCB1 haplotype in OPRM1 118A/A subjects (negatively correlated), and OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects (positively correlated), but not ABCB1 haplotype group or OPRM1 118A > G genotype, as significant predictors of dose (Table SB1).
Table SB1.
Multiple linear regression models for estimation of methadone dose based on weight and ABCB1 and OPRM1 genetic variability
| Model variables | Model adjusted R2 | P value |
|---|---|---|
| Weight | 0.054 | 0.013 |
| Weight, ABCB1 haplotype in OPRM1 118A1A subjects | 0.478 | 0.022 |
| ABCB1 haplotype in OPRM1 118A1A subjects | 0.482 | 0.007 |
| Weight, OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects | 0.201 | 0.082 |
| OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects | 0.229 | 0.03 |
No demographic variable was a significant predictor of Ctrough or Ctrough/dose. ABCB1 haplotype in OPRM1 118A/A subjects (model adjusted R2 = 0.481, P = 0.023), and OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects (model adjusted R2 = 0.458, P = 0.019) were the only significant predictors of Ctrough. Neither ABCB1 haplotype group, nor ABCB1 haplotype group in OPRM1 118A/A subjects, were significant predictors of Ctrough/dose.
Supplementary material C
Supplementary tables
Table SC1.
Frequencies of ABCB1 haplotypesa and OPRM1 118A > G minor allele and genotypes in opioid-dependent methadone maintenance treatment patients
| ABCB1 haplotypeb | % (n) | OPRM1 c.118 A > G | % (n) |
|---|---|---|---|
| AGCGC | 29.8 (68) | MAF | 21.0 (50) |
| AGCGT | 9.6 (22) | A/A | 64.7 (77) |
| AGCTC | 3.1 (7) | A/G | 28.6 (34) |
| AGTGC | 4.8 (11) | G/G | 6.7 (8) |
| AACGC | 2.2 (5) | ||
| AGCTT | 5.7 (13) | ||
| AGTGT | 1.8 (4) | ||
| AGTTC | 2.6 (6) | ||
| AACTC | 0.4 (1) | ||
| GGCGT | 0.4 (1) | ||
| AGTTT | 32.0 (73) | ||
| GGTTT | 7.5 (17) |
Notes: Variant nucleotides in ABCB1 haplotypes are underlined.
Excluding subjects with PHASE call confidence probabilities less than 0.75.
Haplotypes consisting of 61A > G, 1199G > A, 1236C > T, 2677G > T, and 3435C > T single nucleotide polymorphisms of ABCB1.
Abbreviation: MAF, minor allele frequency.
References
- 1.Mattick R, Digiusto E, Doran C, et al. National Evaluation of Pharmacotherapies for Opioid Dependence: Report of Results and Recommendations. Sydney, Australia: National Drug and Alcohol Research Centre; 2001. [Google Scholar]
- 2.Crettol S, Deglon JJ, Besson J, et al. ABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatment. Clin Pharmacol Ther. 2006;80(6):668–681. doi: 10.1016/j.clpt.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Dyer KR, Foster DJ, White JM, Somogyi AA, Menelaou A, Bochner F. Steady-state pharmacokinetics and pharmacodynamics in methadone maintenance patients: comparison of those who do and do not experience withdrawal and concentration-effect relationships. Clin Pharmacol Ther. 1999;65(6):685–694. doi: 10.1016/S0009-9236(99)90090-5. [DOI] [PubMed] [Google Scholar]
- 4.Dyer KR, White JM. Patterns of symptom complaints in methadone maintenance patients. Addiction. 1997;92(11):1445–1455. [PubMed] [Google Scholar]
- 5.Foster DJR, Somogyi AA, White JM, Bochner F. Population pharmacokinetics of (R)-, (S)- and rac-methadone in methadone maintenance patients. Br J Clin Pharmacol. 2004;57(6):742–755. doi: 10.1111/j.1365-2125.2004.02079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell J, Burrell T, Indig D, Gilmour S. Cycling in and out of treatment; participation in methadone treatment in NSW, 1990–2002. Drug Alcohol Depend. 2006;81(1):55–61. doi: 10.1016/j.drugalcdep.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Australian Institute of Health and Welfare . Statistics on Drug Use in Australia 2006. Canberra, Australia: Australian Institute of Health and Welfare; 2007. [Google Scholar]
- 8.Somogyi AA, Barratt DT, Coller JK. Pharmacogenetics of opioids. Clin Pharmacol Ther. 2007;81(3):429–444. doi: 10.1038/sj.clpt.6100095. [DOI] [PubMed] [Google Scholar]
- 9.Eap CB, Crettol S, Rougier JS, et al. Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin Pharmacol Ther. 2007;81(5):719–728. doi: 10.1038/sj.clpt.6100120. [DOI] [PubMed] [Google Scholar]
- 10.Uehlinger C, Crettol S, Chassot P, et al. Increased (R)-methadone plasma concentrations by quetiapine in cytochrome P450s and ABCB1 genotyped patients. J Clin Psychopharmacol. 2007;27(3):273–278. doi: 10.1097/JCP.0b013e3180592ad2. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca F, de la Torre R, Diaz L, et al. Contribution of cytochrome P450 and ABCB1 genetic variability on methadone pharmacokinetics, dose requirements, and response. PLoS One. 2011;6(5):e19527. doi: 10.1371/journal.pone.0019527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coller JK, Joergensen C, Foster DJ, et al. Lack of influence of CYP2D6 genotype on the clearance of (R)-, (S)- and racemic-methadone. Int J Clin Pharmacol Ther. 2007;45(7):410–417. doi: 10.5414/cpp45410. [DOI] [PubMed] [Google Scholar]
- 13.Hassan HE, Myers AL, Coop A, Eddington ND. Differential involvement of P-glycoprotein (ABCB1) in permeability, tissue distribution, and antinociceptive activity of methadone, buprenorphine, and diprenorphine: in vitro and in vivo evaluation. J Pharm Sci. 2009;98(12):4928–4940. doi: 10.1002/jps.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tournier N, Chevillard L, Megarbane B, Pirnay S, Scherrmann JM, Decleves X. Interaction of drugs of abuse and maintenance treatments with human P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) Int J Neuropsychopharmacol. 2009:1–11. doi: 10.1017/S1461145709990848. [DOI] [PubMed] [Google Scholar]
- 15.Lin JH, Yamazaki M. Clinical relevance of P-glycoprotein in drug therapy. Drug Metab Rev. 2003;35(4):417–454. doi: 10.1081/dmr-120026871. [DOI] [PubMed] [Google Scholar]
- 16.Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin Pharmacokinet. 2003;42(1):59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- 17.Tournier N, Decleves X, Saubamea B, Scherrmann JM, Cisternino S. Opioid transport by ATP-binding cassette (ABC) transporters at the blood-brain barrier: implications for neuropsychopharmacology. Curr Pharm Des. 2011;17(26):2829–2842. doi: 10.2174/138161211797440203. [DOI] [PubMed] [Google Scholar]
- 18.Coller JK, Barratt DT, Dahlen K, Loennechen MH, Somogyi AA. ABCB1 genetic variability and methadone dosage requirements in opioid-dependent individuals. Clin Pharmacol Ther. 2006;80(6):682–690. doi: 10.1016/j.clpt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Crettol S, Deglon JJ, Besson J, et al. No influence of ABCB1 haplotypes on methadone dosage requirement. Clin Pharmacol Ther. 2008;83(5):668–669. doi: 10.1038/sj.clpt.6100305. author reply 669–670. [DOI] [PubMed] [Google Scholar]
- 20.Levran O, O'Hara K, Peles E, et al. ABCB1 (MDR1) genetic variants are associated with methadone doses required for effective treatment of heroin dependence. Hum Mol Genet. 2008;17(14):2219–2227. doi: 10.1093/hmg/ddn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coller JK, Barratt DT, Somogyi AA. Response to “No influence of ABCB1 haplotypes on methadone dosage requirement”. Clin Pharmacol Ther. 2008;83(5):669–670. doi: 10.1038/sj.clpt.6100305. [DOI] [PubMed] [Google Scholar]
- 22.Crettol S, Besson J, Croquette-Krokar M, et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(7):1722–1727. doi: 10.1016/j.pnpbp.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Oertel BG, Kettner M, Scholich K, et al. A common human micro-opioid receptor genetic variant diminishes the receptor signaling efficacy in brain regions processing the sensory information of pain. J Biol Chem. 2009;284(10):6530–6535. doi: 10.1074/jbc.M807030200. [DOI] [PubMed] [Google Scholar]
- 24.Kasai S, Ikeda K. Pharmacogenomics of the human micro-opioid receptor. Pharmacogenomics. 2011;12(9):1305–1320. doi: 10.2217/pgs.11.68. [DOI] [PubMed] [Google Scholar]
- 25.Lötsch J, Skarke C, Wieting J, et al. Modulation of the central nervous effects of levomethadone by genetic polymorphisms potentially affecting its metabolism, distribution, and drug action. Clin Pharmacol Ther. 2006;79(1):72–89. doi: 10.1016/j.clpt.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Campa D, Gioia A, Tomei A, Poli P, Barale R. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clin Pharmacol Ther. 2008;83(4):559–566. doi: 10.1038/sj.clpt.6100385. [DOI] [PubMed] [Google Scholar]
- 27.Hurley M, White J, Chadderton T, et al. Guide for Pharmacists: Addiction Treatment and Maintenance Pharmacotherapy (Methadone and Buprenorphine) Programs in South Australia. Adelaide, Australia: Drug and Alcohol Services South Australia; 2010. [Google Scholar]
- 28.NSW Department of Health . New South Wales Opioid Treatment Program: Clinical Guidelines for Methadone and Buprenorphine Treatment of Opioid Dependence. North Sydney, Australia: Mental Health and Drug and Alcohol Office, NSW Department of Health; 2006. [Google Scholar]
- 29.Cascorbi I, Gerloff T, Johne A, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69(3):169–174. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 30.Coller JK, Beardsley J, Bignold J, et al. Lack of association between the A118G polymorphism of the mu opioid receptor gene (OPRM1) and opioid dependence: A meta-analysis. Pharmacogenomics Pers Med. 2009;2(1):9–19. [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens M, Donnely P. A comparison of Bayesian methods for haplotype reconstruction. Am J Hum Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens M, Smith N, Donnely P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aird RE, Thomson M, Macpherson JS, Thurston DE, Jodrell DI, Guichard SM. ABCB1 genetic polymorphism influences the pharmacology of the new pyrrolobenzodiazepine derivative SJG-136. Pharmacogenomics J. 2008;8(4):289–296. doi: 10.1038/sj.tpj.6500465. [DOI] [PubMed] [Google Scholar]
- 34.Salama NN, Yang Z, Bui T, Ho RJ. MDR1 haplotypes significantly minimize intracellular uptake and transcellular P-gp substrate transport in recombinant LLC-PK1 cells. J Pharm Sci. 2006;95(10):2293–2308. doi: 10.1002/jps.20717. [DOI] [PubMed] [Google Scholar]
- 35.Gow JM, Hodges LM, Chinn LW, Kroetz DL. Substrate-dependent effects of human ABCB1 coding polymorphisms. J Pharmacol Exp Ther. 2008;325(2):435–442. doi: 10.1124/jpet.107.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 37.Morita N, Yasumori T, Nakayama K. Human MDR1 polymorphism: G2677T/A and C3435T have no effect on MDR1 transport activities. Biochem Pharmacol. 2003;65(11):1843–1852. doi: 10.1016/s0006-2952(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 38.Doehring A, Hentig N, Graff J, et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenet Genomics. 2009;19(6):407–414. doi: 10.1097/FPC.0b013e328320a3fd. [DOI] [PubMed] [Google Scholar]
- 39.Levran O, Peles E, Hamon S, et al. Nerve growth factor beta polypeptide (NGFB) genetic variability: association with the methadone dose required for effective maintenance treatment. Pharmacogenomics J. 2011 Mar 1; doi: 10.1038/tpj.2011.6. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonseca F, Gratacos M, Escaramis G, et al. Response to methadone maintenance treatment is associated with the MYOCD and GRM6 genes. Mol Diagn Ther. 2010;14(3):171–178. doi: 10.1007/BF03256370. [DOI] [PubMed] [Google Scholar]
- 41.Oneda B, Crettol S, Bochud M, et al. beta-Arrestin2 influences the response to methadone in opioid-dependent patients. Pharmacogenomics J. 2011;11(4):258–266. doi: 10.1038/tpj.2010.37. [DOI] [PubMed] [Google Scholar]
- 42.Lotsch J, Pruss H, Veh RW, Doehring A. A KCNJ6 (Kir3.2, GIRK2) gene polymorphism modulates opioid effects on analgesia and addiction but not on pupil size. Pharmacogenet Genomics. 2010;20(5):291–297. doi: 10.1097/FPC.0b013e3283386bda. [DOI] [PubMed] [Google Scholar]
- 43.Hung CC, Chiou MH, Huang BH, et al. Impact of genetic polymorphisms in ABCB1, CYP2B6, OPRM1, ANKK1 and DRD2 genes on methadone therapy in Han Chinese patients. Pharmacogenomics. 2011;12(11):1525–1533. doi: 10.2217/pgs.11.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BanI restriction fragment patterns for 2677G > T genotyping.
Notes: bp: size of marker bands in base pairs; M: pUC19/HpaII DNA molecular weight marker; 1, negative control; 2, homozygous variant genotype (T/T) with 224 bp undigested polymerase chain reaction product; 3, heterozygous genotype (G/T) with 224 bp undigested + 198 bp digested fragment; and 4, homozygous wild-type genotype (G/G) with 198 bp digested fragment. 26 bp digested fragments not visible on gel.
Table SB1.
Multiple linear regression models for estimation of methadone dose based on weight and ABCB1 and OPRM1 genetic variability
| Model variables | Model adjusted R2 | P value |
|---|---|---|
| Weight | 0.054 | 0.013 |
| Weight, ABCB1 haplotype in OPRM1 118A1A subjects | 0.478 | 0.022 |
| ABCB1 haplotype in OPRM1 118A1A subjects | 0.482 | 0.007 |
| Weight, OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects | 0.201 | 0.082 |
| OPRM1 118A > G genotype in ABCB1 AGCGC/AGTTT subjects | 0.229 | 0.03 |
Table SC1.
Frequencies of ABCB1 haplotypesa and OPRM1 118A > G minor allele and genotypes in opioid-dependent methadone maintenance treatment patients
| ABCB1 haplotypeb | % (n) | OPRM1 c.118 A > G | % (n) |
|---|---|---|---|
| AGCGC | 29.8 (68) | MAF | 21.0 (50) |
| AGCGT | 9.6 (22) | A/A | 64.7 (77) |
| AGCTC | 3.1 (7) | A/G | 28.6 (34) |
| AGTGC | 4.8 (11) | G/G | 6.7 (8) |
| AACGC | 2.2 (5) | ||
| AGCTT | 5.7 (13) | ||
| AGTGT | 1.8 (4) | ||
| AGTTC | 2.6 (6) | ||
| AACTC | 0.4 (1) | ||
| GGCGT | 0.4 (1) | ||
| AGTTT | 32.0 (73) | ||
| GGTTT | 7.5 (17) |
Notes: Variant nucleotides in ABCB1 haplotypes are underlined.
Excluding subjects with PHASE call confidence probabilities less than 0.75.
Haplotypes consisting of 61A > G, 1199G > A, 1236C > T, 2677G > T, and 3435C > T single nucleotide polymorphisms of ABCB1.
Abbreviation: MAF, minor allele frequency.