Abstract
Higher plants express several isoforms of vacuolar and cell wall invertases (CWI), some of which are inactivated by inhibitory proteins at certain stages of plant development. We have purified an apoplasmic inhibitor (INH) of tobacco (Nicotiana tabacum) CWI to homogeneity. Based on sequences from tryptic fragments, we have isolated a full-length INH-encoding cDNA clone (Nt-inh1) via a reverse transcriptase-polymerase chain reaction. Southern-blot analysis revealed that INH is encoded by a single- or low-copy gene. Comparison with expressed sequence tag clones from Arabidopsis thaliana and Citrus unshiu indicated the presence of Nt-inh1-related proteins in other plants. The recombinant Nt-inh1-encoded protein inhibits CWI from tobacco and Chenopodium rubrum suspension-cultured cells and vacuolar invertase from tomato (Lycopersicon esculentum) fruit, whereas yeast invertase is not affected. However, only in the homologous system is the inhibition modulated by the concentration of Suc as previously shown for INH isolated from tobacco cells. Highly specific binding of INH to CWI could be shown by affinity chromatography of a total cell wall protein fraction on immobilized recombinant Nt-inh1 protein. RNA-blot analysis of relative transcript ratios for Nt-inh1 and CWI in different parts of adult tobacco plants revealed that the expression of both proteins is not always coordinate.
Suc is the predominant sugar in higher plants. It serves several important functions, including acting as the major carbohydrate transport form, as a storage compound, and as an osmoprotectant (Eschrich, 1989; Frommer and Sonnewald, 1995; Ruan and Patrick, 1995; Stitt and Sonnewald, 1995). Higher plants metabolize Suc either by Suc synthase or via invertases (Stitt and Sonnewald, 1995; Sturm et al., 1995; Zrenner et al., 1995). Suc synthase is localized in the cytosol or bound to the inner side of the plasma membrane (Delmer and Amor, 1995) and catalyzes an equilibrium reaction, whereas invertases, which catalyze an irreversible hydrolysis, exist in isoforms located in the apoplasmic space (CWI), in the vacuole (VI), and in the cytosol (Frommer and Sonnewald, 1995; Sturm et al., 1995). Recently, several CWI and VI isoforms were cloned and their expression was studied with respect to developmental regulation and tissue- or cell-specific expression (Sturm et al., 1995; Weber et al., 1995; Cheng et al., 1996) and in response to wounding and pathogen attack (Sturm and Chrispeels, 1990; Zhang et al., 1996). In particular, CWI may be involved in phloem unloading (Miller and Chourey, 1992; Ruan and Patrick, 1995; Weber et al., 1995), regulation of sink strength (Weil and Rausch, 1990; Roitsch et al., 1995; Ehness and Roitsch, 1997), and hexose production for wound- or pathogen-induced respiration (Sturm and Chrispeels, 1990; Zhang et al., 1996). Recently, hexoses formed in the apoplasmic space were proposed to act, after cellular uptake (Truernit et al., 1996), as metabolic signals strongly affecting the expression of other genes (Herbers et al., 1996; Jang et al., 1997).
Although the regulation of CWI activity by changes in gene expression has been well documented, little is known about possible posttranslational regulation. Since CWI is localized outside the symplasm, a posttranslational modification of its structure leading to enzyme activation or inactivation would have to be operative in the specific ionic environment of the apoplasmic space. Clearly, by lowering the apoplasmic pH, cells may activate CWI because of its low pH optimum (pH 4.5). At lower pH the uptake of the resulting hexoses via H+/monosaccharide co-transporters is also stimulated (Sauer and Stadler, 1993; Truernit et al., 1996). Thus, it is noteworthy that for Chenopodium rubrum suspension-cultured cells a coordinate induction of transcripts for CWI and a specific Glc transporter isoform has been demonstrated (Ehness and Roitsch, 1997).
Recently, we characterized an entirely different regulatory mechanism based on interaction of CWI with an inhibitory polypeptide co-localized with CWI in the apoplasmic space (Weil and Rausch, 1994; Weil et al., 1994; Krausgrill et al., 1996). The inhibitory protein shares some characteristics with several INHs previously described (Schwimmer et al., 1961; Pressey, 1968, 1994; Jaynes and Nelson, 1971; Bracho and Whitaker, 1990; Ovalle et al., 1995). The tobacco (Nicotiana tabacum) INH is a heat-stable nonglycosylated protein (Weil et al., 1994). Inhibition of CWI by INH is modulated by the Suc concentration, the substrate apparently protecting the enzyme by binding to CWI and/or INH. Whereas the Km value for Suc hydrolysis was 0.6 mm (Weil and Rausch, 1990), the half-maximum substrate protection was observed with 1.2 mm Suc (Weil et al., 1994). The N-terminal sequence obtained from the purified tobacco INH shows similarity with the N terminus of an INH expressed in tomato (Lycopersicon esculentum) fruit (Pressey, 1994; Weil et al., 1994). Partially purified inhibitors isolated from tobacco suspension-cultured cells and from tomato fruit both inhibit tobacco CWI and tomato VI; however, no substrate protection was found for tomato VI (Sander et al., 1996).
A developmental study of the expression of CWI and INH in tobacco suspension-cultured cells has shown that both proteins are expressed during the entire culture period, although CWI inhibition has been observed only after the medium was Suc depleted (Krausgrill et al., 1998). The observed binding of the CWI–INH complex to concanavalin A-Sepharose (free INH does not bind) suggests that both proteins are in physical contact throughout the entire culture period, an assumption confirmed by co-migration of CWI and INH as a stable complex during native cathodic PAGE. These and other observations have led to the hypothesis that in suspension-cultured tobacco cells INH operates as a regulatory switch for CWI, with INH always being bound to CWI but inducing the inhibitory conformational change only when Suc concentration decreases below a threshold level (Weil et al., 1994; Krausgrill et al., 1998).
To further characterize the structure of the CWI–INH complex and the physiological role of INH for CWI regulation during plant development, we have now cloned the tobacco inhibitor. We present proof of function for the Nt-inh1-encoded protein, after heterologous expression in Escherichia coli, and demonstrate the specificity of INH binding to CWI. To our knowledge, the Nt-inh1 clone represents the first fully characterized plant INH, a member of a novel protein group with moderate but significant sequence conservation. A comparison of transcript levels for CWI and Nt-inh1 in different tissues reveals that during plant development their expression is not always coordinate.
MATERIALS AND METHODS
The origin and growth conditions of Agrobacterium tumefaciens-transformed tobacco (Nicotiana tabacum cv Petit Havana) cells were previously described (Weil and Rausch, 1990, 1994). For the analysis of transcript amounts in different plant organs, 7-week-old nonflowering and 15-week-old flowering tobacco (N. tabacum cv SNN) plants from the greenhouse were used. Tomato (Lycopersicon esculentum) fruits were purchased at the local market. The photoautotrophic Chenopodium rubrum cell-suspension culture was a gift from M. Stitt (Heidelberg, Germany). Fungal invertases (from Saccharomyces cerevisiae and Candida utilis) were obtained from Sigma.
Purification and Tryptic Digest of INH Protein
INH protein was isolated and purified from transformed tobacco cells according to the work of Weil et al. (1994). After fractionated ammonium sulfate precipitation, the 40 to 85% fraction was desalted and chromatographed twice on sulfopropyl Sephadex using a pH-gradient elution followed by an NaCl-gradient elution. After the NaCl-gradient peak fraction was separated by SDS-PAGE, the homogeneous INH protein was electroeluted and subjected to a second semipreparative SDS-PAGE run. The Coomassie-stained INH band was excised, and after destaining/shrinking in acetonitrile, it was subjected to in situ proteolytic digestion with trypsin in 25 mm ammonium bicarbonate for 6 h at room temperature. The released peptides were recovered from the supernatant and separated by reverse-phase HPLC. Purified peptide fractions were dotted on glass fiber membranes and subjected to automated Edman degradation on a sequencer (model 473A, Applied Biosystems).
Cloning of a Nt-inh1 cDNA
Total RNA was prepared from transformed tobacco cells according to the method of Logemann et al. (1987), from which the poly(A+) fraction was isolated with the Dynabeads biomagnetic separation system (Dynal, Oslo, Norway). cDNA was synthesized using the ZAP-Express cDNA-synthesis kit (Stratagene) and used for reverse-transcriptase PCR. Antisense primers were designed according to all peptide sequences obtained from the tryptic digest and used for PCRs in combination with a sense primer derived from the previously sequenced N terminus of INH (Weil et al., 1994). For reverse-transcriptase PCR approximately 1 ng of cDNA was used with Goldstar DNA polymerase (Eurogentec, Seraing, Belgium). With the following primers a 300-bp large partial INH cDNA was amplified, which was later used as a probe for library screening and Southern- and RNA-blot analysis (see below): sense primer 5′-AAGAACACACCIAAC/TTAC/TCA-3′ and antisense primer 5′-CCAACCATA/TCCATCC/TTCA/TGC-3′.
The preparation of the cDNA library from transformed tobacco cells was previously described (Greiner et al., 1995). The library contained 2 × 106 independent clones. Screening and in vivo excision of the phagemid were performed according to the manufacturer's instructions (ZAP-Express cDNA-synthesis kit). Plaques that were 5 × 105 were screened and five independent clones were isolated. The clones were sequenced by automatic sequencing (ABI Prism 377 [Applied Biosystems]; TopLab Laboratories, Munich, Germany).
RNA-Blot and Southern-Blot Analyses
The preparation of total RNA followed the protocol of Logemann et al. (1987). Genomic DNA was isolated from tobacco leaves (N. tabacum cv SNN) according to the method of Murray and Thompson (1980). Nonradioactive Southern- and RNA-blot procedures with biotinylated probes and chemiluminescence detection were performed according to the method of Löw and Rausch (1996), except that for RNA blots transfer was overnight in 20× SSC. For Southern-blot analysis the conditions for the high-stringency wash were based on approximately 85% homology.
Immunoblot Analysis
Immunoblotting (Towbin et al., 1979) was performed as described by Weil and Rausch (1994), using the semidry procedure. After proteins were transferred to Immobilon P membranes (Millipore) for 1 h, the membranes were blocked with 8% BSA and incubated in the different antisera for 12 h at 4°C. Immunoblots were developed with anti-rabbit IgG-alkaline phosphatase conjugate (Sigma).
Expression of Nt-inh1 Protein in Escherichia coli
The Nt-inh1 protein was expressed as a fusion protein using the pQE-30 vector from Qiagen (Hilden, Germany), which provides an N-terminal 6× His tag. For this purpose, the Nt-inh1 cDNA was amplified from the cDNA clone (pBK-CMV vector) with Pfu polymerase (Stratagene) using the following primers: sense primer 5′-TATATGGATCCAATAATCTAGTAGAAACTA-3′ (BamHI site underlined) and antisense primer 5′-ACATAGTCGACTCACAATAAATTTCTGACAATA-3′ (SalI site underlined). The amplification product was cloned into the EcoRV-restricted pBluescript SKII vector (Stratagene) from which it was released with BamHI and SalI and subsequently ligated with the BamHI/SalI-restricted pQE-30 vector. The sequence of the cloned construct was confirmed by automatic sequencing (see above). Transformation of host cells was performed according to the manufacturer's instructions (Qiagen).
For induction the following protocol was used. An overnight culture (37°C) was started from a single colony. After bacteria were diluted 1:500 with Luria-Bertani medium, bacteria were grown to a density of 0.6, induced by adding IPTG to 1.5 mm, and further grown for 4 h at 37°C. Thereafter, cells were pelleted for 5 min at 10,000g and extracted with lysis buffer (1/20 volume of initial culture volume: 8 m urea, 0.1 m sodium phosphate, and 0.01 m Tris-HCl, pH 8.0). After the sample was centrifuged for 10 min at 15,000g, the supernatant was mixed with 1/5 volume of a 50% slurry of Ni-NTA resin (Qiagen) and stirred at room temperature for 45 min before loading into a column. The column was first washed with 10 volumes of lysis buffer and then with 10 volumes of washing buffer (lysis buffer adjusted to pH 6.3 with 1 n HCl). For on-column renaturation of the bound recombinant Nt-inh1 protein the column was first washed with 50 volumes of TBS (20 mm Tris-HCl, 100 mm NaCl, and 1 mm PMSF, pH 7.5) and then eluted with 3 column volumes of the same buffer including 50 mm EDTA.
Affinity Purification of the INH Antiserum
The recombinant Nt-inh1 protein was used for affinity purification of the previously prepared antiserum directed against purified plant INH protein (Krausgrill et al., 1996). Five-hundred micrograms of purified recombinant Nt-inh1 protein was diluted in 10 mL of transfer buffer (48 mm Gly, 40 mm Tris base, 0.038% [v/v] SDS, and 20% [v/v] methanol) and bound to a 60-cm2 Immobilon P membrane (Millipore). After a 15-min incubation the membrane was washed in 50 mL of TBST (Tris-buffered saline plus Tween-20) for 5 min, blocked with 5% defatted milk powder in TBST, and washed again three times in TBST. Thereafter, INH antiserum, diluted 1:100 in 1% defatted milk powder/TBST, was bound to the Nt-inh1 protein-coated membrane for 1 h at room temperature. The membrane was then washed three times with TBST, and bound antibodies were eluted with 2 mL of Gly buffer (0.1 m Gly, 0.5 m NaCl, and 0.05% Tween 20, adjusted to pH 2.8 with 1 N HCl) during a 3-min incubation. The eluate was immediately mixed with 300 μL of 1 m Tris-HCl, pH 8.1, and the elution procedure was repeated. To the combined eluted fractions BSA and sodium azide were added at 1.5% and 1 mm final concentrations, respectively.
Chromatography of Total Cell Wall Protein on Ni-NTA-Bound Nt-inh1 Protein
Expression of Nt-inh1 in E. coli, binding of the solubilized recombinant Nt-inh1 protein to the Ni-NTA column, and on-column renaturation were performed as described above. To the Ni-NTA column with bound renatured inhibitor protein we applied 100 μg of total cell wall protein, diluted in 4 volumes of TBS and adjusted to pH 6.5, and the flow-through was passed through the column two additional times. After this binding step, the column was first washed with at least 25 column volumes of TBS and then eluted with 2 volumes of TBS including 50 mm EDTA. The different fractions were analyzed by SDS-PAGE and Coomassie staining for their polypeptide patterns. For immunoblot analysis all fractions were adjusted to 100 μg protein/mL by adding BSA to ensure complete protein precipitation by TCA prior to SDS-PAGE.
Assay for Inhibitor Function of Recombinant Nt-inh1
Tobacco CWI and tomato fruit VI were prepared as previously described (Weil and Rausch, 1994; Sander et al., 1996). Inhibition assays followed the protocol of Weil et al. (1994). Invertase preparations and Nt-inh1 protein preparations were mixed and incubated for 60 min at 37°C in the absence or presence of 20 mm Suc. After this preincubation 20 mm Suc was also added to the minus-Suc sample, and the Glc released during a subsequent 60-min incubation at 37°C was determined enzymatically. The concentration of the recombinant Nt-inh1 protein was determined according to the method of Peterson (1977).
RESULTS
Cloning of a Tobacco INH-Encoding cDNA
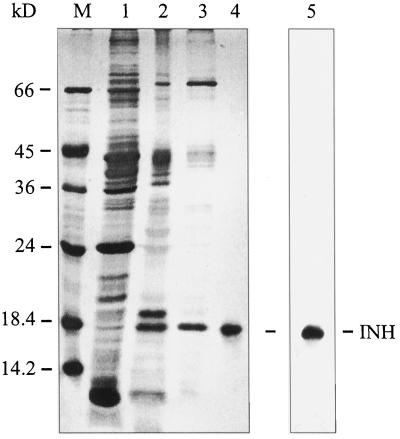
The INH protein expressed in transformed tobacco cells (Weil et al., 1994) was purified from a cell wall protein fraction obtained by nondisruptive salt elution as previously described (Weil and Rausch, 1994). After fractionated ammonium sulfate precipitation and ion-exchange chromatography on sulfopropyl Sephadex using sequentially pH-gradient and NaCl-gradient elution, the purified INH protein was electroeluted from the gel (Fig. 1). The homogeneous INH protein was then subjected to a tryptic digest. Four peptides could be separated by HPLC and their sequences were determined via Edman degradation. The locations of all peptide sequences in the Nt-inh1 full-length clone are indicated in Figure 2 (see below).
Figure 1.
Purification of tobacco INH from a salt-eluted cell wall protein fraction from transformed tobacco cells. Lane M, Marker proteins; lane 1, ammonium sulfate fraction (40–85% saturation); lanes 2 and 3, CWI activity peak fractions from cation-exchange chromatography, pH gradient (lane 2) and NaCl gradient (lane 3; Weil and Rausch, 1994); lane 4, electroeluted INH protein from the NaCl-gradient peak fraction (see lane 3); lane 5, immunoblot of ammonium sulfate fraction (see lane 1) with affinity-purified polyclonal antiserum directed against INH.
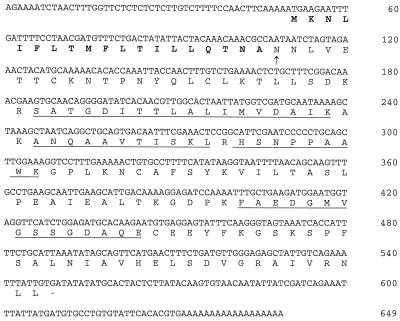
Figure 2.
cDNA sequence of the Nt-inh1 clone. The putative signal peptide is in bold type. The arrow indicates the predicted N terminus of the mature protein and is identical to the N terminus from direct INH protein sequencing (Weil et al., 1994). The peptide sequences obtained from INH tryptic digest are underlined.
Antisense primers were designed according to the obtained peptide sequences and used for PCR in combination with a sense primer deduced from the N-terminal protein sequence previously determined (Weil et al., 1994). With cDNA from transformed tobacco cells as the template, the longest specific amplification product obtained had a size of 300 bp. The sequence of this cDNA fragment contained a continuous open reading frame comprising all five peptide sequences obtained directly from the INH protein.
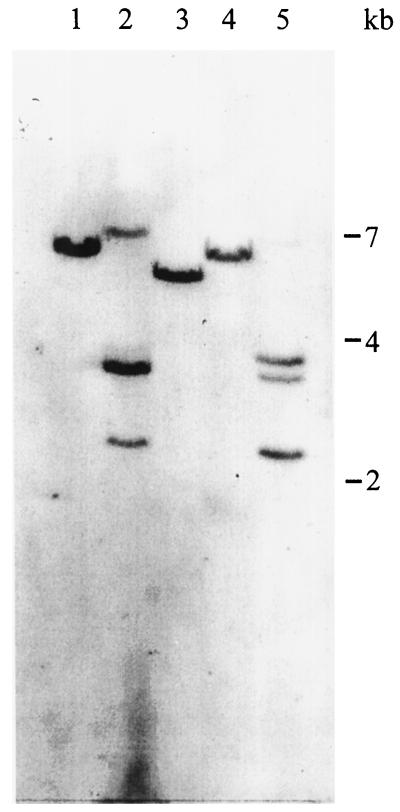
The screening of a cDNA library from transformed tobacco cells (Greiner et al., 1995) yielded five independent positive clones hybridizing with the 300-bp partial cDNA obtained by reverse-transcriptase PCR (see above). The cDNA sequence of one of the clones, Nt-inh1, has a total length of 631 bases, excluding the poly(A+) tail, and predicts an open reading frame of 182 amino acids (Fig. 2). The cDNA fragment used as a probe extends from position 134 to 433 of the full-length clone. All other cDNA clones showed the identical sequence except for the length of the 5′ untranslated region, which was 49 bases long in four of the clones and 13 bases shorter in one of the clones. Southern-blot analysis performed with the same 300-bp coding region probe indicated that INH is encoded by a single- or low-copy gene (Fig. 3), making the existence of closely related isoforms unlikely.
Figure 3.
Southern-blot analysis of INH sequences in the tobacco genome. Ten micrograms of genomic DNA was digested with BamHI (lane 1), DraI (lane 2), EcoRI (lane 3), EcoRV (lane 4), and HindIII (lane 5). The blot was hybridized with a 300-bp Nt-inh1-coding region probe.
Characteristics of the INH Protein Predicted by the Nt-inh1 Sequence
Since the Nt-inh1 clone contains all peptide sequences obtained from the tryptic digest of INH protein (see above), it certainly encodes the previously purified inhibitor protein (Weil et al., 1994). Furthermore, when E. coli cells harboring the pBK-CMV-Nt-inh1 plasmid were induced with IPTG, a protein of approximately 18 kD was induced that strongly reacted with the antiserum directed against the purified INH protein (data not shown).
A comprehensive database search did not reveal any homologous sequence except for two EST clones, one from Arabidopsis thaliana (accession no. T88540) and one from Citrus unshiu (accession no. C22245). The full cDNA sequence of the A. thaliana clone was determined (accession no. Y12807) and will be referred to as At-inhh (A. thaliana inhibitor homolog). Its predicted protein sequence has been aligned with Nt-inh1, together with part of the C. unshui EST clone (Fig. 4). At the protein level Nt-inh1 and At-inhh shared 25% sequence identity when the putative signal peptides of both proteins were omitted. It is noteworthy that the N-terminal parts showed a stronger sequence conservation than the C-terminal parts. When partial protein sequences of all three clones (corresponding to positions Asn-20 to Trp-85 in the Nt-inh1 clone) were aligned, At-inhh showed a higher similarity with the C. unshiu EST clone (53% identity) than with Nt-inh1 (30% identity). Whereas the protein sequences of Nt-inh1 and At-inhh both predicted signal peptides with a high score (Von Heijne, 1986), the C. unshiu EST clone produced a potential signal peptide with only a weak score. The most likely cleavage sites, as deduced from the -1/-3-rule (Von Heijne, 1986), are indicated in Figure 4; however, only for the Nt-inh1 clone was the cleavage site confirmed by N-terminal sequencing of the mature protein.
Figure 4.
Protein sequence alignment of Nt-inh1 with putative homologs from A. thaliana (A.t.) and C. unshiu (C.u.). The A. thaliana cDNA clone At-inhh (A.t.; EMBL nucleotide sequence database, accession no. Y12807) is the full-length clone corresponding to the A. thaliana EST clone with the accession no. T88450. The partial C. unshiu cDNA sequence (C.u.) represents an EST clone with the accession no. C22245. The putative signal sequences are underlined, and the most likely predicted cleavage sites are marked by upward arrows. Note that the putative signal peptide of the C. unshiu EST clone shows a very low score (Von Heijne, 1986). Conserved Cys residues are indicated by downward arrows. Amino acid residues identical between at least two sequences are in bold type. Except for the N-terminal sequences (upstream from Asn-20 in Nt-inh1), the predicted mature protein sequences were aligned using the CLUSTAL program from PC Gene (Intelligenetics, Mountain View, CA).
The nucleotide sequence of Nt-inh1 (Fig. 2) encodes a hydrophilic protein with an N-terminal signal peptide, in agreement with the apoplasmic location of the INH protein. The predicted cleavage site between Ala-19 and Asn-20 (Von Heijne, 1986) would yield an N terminus of the mature protein identical to the sequence obtained from INH protein isolated from tobacco suspension-cultured cells (Weil et al., 1994). The protein contains four Cys residues, the locations of which are conserved when compared with the At-inhh clone. Recently, a tobacco cDNA encoding a putative cytosolic homolog of Nt-inh1 was isolated, which also had four Cys residues at the same positions (S. Greiner and T. Rausch, unpublished data). Thus, it appears that these Cys residues are essential for Nt-inh1 function, an assumption supported by the observation that treatment with DTT alleviates the inhibitory action of the INH protein (R. Vogel and T. Rausch, unpublished results). It is noteworthy that the equally conserved Thr-42, neighboring the first Cys residue, is predicted to be a phosphorylation site for protein kinase C.
Whereas the Nt-inh1 sequence does not predict N-glycosylation of the encoded protein, in agreement with the previous characterization of the INH protein isolated from tobacco cells (Weil et al., 1994), one of the putative N-glycosylation sites of At-inhh (Asn-73) is located in a sequence motif of 20 amino acids (Asp-58 to Thr-77), which is highly conserved (80% identities) between At-inhh and the C. unshiu EST clone.
Heterologous Expression in E. coli and Proof of Nt-inh1 Function
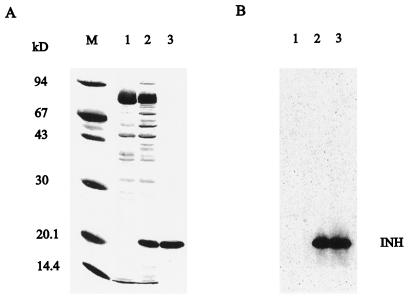
To unequivocally demonstrate the function of the Nt-inh1-encoded protein as a bona fide INH, we analyzed the effect of E. coli-expressed Nt-inh1 protein on different plant and fungal invertase preparations. For this purpose we cloned the open reading frame of the Nt-inh1 cDNA into an expression vector providing an N-terminal His tag. After IPTG induction the His-tagged fusion protein could be recovered from the insoluble fraction by urea solubilization (Fig. 5). The fusion protein had the predicted size and was purified to homogeneity by Ni-affinity chromatography on an Ni-NTA resin. After on-column renaturation the recombinant Nt-inh1-encoded protein was eluted by EDTA and used for inhibition studies.
Figure 5.
Expression of the recombinant His-tagged Nt-inh1-encoded protein in E. coli and purification by Ni-affinity chromatography. The open reading frame of the Nt-inh1 cDNA was amplified by PCR and cloned into the pQE-30 vector (Qiagen). Expression of the fusion protein was induced with IPTG. A, Coomassie-stained SDS-PAGE gel. Lane M, Marker proteins; lane 1, protein from uninduced bacteria; lane 2, protein from IPTG-induced bacteria; lane 3, purified recombinant protein after Ni-affinity chromatography. B, Immunoblot of fractions 1 to 3 from A developed with the polyclonal antiserum directed against the INH protein (Krausgrill et al., 1996).
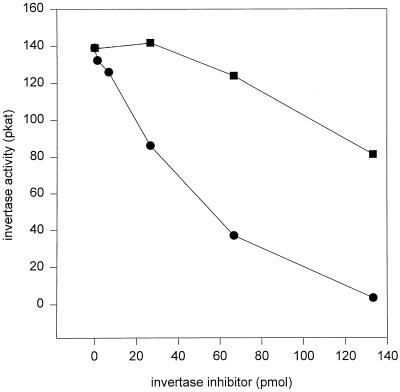
The results of inhibition of tobacco CWI by the recombinant inhibitor protein are depicted in Figure 6. After a 60-min preincubation in the absence of substrate a tobacco CWI preparation was completely inhibited at an inhibitor concentration of 650 nm. However, the effective inhibitor concentration required for full inhibition could de facto be significantly lower since the degree of on-column renaturation of the recombinant Nt-inh1-protein could not yet be determined. A significant substrate protection was observed when Suc was present during the preincubation, in agreement with the inhibition characteristics determined earlier for the partially purified INH protein isolated from transformed tobacco cells (Weil et al., 1994; Sander et al., 1996). To further confirm the specificity of invertase inhibition, we analyzed the effect of the recombinant inhibitor on other plant and fungal invertase preparations. CWI from C. rubrum cells and VI from tomato fruits were also strongly inhibited at similar INH concentrations, but no substrate protection was observed for these invertase preparations (Table I). In marked contrast to plant CWI and VI preparations, fungal invertases from S. cerevisiae and C. utilis were not affected by INH (Table I).
Figure 6.
In vitro inhibition of tobacco CWI by the recombinant Nt-inh1 protein. Partially purified CWI (Weil and Rausch, 1994) from transformed tobacco cells was preincubated with the recombinant Nt-inh1 protein in the absence (•) or presence (▪) of 20 mm Suc for 1 h in a volume of 200 μL, and its activity was determined in a subsequent 1-h incubation.
Table I.
Effect of the recombinant Nt-inh1 protein on plant and fungal invertases
| Organism | Invertase | Enzyme Activity
|
||
|---|---|---|---|---|
| Control | +INH−Suc | +INH+Suc | ||
| % | ||||
| N. tabacum | CWI | 100 | 1 | 59 |
| L. esculentum | VI | 100 | 12 | 7 |
| C. rubrum | CWI | 100 | 17 | 15 |
| S. cerevisiae | Inv | 100 | 90 | 86 |
| C. utilis | Inv | 100 | 104 | 85 |
Invertase preparations were preincubated with the recombinant Nt-inh1 protein in the absence (+INH-Suc) or presence (+INH+Suc) of 20 mm Suc for 1 h in 200 μL, and their activities were determined in a subsequent 1-h incubation. Enzyme amounts were adjusted to give 140 pKat per assay, and 133 pmol of recombinant Nt-inh1 protein was added. Fungal invertases (Inv) were obtained from Sigma.
Specificity of Nt-inh1 Protein Binding to CWI
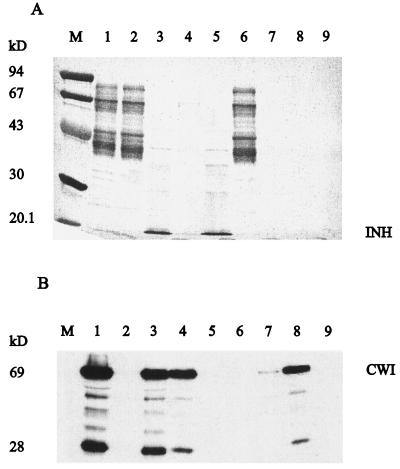
To determine the specificity of the binding of the Nt-inh1-encoded protein to tobacco CWI, we chromatographed a total cell wall protein fraction on Ni-NTA resin-bound recombinant Nt-inh1 protein (Fig. 7). Most of the cell wall protein was recovered in the nonbound eluate; however, CWI was absent in this fraction from both the control and the recombinant inhibitor-bearing Ni-NTA resin. Only after an extensive washing step did CWI elute from both columns, suggesting some inhibitor-independent weak binding of CWI to the Ni-NTA resin. However, only from the recombinant inhibitor-bearing Ni-NTA resin did a significant amount of CWI protein co-elute with the inhibitor protein during the subsequent EDTA elution, which releases the recombinant Nt-inh1 protein from the Ni-NTA resin (see above). From the control Ni-NTA resin EDTA did not release any CWI protein. These results indicate a specific binding of INH to CWI.
Figure 7.
Specific binding of CWI to recombinant Nt-inh1 protein immobilized on a Ni-affinity column. A total cell wall protein fraction from tobacco cells was passed through a column packed with recombinant Nt-inh1 protein bound to the Ni-NTA matrix (Qiagen). After extensive washing the recombinant Nt-inh1 protein with bound CWI was eluted with EDTA buffer. A, Analysis by SDS-PAGE and Coomassie staining. Total cell wall protein (lane 1) was passed through Ni-NTA columns with bound Nt-inh1 protein (lanes 2–5) or without Nt-inh1 protein (lanes 6–9); lanes 2 and 6, flow-through; lanes 3 and 7, EDTA eluate; lanes 4 and 8, washing fractions before EDTA elution; lane M, marker proteins. B, Immunoblot analysis of the same fractions as in A developed with an antiserum directed against CWI. The 28-kD splitting product results from intrinsic CWI lability (Weil and Rausch, 1994).
Expression of CWI and Nt-inh1 during Plant Development
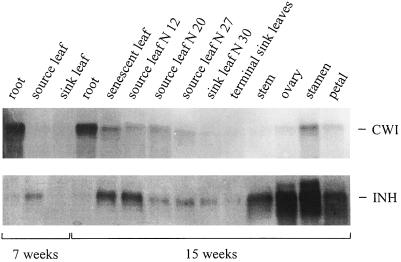
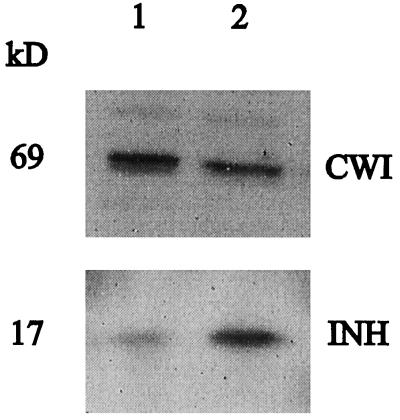
We previously showed that CWI protein and INH protein in transformed tobacco cells are both expressed at fairly constant amounts throughout the entire culture period (Krausgrill et al., 1998). Using cDNA probes for CWI (Greiner et al., 1995) and Nt-inh1 we have now compared the expression of CWI and INH at the transcript level in different organs of adult tobacco plants (Fig. 8). In both 7-week-old nonflowering and 15-week-old flowering plants, transcript amounts for CWI and INH appeared to be independently regulated. Whereas in plants of both ages CWI transcripts were abundant in roots, this was not the case for INH transcripts. In internodes, petals, ovaries, and stamens of 15-week-old plants INH transcript amounts were high, whereas CWI transcripts were abundant only in the stamen. It is noteworthy that in total RNA from stamen the Nt-inh1 probe apparently hybridized with two transcripts, one of which had the expected size and the other appeared to be approximately 200 bases larger. Only in leaves did the transcript amounts for both proteins show some correlation, with the highest amounts in senescent leaves and the lowest amounts in young sink leaves. Immunoblot analysis of leaf protein samples indicated that both proteins are expressed in mature and senescent leaves (Fig. 9).
Figure 8.
Transcript amounts for CWI and Nt-inh1 in different tissues of tobacco plants as determined by RNA-blot analysis. Total RNA was isolated from different tissues of 7- and 15-week-old tobacco (cv SNN) plants. For 15-week-old plants leaves were numbered starting from the oldest (senescent) leaf. For each tissue 10 μg of total RNA was loaded per lane. Blots were hybridized with coding region probes for CWI (Krausgrill et al., 1996) and Nt-inh1.
Figure 9.
Expression of CWI and INH protein in tobacco leaf tissue as determined by immunoblot analysis. Cell wall protein fractions from source (lane 1) and senescent (lane 2) leaves were separated by SDS-PAGE and subsequently analyzed by immunoblot. The polyclonal antisera were directed against CWI and the recombinant Nt-inh1 protein (affinity-purified antiserum).
DISCUSSION
Cloning of the First Plant INH
To our knowledge, the protein encoded by the Nt-inh1 cDNA (Fig. 2) represents the first plant INH for which the molecular structure has been determined (Weil et al., 1994; Krausgrill et al., 1996; this paper). The perfect identity of five different peptide sequences independently obtained from the plant INH protein leaves no doubt that Nt-inh1 encodes the previously characterized tobacco INH expressed in A. tumefaciens-transformed cells (Fig. 2; Weil et al., 1994; Krausgrill et al., 1996). In particular, the predicted N terminus of the mature Nt-inh1 protein is identical to the N terminus determined for a homogeneous INH preparation of tobacco cells (Weil et al., 1994). Further confirmatory features are the absence of glycosylation sites (Weil et al., 1994) and the cross-reaction of the Nt-inh1 protein with an antiserum directed against INH isolated from tobacco cells (Krausgrill et al., 1996, 1997). The high stability of the tobacco INH protein with respect to high temperature and low pH may result from intramolecular disulfide bridges, since the Nt-inh1 sequence predicts four Cys residues, the positions of which are conserved between Nt-inh1 and At-inhh (Fig. 4). The presence of structure-stabilizing disulfide bridges is a common feature of other plant apoplasmic proteins, such as peroxidases and the chitin-binding domain of several defense-related proteins (Raikhel et al., 1993). Whether the conserved Cys residues are also involved in the regulation of INH activity is not known; however, preliminary evidence suggests that the activity of at least some INHs may be reduced by treatment with DTT (R. Vogel and T. Rausch, unpublished results). The presence of a putative phosphorylation site, a conserved Thr residue (Thr-42 in Nt-inh1), is intriguing, but to our knowledge, regulation of apoplasmic proteins by phosphorylation has not yet been demonstrated.
A comparison of Nt-inh1 with (a) the complete sequence of the A. thaliana clone At-inhh, (b) the N terminus of a tomato fruit INH (Pressey, 1994), (c) an EST clone from C. unshiu, and (d) a putative cytosolic tobacco homolog of Nt-inh1 (not shown) reveals a new protein family with limited sequence conservation except for specific sequence motifs, including the four Cys residues at almost identical positions (Fig. 4). However, at present only the Nt-inh1-encoded protein and the tomato protein have been characterized as INHs, whereas the function(s) of the other members of this new protein family remain as yet unknown.
Whereas tobacco INH, the Nt-inh1-encoded protein, has been previously localized in the apoplasmic space (Weil and Rausch, 1994), it is not yet known whether apoplasmic and/or vacuolar isoforms exist for Nt-inh1. Although the tobacco INH has been shown to inhibit both VI and CWI isoforms in vitro (Sander et al., 1996), the existence of VI inhibitors cannot be excluded. Whereas Southern-blot analysis (Fig. 3) suggested that Nt-inh1 may not have closely related isoforms, the RNA blot of tobacco stamen (Fig. 8) revealed transcripts of different sizes. Whether the different transcripts detected in stamen result from differential splicing, different transcription initiation, or polyadenylation sites, or, alternatively, represent bona fide isoforms, is not yet known.
The inhibition of tobacco CWI and tomato fruit VI by tobacco INH isolated from tobacco cells (Weil et al., 1994; Sander et al., 1996) could be faithfully reproduced by the recombinant Nt-inh1 protein expressed in E. coli (Figs. 5 and 6; Table I). In particular, the specific effect of strong substrate protection, which was confined to tobacco CWI and absent for tomato VI (Sander et al., 1996), was also observed for the recombinant Nt-inh1 protein. Tobacco CWI and tomato fruit VI preparations of identical enzyme activity were completely inactivated with similar amounts of Nt-inh1 protein. Thus, if we assume similar specific activities for both invertases, the molar ratio of inhibitor protein to invertase would be identical. It is interesting that a CWI preparation from C. rubrum cells was completely inhibited in the presence of 20 mm Suc, showing no substrate protection (Table I). For several plant species, different CWI isoforms have been cloned with sometimes considerable structural differences (Weber et al., 1995). This discrepancy with tobacco CWI may indicate that different CWI isoforms are differentially affected. Alternatively, the substrate protection phenomenon may depend on specific structural features of the protein-protein interaction in the homologous system. The observation that two fungal invertases were not affected by INH indicates that an inhibitory effect on fungal pathogen invertases is at least unlikely.
The binding of tobacco INH to CWI, which has been previously characterized by immunocoprecipitation (Weil et al., 1994), co-chromatography of CWI and INH on concanavalin A-Sepharose, and native cathodic PAGE (Krausgrill et al., 1998), appears to be specific as demonstrated by the results of affinity chromatography of a complex cell wall protein fraction on a recombinant Nt-inh1 protein-bearing Ni-NTA resin (Fig. 7). Although it is not yet clear why CWI shows some weak binding to the control Ni-NTA resin, resulting in its delayed elution, the EDTA-induced co-release of Nt-inh1 protein and CWI leaves little doubt about the specificity of CWI binding to INH. Since the percentage of the on-column renaturation of the Nt-inh1 protein is not known, the binding ratio of both proteins is yet to be determined.
Regulation of CWI-INH Complex Formation and Physiological Role(s) of INH during Plant Development
The previous characterization of in vitro binding of INH to CWI (Weil et al., 1994) had shown that the concentrations of Suc and divalent cations, as well as the pH in the apoplasmic space, were all likely to affect CWI inhibition in vivo. When partially purified INH protein was added to intact suspension-cultured tobacco cells, an in vivo inhibition of CWI was observed (Sander et al., 1996), suggesting that the INH protein could freely move in the cell wall to reach CWI, which is thought to be ionically bound to the cell wall matrix. Both the charge of Nt-inh1 protein (pI 8.2) and its size (17 kD) support the assumption that the inhibitor is mobile in the apoplasmic space (Gogarten, 1988).
Whether CWI and INH are assembled to form a complex during their transit through the secretory pathway or, alternatively, associate only in the apoplasmic space is an intriguing question. In the former case a coordinate expression of both proteins would be required, which has been observed for transformed tobacco cells (Krausgrill et al., 1997). However, the results from RNA-blot analysis of CWI and Nt-inh1 transcript amounts in different parts of adult tobacco plants do not support a strict coordination of their expression. In adult tobacco plants the coordinate expression of Nt-inh1 and CWI appears to be confined to the different stages of leaf development (Figs. 8 and 9). Conversely, in roots a particularly high expression of CWI coincides with a very low expression of Nt-inh1 (Fig. 8), whereas in stem and certain parts of the flower the reverse situation is observed. Some of these discrepancies may be explained by the fact that transcript amounts do not necessarily reflect protein amounts. Thus, in roots, in which significant INH expression is detected at the protein level (Krausgrill et al., 1998), transcript amounts are very low. However, the high transcript levels of Nt-inh1 in internodes and in different parts of the flower with low or not detectable CWI transcript levels have been confirmed by immunoblot analysis (Krausgrill et al., 1998), suggesting that expression of the Nt-inh1-encoded protein may indeed be up-regulated independently from CWI. This phenomenon could be explained in different ways: (a) INH may inhibit other CWI isoforms going undetected with the probe/antiserum used in this study; (b) because the Nt-inh1-encoded protein may interact with CWI and VI, INH may be required in these tissues for regulation of VI; or (c) INH may have additional, as yet unknown functions.
On the premise that complex formation between INH and CWI or VI occurs already during passage through the secretory pathway, specific isoforms for both invertase types may not be required (see above). It is noteworthy that in potato, another solanacean species, a specific VI isoform has been shown by RNA blot to exhibit an expression pattern similar to Nt-inh1, including particularly high transcript amounts in flowers (Zrenner, 1993). Transgenic tobacco plants with constitutive Nt-inh1 overexpression have been raised to test this hypothesis.
Whereas the physiological role(s) of INH during plant development is not yet fully understood, some data are available suggesting that INH may act as an apoplasmic switch to turn off (or on) invertases under specific conditions. In a study of CWI and INH protein expression in A. tumefaciens-transformed tobacco cells, it could be shown that when the culture medium became Suc depleted the apparent CWI activity was strongly reduced, although the amounts of CWI and INH protein remained fairly constant throughout the entire culture period (Krausgrill et al., 1998). These data indicate that the nutritional status (feast versus famine [Koch, 1996]) of the cells affects CWI inhibition by INH.
By cloning the first plant INH, exploring its characterization, and analyzing its expression, we have been allowed to address several longstanding questions about invertase inhibitors, the structure and function of which have remained an enigma for more than 30 years. In particular, the aspect of binding specificity can now be rigorously confirmed. Although for suspension-cultured tobacco cells a specific role could be assigned to the Nt-inh1-encoded protein, the observed tissue-specific differences in the relative expression level of CWI and INH raise a number of new questions concerning the in vivo functions of INH during plant development. Transgenic tobacco plants expressing Nt-inh1 in sense or antisense orientation have been raised and will be studied to further elucidate the role of invertase inhibitors during plant development. Furthermore, the molecular nature of the CWI–INH complex is now being studied with pure recombinant CWI and INH preparations.
ACKNOWLEDGMENT
The antiserum against CWI was a gift from Arnd Sturm.
Abbreviations:
- CWI
cell wall invertase(s)
- EST
expressed sequence tag
- INH
invertase inhibitor
- IPTG
isopropylthio-β-galactoside
- Ni-NTA
Ni-nitrilotriacetic acid
- VI
vacuolar invertase(s)
Footnotes
This work was supported by grant no. SFB 199, TP-C3 from the Deutsche Forschungsgemeinschaft to T.R.
LITERATURE CITED
- Bracho GE, Whitaker JR. Purification and partial characterization of potato (Solanum tuberosum) invertase and its endogenous proteinaceous inhibitor. Plant Physiol. 1990;92:386–394. doi: 10.1104/pp.92.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS. The miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in pedicel. Plant Cell. 1996;8:971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness R, Roitsch T. Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 1997;11:539–548. doi: 10.1046/j.1365-313x.1997.11030539.x. [DOI] [PubMed] [Google Scholar]
- Eschrich W (1989) Phloem unloading of photoassimilates. In DA Baker, JA Milburn, eds, Transport of Photoassimilates. Longman Scientific & Technical, New York, pp 206–263
- Frommer WB, Sonnewald U. Molecular analysis of carbon partitioning in solanaceous species. J Exp Bot. 1995;46:587–607. [Google Scholar]
- Gogarten JP. Physical properties of the cell wall of photoautotrophic suspension cells from Chenopodium rubrum L. Planta. 1988;174:333–339. doi: 10.1007/BF00959518. [DOI] [PubMed] [Google Scholar]
- Greiner S, Weil M, Krausgrill S, Rausch T. A tobacco cDNA coding for cell-wall invertase. Plant Physiol. 1995;108:825–826. doi: 10.1104/pp.108.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Meuwly P, Frommer WB, Metraux JP, Sonnewald U. Systemic acquired resistance mediated by the ectopic expression of invertase: possible hexose sensing in the secretory pathway. Plant Cell. 1996;8:793–803. doi: 10.1105/tpc.8.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes TA, Nelson OE. An invertase inactivator in maize endosperm and factors affecting inactivation. Plant Physiol. 1971;47:629–634. doi: 10.1104/pp.47.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Krausgrill S, Greiner S, Köster U, Vogel R, Rausch T (1998) In transformed tobacco cells the apoplasmic invertase inhibitor operates as a regulatory switch of cell wall invertase. Plant J (in press)
- Krausgrill S, Sander A, Greiner S, Weil M, Rausch T. Regulation of cell wall invertase by a proteinaceous inhibitor. J Exp Bot. 1996;47:1193–1198. doi: 10.1093/jxb/47.Special_Issue.1193. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Löw R, Rausch T (1996) Detection of nucleic acids with biotinylated PCR-amplified probes. In T Meier, F Fahrenholz, eds, A Laboratory Guide to Biotin-Labelling in Biomolecule Analysis. Birkhäuser Verlag, Basel, Switzerland, pp 201–213
- Miller ME, Chourey PS. The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell. 1992;4:297–305. doi: 10.1105/tpc.4.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovalle R, Keyes AC, Ewing EE, Quimby FW. Purification and characterization of the acid-stable proteinaceous inhibitor of potato tuber invertase by nonideal size exclusion chromatography. J Plant Physiol. 1995;147:334–340. [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Pressey R. Invertase inhibitors from red beet, sugar beet, and sweet potato roots. Plant Physiol. 1968;43:1430–1434. doi: 10.1104/pp.43.9.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey R. Invertase inhibitor in tomato fruit. Phytochemistry. 1994;36:543–546. [Google Scholar]
- Raikhel NV, Lee HI, Broekaert WF. Structure and functions of chitin-binding proteins. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:591–615. [Google Scholar]
- Roitsch T, Bittner M, Godt DE. Induction of apoplastic invertase of Chenopodium rubrum by d-glucose and a glucose analog and tissue-specific expression suggest a role in sink-source regulation. Plant Physiol. 1995;108:285–294. doi: 10.1104/pp.108.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RuanYL, Patrick JW. The celllular pathway of postphloem sugar transport in developing tomato fruit. Planta. 1995;196:434–444. [Google Scholar]
- Sander A, Krausgrill S, Greiner S, Weil M, Rausch T. Sucrose protects cell wall invertase but not vacuolar invertase against proteinaceous inhibitors. FEBS Lett. 1996;385:171–175. doi: 10.1016/0014-5793(96)00378-x. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stadler R. A sink-specific H+/monosaccharide cotransporter from Nicotiana tabacum: cloning and heterologous expression in baker's yeast. Plant J. 1993;4:601–610. doi: 10.1046/j.1365-313x.1993.04040601.x. [DOI] [PubMed] [Google Scholar]
- Schwimmer S, Makower RU, Romem ES. Invertase and invertase inhibitor in potato. Plant Physiol. 1961;36:313–316. doi: 10.1104/pp.36.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Sonnewald U. Regulation of metabolism in transgenic plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:341–368. [Google Scholar]
- Sturm A, Chrispeels MJ. cDNA cloning of carrot extracellular β-fructosidase and its expression in response to wounding and bacterial infection. Plant Cell. 1990;2:1107–1119. doi: 10.1105/tpc.2.11.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Sebkova V, Lorenz K, Hardegger M, Lienhard S, Unger C. Development- and organ-specific expression of the genes for sucrose synthase and three isoenzymes of acid β-fructofuranosidase in carrot. Planta. 1995;195:601–610. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Schmid J, Epple P, Illig J, Sauer N. The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell. 1996;8:2169–2182. doi: 10.1105/tpc.8.12.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. Seed coat-associated invertases of fava bean control both unloading and storage functions: Cloning of cDNAs and cell type-specific expression. Plant Cell. 1995;7:1835–1846. doi: 10.1105/tpc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M, Krausgrill S, Schuster A, Rausch T. A 17 kDa Nicotiana tabacum cell-wall peptide acts as an in-vitro inhibitor of the cell-wall isoform of acid invertase. Planta. 1994;193:438–445. doi: 10.1007/BF00201824. [DOI] [PubMed] [Google Scholar]
- Weil M, Rausch T. Cell wall invertase in tobacco crown gall cells: enzyme properties and regulation by auxin. Plant Physiol. 1990;94:1575–1578. doi: 10.1104/pp.94.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M, Rausch T. Acid invertase in Nicotiana tabacum crown-gall cells: molecular properties of the cell-wall isoform. Planta. 1994;193:430–437. doi: 10.1007/BF00201824. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cohn NS, Mitchell JP. Induction of a pea cell-wall invertase gene by wounding and its localized expression in the phloem. Plant Physiol. 1996;112:1111–1117. doi: 10.1104/pp.112.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R (1993) Klonierung und funktionelle Analyse von Genen kodierend für am Saccharosestoffwechsel der Kartoffel beteiligte Proteine. PhD thesis. Freie Universität Berlin, Berlin, Germany
- Zrenner R, Salanoubat M, Willmitzer L, Sonnnewald U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.) Plant J. 1995;7:97–107. doi: 10.1046/j.1365-313x.1995.07010097.x. [DOI] [PubMed] [Google Scholar]