Abstract
Presence of microcystin (MC), a predominant freshwater algal toxin and a suspected liver carcinogen, in Florida’s freshwaters poses serious health threat to humans and aquatic species. Being recalcitrant to conventional physical and chemical water treatment methods, biological methods of MC removal is widely researched. Water samples collected from five sites of Lake Okeechobee (LO) frequently exposed to toxic Microcystis blooms were used as inoculum for enrichment with microcystin LR (MC-LR) supplied as sole C and N source. After 20 days incubation, MC levels were analyzed using high performance liquid chromatography (HPLC). A bacterial consortium consisting of two isolates DC7 and DC8 from the Indian Prairie Canal sample showed over 74% toxin degradation at the end of day 20. Optimal temperature requirement for biodegradation was identified and phosphorus levels did not affect the MC biodegradation. Based on 16S rRNA sequence similarity the isolate DC8 was found to have a match with Microbacterium sp. and the DC7 isolate with Rhizobium gallicum (AY972457).
Keywords: Blue-green algae, Microcystin LR, Lake Okeechobee, Rhizobium gallicum, Ochrobactrum, Microbacterium, Bio-degradation
Introduction
Algal blooms have been a persistent problem for Florida’s fresh and marine waters often due to the indiscriminate agricultural use of nitrogen and phosphorus rich fertilizers, poor water and land management practices and inadequate waste water treatment methods (Phlips et al. 2003). The profuse growth of algae on the surface of waters not only increases turbidity, particulate matter and the production of taste and odor causing compounds in water, but blocks sunlight and kills submerged vegetation and fish which also reduces species diversity within the water body (Fleming and Stephan 2001). When the algal cells finally begin to lyse, some species release their toxic cell metabolites directly into the water body (Metcalf and Codd 2004). These metabolites, referred to as cyanotoxins, are potentially dangerous to many organisms, including humans (Codd et al. 1997; WHO 1999).
Microcystins are the most commonly encountered cyanotoxins in freshwaters, and microcystin LR (MC-LR) is the most extensively studied (Fleming et al. 2002). The toxin was first isolated from the cyanobacteria Microcystis aeruginosa, but later from species in the genera Anabaena, Oscillatoria, Nostoc and Anabaenopsis (Burns 2007). MC is toxic to mammals, fish, plants and invertebrates (Codd et al. 1997; Pflugmacher et al. 1999) and is reported to bioaccumulate in several crop plants (irrigated with contaminated water), mussels, crayfish, and fish (grown in contaminated estuarine water) commonly consumed by humans (Figueiredo et al. 2004). Studies have also shown the possible correlation between consumption of surface water contaminated with MC and the occurrence of HCC (Hepato Cellular Carcinoma), the most prevalent type of liver cancer in humans (Falconer 1991; Fleming et al. 2002). MC toxicity poses a great concern for public health safety, and thus The World Health Organization recommends a level of less than 1 μg/l of the toxin in drinking water to be safe for humans (WHO 1998).
Chemically, MC are a group of monocyclic heptapeptides having a general structure composed of five D-amino acids and two variable L-amino acids (which contributes to the two-letter suffix). Adda (3-amino-9 methoxy-2,6,8 trimethyl-10-phenyldeca-4,6-dienoic acid) is the characteristic β-amino acid responsible for acute toxicity (Harada et al. 1990, 2004). The lethal dose (LD50) value for MC in mice ranges from 50 to 300 μg/kg body weight (bw), with MC-LR having an LD50 of 50 μg/kg bw comparable to that of chemical organophosphate nerve agents (Dawson 1998). Liver is the main target organ for MC toxicity (Robinson et al. 1991; Fischer et al. 2000). The major exposure route for humans is the consumption of contaminated water (WHO 1998). Minor routes include recreational use of lakes and rivers (dermal), contaminated food and inhalation (if MC is present as aqueous aerosols in air) (Codd et al. 1997).
Microcystin is recalcitrant to conventional water treatment (Lahiti and Hiisverta 1989) and is often detected even after chemical treatment of waters. Physical methods of MC removal have been tried but are constantly hampered by the presence of natural organic matter (Newcombe et al. 2003). Chemicals used to kill the algae are potentially toxic and may remain in the water to pose greater danger when consumed (Kenefick et al. 1993). These chemicals also kill the algal cells (releasing more toxins into the water body) and the beneficial bacteria that can degrade the toxin (Jones and Orr 1994). Continued and increased use of such chemicals can induce resistance in organisms, making the management practices much more difficult. Biological methods of removal are being widely researched and are gaining more popularity (Cousins et al. 1996; Ho et al. 2007; Edwards and Lawton 2009). Several MC degrading bacteria have been reported so far (Valeria et al. 2006; Tsuji et al. 2006; Ho et al. 2007; Lemes et al. 2007; Pathmalal et al. 2009).
The first study of biological removal of MC was carried out by Jones et al. (1994) in Murrumbidgee River, New South Wales, Australia. The MC-LR concentration of 1 mg l−1 was degraded within 5d using native bacteria previously exposed to the toxin. Cousins et al. (1996) also identified the possibility of using an indigenous mixed bacterial population to degrade low levels of MC-LR in reservoir waters that have been previously exposed to Microcystis blooms. Ho et al. (2007) reported that during MC-LR and MC-LA biodegradation with Sphingopyxis sp. LH21, no cytotoxic by-products were detected, thus demonstrating the possibility of using such bacterial species for biodegradation in drinking or recreational water supplies. While several studies have been carried out around the world, no research to our knowledge regarding isolation of MC degrading bacteria and MC biodegradation process has been published in United States particularly from South Florida’s Lake Okeechobee (LO) region.
Lake Okeechobee (LO) is the largest freshwater lake of Florida providing for majority of drinking water treatment facilities. It is also an ecologically and recreationally important water body (Audubon of Florida 2005) and a potential aquifer storage and recovery zone (ASR), influencing several other lakes, rivers and canals including Caloosahatchee River, St. Lucie Canal and the Kissimmee River in Florida (Steidinger et al. 1999). Phosphorus levels in LO rose from 40 parts per-billion (ppb) in the 1970s to more than 130 ppb by the 1990s due to runoff from nearby agricultural operations, improved pasture, and dairy operations. Elevated levels of phosphorus in LO have increased the occurrence of algal blooms and changed the species composition of the algal community (Audubon of Florida 2005). Species of Anabaena, Microcystis, and Cylindrospermopsis which produce toxins, are dominant in LO, making the water unpalatable and therefore a poor base for the aquatic food chain (Fleming et al. 2002). Chlorophyll a and MC toxin concentrations were at elevated levels during August and October of 2005 with the highest concentrations observed in the Harney Pond, Indian Prairie Canal and Fish Eating Bay area (SFWMD). Sites such as Clewiston, Pahokee and Taylor slough also show raised levels of MC very frequently. Williams et al. (2007) studied the cyanobacterial toxins in Florida’s freshwater systems during 2005 and reported up to 95 μg/l of MC in some zones of LO. Being frequently exposed to such algal bloom scenarios, these locations would have native bacteria adapted to high toxin levels and that could exhibit toxin degradation abilities. The objective of this study was to isolate and characterize MC biodegrading bacteria or bacterial consortium from Florida’s LO.
Materials and methods
Water sample collection
Four sites were selected in LO area that represented high bloom area (HB), low bloom area (LB) and medium bloom area (MB), based on historic MC levels in these sites as reported by the South Florida Water Management District (SFWMD 2008). The sampling sites and their GPS co-ordinates are given in Table 1. Samples were collected in pre-cleaned 125 ml Nalgene bottles. Sub-surface water samples were collected at each site and transferred to sample bottles. The bottles were rinsed with the water sample before collection. Three samples were collected per site (but were later pooled together for experiments and analyses). Samples were collected at two different locations on the Indian Prairie Canal (sites D and E), since floating algal biomass was seen at two different points of Site D. The bottles were labeled with the site name, placed in an insulated cooler, transported to the lab, and stored at 4°C.
Table 1.
Description of LO sampling sites
| Site | Site name | Bloom frequency | GPS co-ordinates | Number of samples collected |
|---|---|---|---|---|
| A | Clewiston | LB | N 26°45.617′ W 80°55.054′ |
3 (A1,A2,A3) |
| B | Fish Eating Creek Canal | HB | N 26°57.752′ W 81°07.276′ |
3 (B1,B2,B3) |
| C | Harney Pond Canal | MB | N 26°59.158′ W 81°03.955′ |
3 (C1,C2,C3) |
| D | Indian Prairie Canal | HB | N 27°02.578′ W 80°56.940′ |
3 (D1,D2,D3) |
| E | Indian Prairie Canal | HB | N 27°03.134′ W 80°57.664′ |
3 (E1,E2,E3) |
MC and enrichment media
Microcystin LR (MC-LR) (from ARCH Core Facility at Florida International University, Miami) stock solution was prepared and stored at 4°C (1 ml stock = 50 μg MC-LR). The initial acclimatization (enrichment) and later experiments was carried out in minimal salts media (MSM) suggested by Valeria et al. (2006). The composition of the media per liter includes 112 mg MgSO4·H2O, 5 mg ZnSO4·H2O, 2.5 mg Na2MoO4·2H2O, 340 mg KH2PO4, 670 mg Na2HPO4·7H2O, 14 mg CaCl2 and 0.13 mg FeCl3. Twenty-five milliliters of MSM (with 0.25 μg ml−1 MC-LR) was dispensed into sterilized Erlenmeyer flasks under sterile condition.
Enrichment study
Water samples collected from each site served as inoculum for enrichment studies. One milliliter of pooled sample from each site was added to 25 ml of sterile MSM with 0.25 μg ml−1 of MC-LR. Duplicate samples were maintained. A control, consisting of media without MC-LR and inoculated with sample was also maintained under similar conditions. Similar procedure was repeated for other sites. The inoculated flasks, controls and duplicates were incubated at 26°C in a rotary shaker incubator for 20 days after which they were sub-cultured with fresh toxin and continued to incubate for another 20 days. The process was repeated one more time at the end of 40 days. After each 20 day sub-culture, the bacterial populations in the samples were checked using dilution plate method on dilute nutrient agar (10%) and the plates were incubated for 3 days at 26°C. Colonies were represented as colony forming units (CFU) per ml of inoculum.
MC biodegradation studies
After three cycles of enrichment studies (60 days period), 1 ml of inoculum from each of the incubated flasks was transferred to fresh 25 ml MSM amended with 0.25 μg ml−1 of MC-LR and incubated at 26°C in a rotary shaker incubator for a series of 10, 20 and 30 days to observe the pattern of MC degradation by the isolates of each site. Separate sets of control flasks without inoculum and without MC-LR were incubated under similar conditions as experimental flasks. Colony counts were carried out at the end of 10, 20 and 30 days and plated on dilute nutrient agar (10%) plates. Colonies from the nutrient agar plate were then sub-cultured and maintained.
MC analyses
At the end of incubation period the samples were analyzed for MC using high performance liquid chromatography (HPLC). A series of steps were carried out before the samples were injected into the HPLC, to remove bacterial cells suspended in the sample that cause turbidity, which would otherwise interfere with the chromatographic process.
Each sample was sonicated to lyse the bacterial cells and release the cell-bound toxins for complete analysis. This was done by inserting the sonicating probe into the sample solution for 1 min. The sonicating probe was sprayed with 70% alcohol after each sample and wiped clean to prevent contamination. The sonicated samples were then centrifuged at 8°C for 10 min at 10,000 rpm. The supernatant was filtered using a 0.22 μm sterile membrane filter. The filtered samples were then allowed to pass through a C18 solid phase extraction (SPE) cartridge (Extract Clean™ C18-HC 1,000 mg from GRACE). The cartridge was first activated with 10 ml of 100% methanol followed by 10 ml of distilled water. To the activated cartridge, the sample was slowly added and collected separately. Ten milliliters of methanol was finally added to the cartridge to elute the adsorbed MC and the eluent was collected separately. The eluent was then evaporated completely by purging nitrogen gas into the sample for 6–8 h. To the evaporated sample 1 ml methanol was added. If samples showed turbidity, they were centrifuged again at 10,000 rpm for 4 min. The supernatant was then transferred to clean HPLC vials and sealed. HPLC was run on a Hewlett-Packard system equipped with a UV–Vis spectrophotometer, using a 4.6 × 25 cm SUPELCO-Discovery C18 HPLC column (5 μm), with acetonitrile: 0.05% and ammonium acetate (20 mM; pH 5) as mobile phase, flow rate: 0.1 ml min−1, column temperature: 20°C and UV detection at 238 nm. Standards of MC-LR (4, 6 and 8 μg) were prepared and analyzed for determining the calibration curve. Trend line and R2 value was generated using Microsoft Excel™ software (Version 1997–2003). The HPLC peak area was used in the trend-line equation to calculate the amount of MC-LR recovered from the sample and controls.
Selection of isolates and optimal condition studies
From the results of HPLC analyses, the sample showing maximum degradation over the 30-day incubation period was identified. Controls with no inoculum were also analyzed for MC-LR content to account for photo-degradation. The amount of toxin degradation was calculated using the difference between initial and final toxin levels divided by the initial toxin concentration expressed as a percentage value.
A consortium of two bacterial isolates DC7 and DC8 from site D responsible for maximum toxin degradation was used for further experiments. The bacterial isolates DC7 and DC8 were grown separately in 250 ml Erlenmeyer flasks containing 50 ml of MSM+ medium (MSM amended with glucose and nitrogen to increase cell yield) on a rotary shaker (200 rpm) at 26°C. Bacterial cells were harvested during exponential growth phase by centrifugation (10,000×g, 10 min, 4°C), washed once with sterile MSM solution and resuspended in MSM medium and used immediately. Each treatment flask received 5 × 105 CFU ml−1 of DC7 and 5 × 105 CFU ml−1 of DC8 separately, adding up to an initial consortium inoculum of 1 × 106 CFU ml−1. The bacterial consortium was grown in MSM media amended with 0.3 μg ml−1 of MC-LR to study their toxin tolerance level at 26°C for 15 and 20 days. The effect of incubation temperature was determined by carrying out the biodegradation at 24, 26 and 28°C. Twenty-five milliliters of MSM (with 0.25 μg ml−1 MC-LR) was inoculated with the bacterial consortium and incubated for 20 days. Since high phosphate levels are quite common in LO the possible interference of phosphate in the biodegradation process was studied by using media rich in phosphate which consists of 112 mg MgSO4·H2O, 5 mg ZnSO4·H2O, 2.5 mg Na2MoO4·2H2O, 340 mg KH2PO4, 1340 mg Na2H-PO4·7H2O, 14 mg CaCl2 and 0.13 mg FeCl3 per 1 l of deionized water. The bacterial consortium was incubated with high phosphate MSM media along with 0.25 μg ml−1 MC-LR at 26°C for 20 days. Control with no MC-LR and duplicates were maintained for all experiments.
DC7 and DC8 isolate 16S rRNA sequence analysis
The site D sample bacterial isolates DC7 and DC8 that showed maximum MC degradation were first characterized using standard bacteriological methods. The 16S rRNA gene was PCR amplified from genomic DNA isolated from pure bacterial colonies. Primers used are universal 16S primers that correspond to positions 0005F and 0531R for a 500 bp sequence, and 0005F and 1513R for the 1,500 bp sequence. Amplification products were purified from excess primers and dNTPs and checked for quality and quantity by running a portion of the products on an agarose gel. Cycle sequencing of the 16S rRNA amplification products was carried out using DNA polymerase and dye terminator chemistry. Excess dye-labeled terminators were then removed from the sequencing reactions. The samples were electrophoresed on ABI 3130 Genetic Analyzer. Sequence analysis was performed using Sherlock® DNA microbial analysis software and database.
Results and discussion
Bacterial population during enrichment
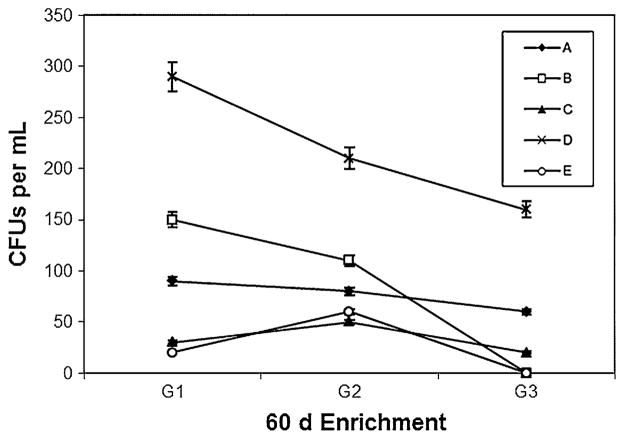
Heterotrophic colony count studies carried out every 20 days (for three cycles) during initial enrichment phase revealed that sample from site D showed high turbidity and constantly higher number of colonies on the dilute nutrient agar plates when compared to samples from other sites (Fig. 1). During first cycle G-1 (at the end of 20 days), number of colonies from site D was as high as 290 × 106 CFUs ml−1 and with third cycle G-3 (at the end of 60 days) the colonies were still as high as 160 × 106 CFUs ml−1. Even though samples from other sites showed higher colony counts during first cycle, the numbers started to dwindle rapidly over the course of 60 days. The constantly high number of colonies surviving from sample D showed the prospective presence of MC assimilating organisms present in the sample.
Fig. 1.
Changes in heterotrophic bacterial population (CFU × 106 ml−1) in MC enrichment cultures derived from LO sampling sites. Interval between each cycle (G1, G2, and G3) was 20 days. A, B, C, and E are sampling sites
MC degradation studies
During the MC degradation study, sample from site D showed highest degradation rate (Table 2). At the end of 10 days of incubation, the recovery of MC from controls averaged to 3.14 μg in 25 ml of medium (initial concentration was 6.25 μg in 25 ml of medium). Loss of MC-LR in controls could be attributed to photo degradation during incubation. Except sample D, all other samples showed lower degradation than the controls. Sample D showed about 6% more degradation than the controls and this pattern continued during 20 and 30 day incubation too. The initial degradation rate predominantly depends on the amount of MC available as well as the population of organisms present to make use of the toxin. Once the population establishes and starts to degrade the toxin, the rate of degradation increases and occurs at a faster rate. Cousins et al. (1996) report that during the first 12 day of MC degradation, the rate was high only when they incubated the sample in raw reservoir waters, while the rate did not change rapidly when similar studies were carried out in deionized water.
Table 2.
MC degradation (%) in enrichment cultures after 10, 20 and 30 day incubation
| Incubation period and sample site | Average toxin recovered (μg) | Degradation (%) |
|---|---|---|
| Day 10 | ||
| A | 3.78 (±0.42) | 39.52 |
| B | 3.88 (±0.05) | 37.92 |
| C | 3.75 (±0.47) | 40 |
| D | 2.80 (±0.09) | 55.2 |
| E | 3.64 (±0.37) | 41.84 |
| Control | 3.14 (±0.09) | 49.76 |
| Day 20 | ||
| A | 2.05 (±0.05) | 67.2 |
| B | 3.26 (±0.55) | 47.44 |
| C | 2.46 (±0.31) | 60.72 |
| D | 1.59 (±0.16) | 74.56 |
| E | 2.93 (±0.66) | 53.20 |
| Control | 3.79 (±0.06) | 39.36 |
| Day 30 | ||
| A | 1.72 (±0.55) | 72.48 |
| B | 2.09 (±0.37) | 66.56 |
| C | 2.17 (±0.04) | 65.36 |
| D | 1.00 (±0.45) | 84 |
| E | 2.29 (±0.13) | 63.28 |
| Control | 3.41 (±0.17) | 45.44 |
Values in parenthesis are SE
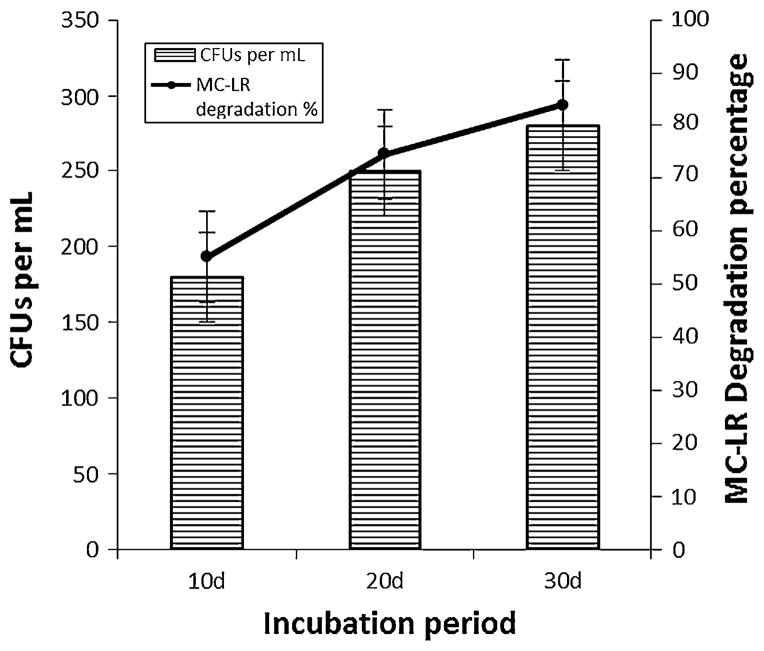
At the end of 20 day incubation, the degradation was much faster with sample D. While 3.79 μg MC per 25 ml of MSM were recovered from the controls, only about 1.59 μg MC were recovered from sample D. There was a corresponding increase in bacterial cell count along with MC degradation (Fig. 2). Bacterial cell count studies during day 0, 11, 21 and 31 confirms that the increasing cell numbers were indeed responsible for subsequent MC degradation (Table 3). Controls (with no MC) did not show any colonies on dilute nutrient agar plates. At the end of 30 day incubation the degradation rate began to get slower, which could be due to dwindling microbial count or the inability of the microbes to breakdown the toxin any further. Analysis of metabolites arising from this degradation would give a clear idea about further progress in degradation.
Fig. 2.
MC-LR biodegradation (%) by site D bacterial consortium and changes bacterial cell count (CFU × 106 ml−1) during incubation period
Table 3.
Changes in heterotrophic bacterial population (CFU × 106 ml−1) during MC enrichment culture
| Sample site | Day 0 | Day 11 | Day 21 | Day 31 |
|---|---|---|---|---|
| CFUa (ml−1) | ||||
| A | 40 | 110 | 150 | 180 |
| B | 40 | 100 | 130 | 200 |
| C | 40 | 170 | 200 | 180 |
| D | 40 | 180 | 250 | 280 |
| E | 40 | 170 | 190 | 210 |
CFU colony forming units, mean three replications
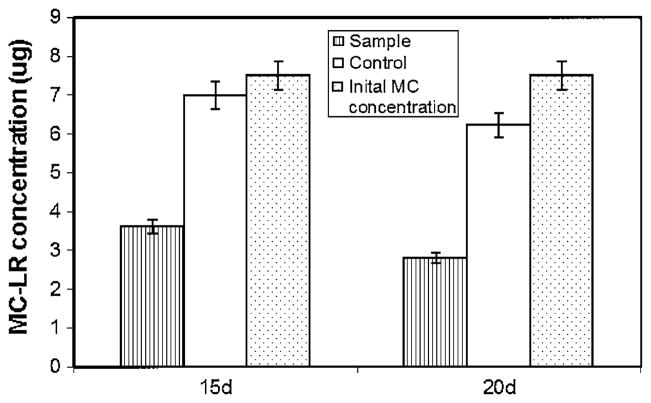
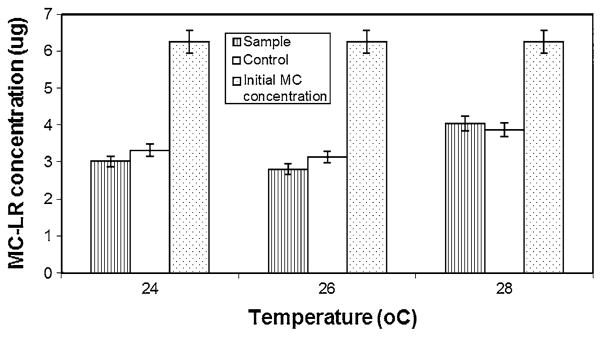
The bacterial population from the site D enrichment was found to be consisting of two bacterial colony types. The two isolates DC7 and DC8, when tested individually in a replicated study showed no significant growth or biodegradation of MC (data not shown), therefore the two isolates were used together as a consortium for further studies. With the HPLC results, it was confirmed that bacterial consortium from site D had maximum MC-LR degradation capacity. The maximum amount of MC was degraded during the first 20 days of incubation with sample D. Hence this consortium was chosen for further characterization studies. When MC concentration was increased from 0.25 to 0.3 μg ml−1, the site D consortium was still able to degrade 51% more toxin compared to the controls in 15 days and 45% more than the control in 20 days (Fig. 3). Temperature seems to have some effect on MC degradation (Fig. 4). While at 28°C, the degradation was poor and at 24 and 26°C, the degradation remained more or less the same. Phosphate level in the media seems to have no effect on MC degrading bacteria (Fig. 5). Even at higher phosphate levels the bacteria was still able to degrade the toxin.
Fig. 3.
MC-LR recovery from site D consortium culture after 20 day incubation. Initial MC content in each flask was 7.5 μg for both sample and control, with no bacterial inoculum in control
Fig. 4.
MC-LR recovery from site D consortium culture after 20 day incubation at different temperatures. Initial MC content in each flask was 6.25 μg for both sample and control, with no bacterial inoculum in control
Fig. 5.
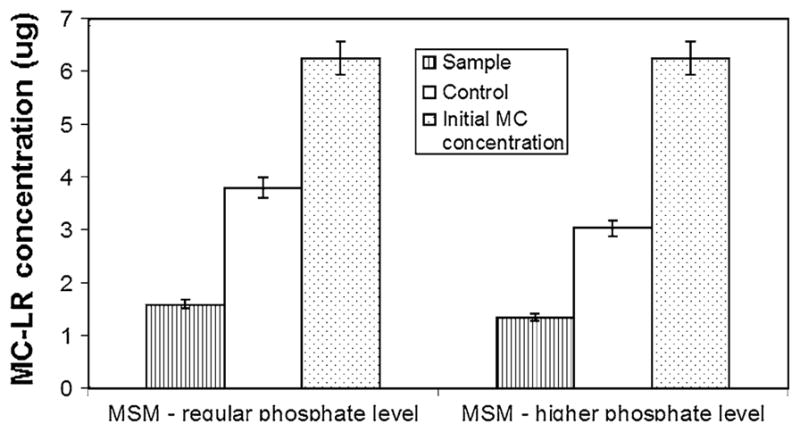
Effect of high phosphate concentration on MC-LR recovery after 20 day incubation of site D consortium culture. Initial MC content in each flask was 6.25 μg for both sample and control, with no bacterial inoculum in control
Identification of MC degrading bacteria
The site D consortium consisted of two colony types. DC8: large, more or less circular, pale yellow, glistening colonies with slightly uneven margins, Gram positive rod, non-motile.
DC7: Small, circular, bright white, mucoid colonies with smooth edges, Gram negative bacilli, actively motile.
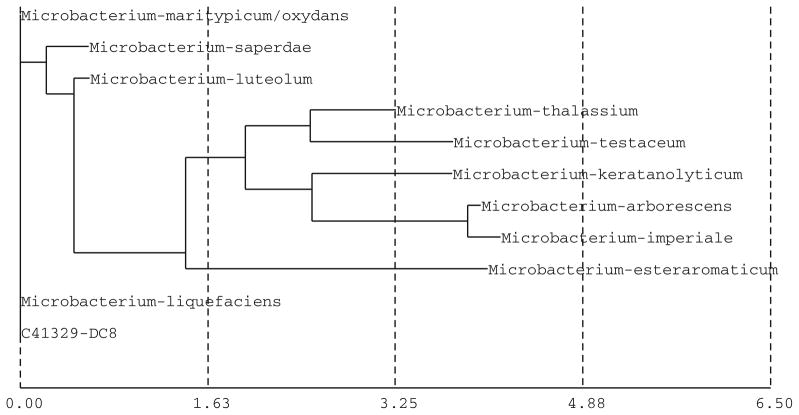
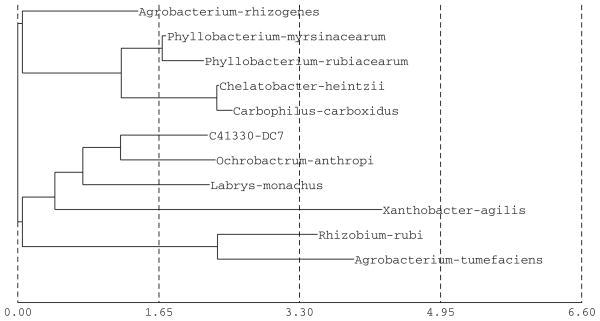
The partial sequence of the 16S rRNA gene for the bacterial isolates DC8 and DC7 was used for database search (Figs. 8, 9) and construction of phylogenetic tree using neighbor joining method (Figs. 6, 7). Based on the results, the isolate DC8 belongs phylogenetically to the genus Microbacterium, while DC7 has its closest match with Ochrobactrum anthropi. The bacterial isolate DC8 has in fact 0% difference with two sp. of Microbacterium (Microbacterium liquefaciens and Microbacterium maritypicum/oxydans). The bacterial strains presenting 16S rRNA gene similarities lower than 99.5% and higher than 97% may belong to different species; a full 16S rDNA sequence analysis and other tests are needed for definitive taxonomic identity (Tindall et al. 2010). Definitive identification of isolate DC8 may require additional biochemical tests or sequencing of additional gene loci. The second bacterial isolate DC7 has sequence matches with multiple genera, with O. anthropi as the closest match (2.13% difference). However, based on the online homology BLAST search in the openly available Genbank data, the DC7 isolate was found to be 100% matching with Rhizobium gallicum (AY972457). Definitive identification of isolate DC7 may require additional biochemical tests or sequencing of additional gene loci. There are problems in identifying groups of bacteria for which 16S rRNA gene sequences are not discriminative enough; in addition the use of unverified public databases such as GenBank can be problematic in that errors in sequence data are not uncommon (Woo et al. 2009).
Fig. 8.
Partial 16S rRNA gene sequences of bacterial isolate DC8
Fig. 9.
Partial 16S rRNA gene sequences of bacterial isolate DC7
Fig. 6.
Construction of phylogenetic tree using neighbor joining method for identification of the DC8 isolate
Fig. 7.
Construction of phylogenetic tree using neighbor joining method for identification of the DC7 isolate
Microbacterium species are usually Gram-positive, aerobic or facultatively anaerobic, non-motile, non-spore-forming rods. It grows between 15 and 37°C. The pH range for growth is 5–8. O. anthropi is a gram negative aerobic, peritrichously flagellated, nonfermentative, nonfastidious motile rods that are positive for oxidase, catalase and urease production. The organism is ubiquitous in nature but most frequently encountered in aquatic bodies. Laura et al. (1996) report the aerobic degradation of atrazine using this microbe, isolated from an activated sludge. Rhizobium species are Gram negative, aerobic, motile non-spore forming rods. Rhizobium sp. are common soil bacteria that form plant nodules and have the enzyme nitrogenase for fixing nitrogen. Biodegradation of sulfonated phenylazonaphthol dye (Ruiz-Arias et al. 2010) and phenol (Wei et al. 2008) was found to be associated with Rhizobium sp. While Microbacterium sp. has never been reported so far in biodegradation or bioremediation studies, it could still play an important role in this particular study. Further research is needed to understand the role of each of these bacteria in degrading the toxin, whether they act together or separately, which organism precedes the other, and determining the optimal concentration (bacterial count) needed for effective degradation. This could be an important next step in commercial applications to remove MC from drinking water supply.
The results from the present study showed that a bacterial consortium consisting of Microbacterium sp. and R. gallicum (AY972457) isolated from a high frequency algal bloom site at LO (Indian Prairie canal), was effective in degrading up to 74% of MC within 20 days at 26°C. The time frame observed for such an effective degradation seems to be similar to other MC biodegradation studies. Matthiensen et al. (2000) report 70% of MC-LR degradation within 10–15 days using samples from Patos Lagoon, Brazil (degradation rate 0.03–0.06 mg MC-LR ml−1 day−1). The time period for MC-LR removal showed that the degradation was highest during the initial 20 days and then decreased gradually. While many authors (Jones et al. 1994; Lam et al. 1995) report an initial lag phase during biodegradation, Christofferson et al. (2002) did not observe a lag phase. These differences in the presence and absence of lag phase as well as its duration could be the result of different bacterial community composition and its degree of adaptation (Valeria et al. 2006). But most authors agree that native microbial community can be utilized to perform biodegradation of these cyanotoxins quite rapidly. The degradation also seemed to have been influenced by the bacterial cell density, with higher the bacterial cell density yielding higher degradation rates. Even though samples from other sites showed some amount of degradation, isolates from site D showed the highest. Temperature had an effect on MC-LR degradation. Optimal degradation was achieved at 24 and 26°C and any further increase resulted in decreased degradation rate. Similar results have been reported by Park et al. (2001) during their biodegradations studies using Y2 strain isolated from a hypertrophic lake in Lake Suwa, Japan. They report that maximum biodegradation occurred between 20 to 25°C. It is interesting to note that higher phosphate levels in the media did not hamper the biodegradation process in our studies.
With Florida’s growing population, subtropical environment, climate change, and growing agricultural operations all over the state, it is quite natural to expect increasing bloom scenarios (and subsequent toxin release) in its lakes, rivers and estuaries. Adopting suitable water treatment technologies, preferably a combination of physical and biological methods, would effectively remove such toxins from drinking and recreational water supply and will help safe-guard Floridian’s health and protect the lives of aquatic species.
Acknowledgments
We thank the ARCH Core Facility at Florida International University, Miami for providing MC-LR and helping with the HPLC analysis.
Contributor Information
A. Ramani, Department of Earth and Environment, Florida International University, 11200 SW 8th Street, Miami, FL 33199-001, USA
K. Rein, Department of Chemistry and Biochemistry, Florida International University, Miami, FL 33199, USA
K. G. Shetty, Department of Earth and Environment, Florida International University, 11200 SW 8th Street, Miami, FL 33199-001, USA
K. Jayachandran, Email: jayachan@fiu.edu, Department of Earth and Environment, Florida International University, 11200 SW 8th Street, Miami, FL 33199-001, USA
References
- Gray PN, Farrell CJ, Kraus ML, Gromnicki AH, editors. Audubon of Florida. Lake Okeechobee: A synthesis of information and recommendations for its restoration. Audubon of Florida; Miami: 2005. p. 106. [Google Scholar]
- Burns J. Toxic cyanobacteria in Florida waters. In: Kenneth Hudnell H, editor. Adv Exp Med Biol; Proceedings of the interagency, international symposium on cyanobacterial harmful algal blooms, Chapter 5; 2007. pp. 127–137. [Google Scholar]
- Christofferson K, Lyck S, Winding A. Microbial activity and bacterial community structure during degradation of microcystins. Aquat Microb Ecol. 2002;27:125–136. [Google Scholar]
- Codd GA, Ward CJ, Bell SG. Cyanobacterial toxins: occurrence, modes of action, health effects and exposure routes. Arch Toxicol Suppl. 1997;19:399–410. doi: 10.1007/978-3-642-60682-3_38. [DOI] [PubMed] [Google Scholar]
- Cousins IT, Bealing DJ, James HA, Sutton A. Biodegradation of MC-LR by indigenous mixed bacterial populations. Water Res. 1996;30:481–485. [Google Scholar]
- Dawson RM. Toxicology of microcystins. Toxicon. 1998;36:953–962. doi: 10.1016/s0041-0101(97)00102-5. [DOI] [PubMed] [Google Scholar]
- Edwards C, Lawton LA. Bioremediation of cyanotoxins. In: Laskin AI, Gadd GM, Sariaslani S, editors. Advances in applied microbiology. Academic Press; New York: 2009. pp. 109–129. [DOI] [PubMed] [Google Scholar]
- Falconer IR. Tumor promotion and liver injury caused by oral consumption of cyanobacteria. Environ Toxicol Water Qual. 1991;6:177–184. [Google Scholar]
- Figueiredo DR, Azeiteiro UM, Esteves SM, Goncalves FJM, Pereira MJ. Microcystin producing blooms—a serious global public health issue. Ecotoxicol Environ Saf. 2004;59:151–163. doi: 10.1016/j.ecoenv.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Fischer WJ, Hitzfeld BC, Tencalla F, Eriksson JE, Mikhailov A, Dietrich DR. Microcystin-LR toxicodynamics, induced pathology, and immunohistochemical localization in livers of blue-green algae exposed rainbow trout (Oncorhynchus mykiss) Toxicol Sci. 2000;54:365–373. doi: 10.1093/toxsci/54.2.365. [DOI] [PubMed] [Google Scholar]
- Fleming LE, Stephan W. Report to the Florida Harmful Algal Bloom Taskforce. NIEHS Marine and Freshwater Biomedical Sciences Center, University of Miami; Miami: 2001. Blue green algae, their toxins and public health issues. [Google Scholar]
- Fleming LE, Rivero C, Burns J, Williams C, Bean JA, Shea KA, Shin J. Blue green algal (cyanobacterial) toxins, surface drinking water and liver cancer in Florida. Harmful Algae. 2002;1:157–168. [Google Scholar]
- Harada KI, Matsuura K, Suzuki M, Watanabe MF, Oishi S, Dahlem AM, Beasley VR, Carmichael WW. Isolation and characterization of the minor components associated with microcystins LR and RR in the cyanobacterium (blue-green algae) Toxicon. 1990;28:55–64. doi: 10.1016/0041-0101(90)90006-s. [DOI] [PubMed] [Google Scholar]
- Harada KI, Imanishi S, Kato H, Masayoshi M, Ito E, Tsuji K. Isolation of Adda from microcystin-LR by microbial degradation. Toxicon. 2004;44:107–109. doi: 10.1016/j.toxicon.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Ho L, Gaudieux AL, Fanok S, Newcombe G, Humpage AR. Bacterial degradation of microcystin toxins in drinking water eliminates their toxicity. Short communication. Toxicon. 2007;50:438–441. doi: 10.1016/j.toxicon.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Jones GJ, Orr PT. Release and degradation of microcystin following algicide treatment of a Microcystis aeruginosa bloom in a recreational lake, as determined by HPLC and protein phosphatase inhibition assay. Water Res. 1994;28:871–876. [Google Scholar]
- Jones GJ, Bourne DG, Blakeley RL, Doelle H. Degradation of the cyanobacterial hepatotoxin microcystin by aquatic bacteria. Nat Toxins. 1994;2:228–235. doi: 10.1002/nt.2620020412. [DOI] [PubMed] [Google Scholar]
- Kenefick SL, Hrudey SE, Peterson HG, Prepas EE. Toxin release from Microcystis aeruginosa after chemical treatment. Water Sci Technol. 1993;27:433–440. [Google Scholar]
- Lahiti K, Hiisverta L. Removal of cyanobacterial toxins in water treatment processes—review of studies conducted in Finland. Water Supply. 1989;7:149–154. [Google Scholar]
- Lam AKY, Fedorak PM, Prepas EE. Biotransformation of the cyanobacterial hepatotoxin microcystin LR, as determined by HPLC and protein phosphatase bioassay. Environ Sci Technol. 1995;29:242–246. doi: 10.1021/es00001a030. [DOI] [PubMed] [Google Scholar]
- Laura D, De Socio G, Frassanito R, Rotilio D. Effects of atrazine on Ochrobactrum anthropi membrane fatty acids. Appl Environ Microbiol. 1996;62:2644–2646. doi: 10.1128/aem.62.7.2644-2646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemes GAF, Kersanach R, Pinto LS, Dellagostin OA, Yunes JS, Mattheinsen A. Biodegradation of microcystins by aquatic Burkholderia sp. from a south Brazilian coastal lagoon. Ecotoxicol Environ Saf. 2007;69:358–365. doi: 10.1016/j.ecoenv.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Matthiensen A, Beattie KA, Yunes JS, Kaya K, Codd GA. [DLeu] microcystin-LR, from the cyanobacterium Microcystis RST 9501 and from a Microcystis bloom in the Patos Lagoon estuary, Brazil. Phytochemistry. 2000;55:383–387. doi: 10.1016/s0031-9422(00)00335-6. [DOI] [PubMed] [Google Scholar]
- Metcalf JS, Codd GA. Cyanobacterial toxins in the water environment. A review of current knowledge. Foundation for Water Research. University of Dundee; Marlow, Bucks: 2004. [Google Scholar]
- Newcombe G, Cook D, Brooke S, Ho L, Slyman N. Treatment options for microcystin toxins: similarities and differences between variants. Environ Toxicol. 2003;24:299–308. doi: 10.1080/09593330309385562. [DOI] [PubMed] [Google Scholar]
- Park HD, Sasaki Y, Maruyama T, Yanagisawa E, Hiraishi A, Kato K. Degradation of cyanobacterial hepatotoxin microcystin by a new bacterium isolated from a hypertrophic lake. Environ Toxicol. 2001;16:337–343. doi: 10.1002/tox.1041. [DOI] [PubMed] [Google Scholar]
- Pathmalal MM, Edwards C, Singh BK, Lawton LA. Isolation and identification of novel microcystin-degrading bacteria. Appl Environ Microbiol. 2009;75:6924–6928. doi: 10.1128/AEM.01928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugmacher S, Codd GA, Steinberg CEW. Effects of the cyanobacterial toxin microcystin-LR on detoxication enzymes in aquatic plants. Environ Toxicol. 1999;14:111–115. [Google Scholar]
- Phlips EJ, Bledsoe E, Cichra M, Badylak S. The distribution of potentially toxic cyanobacteria in Florida. In: Johnson D, Harbison RD, editors. Proceedings of health effects of exposure to cyanobacterial toxins: State of the Science. Florida Department of Health and the Center for Disease Control; St. Petersburg, Florida: 2003. [Google Scholar]
- Robinson NA, Pace J, Matson CF, Miura GA, Lawrence WB. Characterization of chemically titrated microcystin-LR and its distribution in mice. Toxicol In Vitro. 1991;5:341–345. [Google Scholar]
- Ruiz-Arias A, Juarez-Ramirez C, de los Cobos-Vasconcelos D, Ruiz-Ordaz N, Salmeron-Alcocer A, Ahuatzi-Chacon D, Galindez-Mayer J. Aerobic biodegradation of a sulfonated phenylazonaphthol dye by a bacterial community immobilized in a multistage packed-bed BAC reactor. Appl Biochem Biotechnol. 2010;162:1689–1707. doi: 10.1007/s12010-010-8950-z. [DOI] [PubMed] [Google Scholar]
- [Accessed 9 Oct 2008];SFWMD (South Florida WATER Management District) Lake Okeechobee: Algal Bloom Monitoring Program. http://www.sfwmd.gov/portal/page/portal/pg_grp_sfwmd_watershed/pg_sfwmd_watershed_inlake?_piref2294_4946961_2294_4946960_4946960.tabstring=tab20940638&_piref2294_4946961_2294_4946960_49469602294_20940817_2294_4946514_20940639.tabstring=tab20940808.
- Steidinger KA, Landsberg JH, Tomas CR, Burns JW. Harmful algal blooms in Florida. Florida Marine Research Institute; St. Petersburg, FL: 1999. Unpublished technical report submitted to the Florida Harmful Algal Bloom Task Force. [Google Scholar]
- Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kampfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- Tsuji K, Asakawa M, Anzai Y, Sumino T, Harada K. Degradation of microcystin using immobilized microorganisms isolated in a eutrophic lake. Chemosphere. 2006;65:1117–1124. doi: 10.1016/j.chemosphere.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Valeria AM, Ricardo EJ, Stephan P, Alberto WD. Degradation of microcystin-RR by Sphingomonas sp. CBA4 isolated from San Roque reservoir (Cordoba—Argentina) Biodegradation. 2006;17:447–455. doi: 10.1007/s10532-005-9015-9. [DOI] [PubMed] [Google Scholar]
- Wei G, Yua J, Zhua Y, Chena W, Wanga L. Characterization of phenol degradation by Rhizobium sp. CCNWTB 701 isolated from Astragalus chrysopteru in mining tailing region. J Hazard Mater. 2008;151:111–117. doi: 10.1016/j.jhazmat.2007.05.058. [DOI] [PubMed] [Google Scholar]
- Williams CD, Aubel MT, Chapman AD, Aiuto PE. Identification of cyanobacterial toxins in Florida’s Freshwater systems. Lakes Reserv Manag. 2007;23:144–152. [Google Scholar]
- Woo PCY, Teng JLL, Wu JKL, Leung FPS, Herman Tse H, Fung AMY, Lau SKP, Yuen K. Guidelines for interpretation of 16S rRNA gene sequence-based results for identification of medically important aerobic Gram-positive bacteria. J Med Microbiol. 2009;58:1030–1036. doi: 10.1099/jmm.0.008615-0. [DOI] [PubMed] [Google Scholar]
- World Health Organization, Geneva. Addendum to vol 2. Health criteria and other supporting information. 2. 1998. Guidelines for drinking-water quality; pp. 95–110. [Google Scholar]
- World Health Organization. Cyanobacterial toxins. In: Chorus I, Bartram J, editors. Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. 1999. [Google Scholar]