Abstract
The design and synthesis of an agarose resin functionalized with a Gram-negative quorum sensing (QS) signaling molecule analogue is described. The modified resin was utilized in affinity pull-down assays to successfully isolate QscR, a LuxR-type QS receptor from Pseudomonas aeruginosa. This resin may facilitate the identification of novel QS signal receptors using affinity chromatography techniques.
Keywords: Affinity chromatography, AHL, N-Acylated L-homoserine lactone, LuxR-type receptors, QscR, Quorum sensing
Bacteria can regulate specific phenotypes as a function of their cell density by sensing the concentration of small molecule chemical signals. This small molecule signaling process is known as quorum sensing (QS), and has attracted considerable interest from the microbiology, chemical biology, and drug discovery communities.1–5 Gram-negative bacteria use N-acylated L-homoserine lactones (AHLs, Fig. 1A) as their primary QS signals,6–8 which are produced by synthase proteins (LuxI-type proteins) and are perceived by cytoplasmic transcription factors (LuxR-type proteins).9 In general, AHLs are cell permeable and their concentration increases with increasing cell density. Once a threshold AHL concentration is achieved, productive AHL:LuxR-type receptor binding occurs. These complexes typically dimerize, associate with DNA, and initiate the transcription of the genes critical for density-dependent phenotypes. Among the diverse phenotypes regulated by this process are biofilm formation in pathogens like Pseudomonas aeruginosa,10,11 bioluminescence in the symbiont Vibrio fischeri,12,13 and root nodulation in mutualists like the Rhizobia spp.14 Due to the impact of these QS-related phenotypes in fields from healthcare to agriculture, there is considerable interest in developing new approaches for modulating and evaluating the chemical dialogue among bacteria.15,16 Chemical approaches to intercept bacterial QS, whether by blocking AHL:LuxR-type receptor binding or sequestering/degrading AHL signals, have become prominent techniques in the field.17–21
Figure 1.

(A) Generic structure of an N-acylated L-homoserine lactone (AHL); (B) N-(3-oxo-dodecanoyl)-L-homoserine lactone (OdDHL), a primary QS signal in P. aeruginosa.
Chemical tools for evaluating the biochemical interactions between AHLs and LuxR-type proteins, as well as for the identification of novel AHL receptors, would be useful for a broad range of experiments.16,22,23 LuxR-type proteins have proven challenging to characterize using standard biochemical and structural techniques (largely due to their instability in the absence of AHL ligands), and this has limited our understanding of how they perceive both native and nonnative AHL signals.9,24 In addition, increasing evidence suggests that AHLs from one bacterial species can induce phenotypic changes in neighboring species, including other Gram-negative bacteria25 and potentially even their eukaryotic hosts.26 This implies that bacteria may have more extensive networks of communication pathways, both at the inter-species and the inter-kingdom levels. Therefore, identifying new receptors for AHLs may lead to a deeper understanding of these phenomena.
Affinity chromatography could provide solutions for several of the challenges described above.27 In this methodology, small molecules of interest are chemically modified and covalently bound to an insoluble solid support, typically agarose or sepharose resin. These modified resins are then incubated with samples containing putative biological targets. Theoretically, small molecule moieties on the resin should only bind to their specific protein partners, permitting the latter to be retained throughout resin washing steps. Isolated proteins can then be eluted, analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), and identified using mass spectrometry (MS) or other techniques. This strategy has successfully identified several small-molecule target proteins, including histone deacetylase28 and the FK-506 binding protein (FKBP).29 Follow-up studies with these molecular targets have contributed to the overall understanding of their associated biological processes.
To date, the design of affinity matrices to assist in the identification of QS-related protein targets has been limited. Certain matrices and related chemical tools are relevant, however, and are introduced briefly here. Spring and co-workers have utilized an azithromycin-modified sepharose resin to identify its biological targets in P. aeruginosa.30 Sub-bacteriocidal doses of azithromycin have been shown to inhibit biofilm formation in P. aeruginosa, and because biofilm formation and QS are linked in this pathogen, QS pathways would appear to be a plausible target for azithromycin. Interestingly, analysis of the azithromycin–sepharose resin showed that many of the captured targets were ribosomal proteins, which suggests P. aeruginosa biofilm inhibition via azithromycin occurs through the ribosome as opposed to QS pathways. Concurrently, Spring and co-workers have developed an elegant 3D small-molecule microarray system31 that allows for the printing of AHL derivatives in a spatially addressable format.32 These arrays consist of reactive polymer gel matrices, and can be readily derivatized with AHL analogues containing nucleophilic groups at their acyl chain termini. The 3D arrays have proven useful for the screening of new AHL analogues for LuxR-type protein binding. In related work, the Meijler group has synthesized an AHL probe equipped with both a photoactive diazirine and ‘click’ chemistry reactive alkyne.22 In this proof-of-concept work, they demonstrated that their AHL probe binds to LasR, a prominent LuxR-type protein in P. aeruginosa, with only a slight decrease in activity relative to LasR’s native AHL, N-(3-oxo-dodecanoyl)-L-homoserine lactone (OdDHL; Fig. 1B). The authors also demonstrated that UV exposure leads to covalent attachment of the AHL probe to LasR through the diazirine moiety. These results suggest that the alkyne moiety could be used to further react the AHL-LasR complex with an affinity resin via click chemistry, and the authors highlight this possibility as a goal for future work. Lastly, motivated by reports of the immunomodulatory effects of OdDHL in humans,33 Seabra et al. prepared two OdDHL-derivatized resins and applied them for the identification of human receptors for AHLs.34 The OdDHL ligand was appended to the matrix through either the acyl chain termini or a mid-chain branch point using a piperazine linker. Two proteins were isolated using these matrices, but neither appeared to have immunological relevance.
Inspired by these recent studies, we sought to apply the affinity chromatography technique in our own QS research with the ultimate goal of developing a new tool to aid in the discovery of novel QS targets. To the best of our knowledge, the application of AHL-derivatized affinity resins for the identification of LuxR-type receptors has not been reported to date. Herein, we describe our initial studies toward the design, synthesis, and application of an AHL-derivatized agarose matrix. Affinity pull-down assays using this new AHL-functionalized matrix confirmed the binding of QscR, an ‘orphan’ LuxR-type protein from P. aeruginosa,35 to the matrix. These results serve to demonstrate the feasibility of utilizing AHL-derivatized affinity matrices for LuxR-type receptor isolation and characterization.
To guide our design of an appropriate ligand for matrix derivatization, we first scrutinized the AHL:receptor binding interactions in the three reported structures of AHL:LuxR-type proteins (TraR, LasR, and SdiA).36–40 Each of these structures shows a series of conserved hydrogen-bonding interactions between the AHL lactone ring and residues in the binding pocket of the receptor. To prevent disruption of these key interactions, we decided that attachment of an AHL to the resin matrix should occur through the acyl tail as opposed to the lactone head group. Such an attachment strategy was successfully utilized by Spring32 and Seabra34 (see above). For our AHL scaffold, we chose to synthesize an analogue of OdDHL (Fig. 1B). We note that this molecule is the native ligand for both LasR and QscR in P. aeruginosa. QscR is a unique QS receptor, as it appears to bind OdDHL, the native ligand for LasR, and repress the activity of LasR.35 Notably, QscR can be readily produced in full-length form (in P. aeruginosa), 41 and we have considerable experience in its manipulation. 42 We reasoned these capabilities would enable proof-of-concept resin-QscR binding studies. We selected Affigel-10 agarose resin (Bio-Rad; derivatized with N-succinimidyl (N-OSu) esters) as our matrix due to its wide and successful usage in other affinity chromatography applications.27 To facilitate coupling to the activated esters of Affigel-10, we designed an OdDHL analogue (6) with a primary amino group at the end of the acyl tail (Scheme 1).
Scheme 1.
Synthesis of OdDHL-derivatized resin 7. Abbreviations: Boc = tert-butoxycarbanoyl, DCC = N,N′-dicyclohexylcarbodiimide, DMAP = 4-(dimethylamino)pyridine; TFA = trifluoroacetic acid; DIEA = diisopropylethylamine.
The synthesis of AHL 6 commenced with nucleophilic substitution of NH3 onto 10-bromodecanoic acid 1 at ambient temperature, followed by N-Boc protection to generate acid 2.23 Thereafter, 2 was allowed to react with Meldrum’s acid 3 under standard conditions and coupled to homoserine lactone (4) to afford N-Boc AHL 5 in 78% yield over two steps. Cleavage of the N-Boc group with TFA yielded amino-AHL 6 as the TFA salt in 56% yield after preparative HPLC purification.
Next, AHL 6 was mixed with Affigel-10 resin (N-OSu loading = 7.5 μmol/mL of slurry) and DIEA at ambient temperature for 2 h to affect coupling. Using AHL 6 as the limiting reagent (0.5 equiv relative to N-OSu available on resin) gave quantitative resin coupling (i.e., 50% coverage), as indicated by LC–MS analysis (see Supplementary data). Finally, ethanolamine was used to block all remaining activated esters on the resin (to ensure that they would not interfere with the subsequent experiments), and afforded OdDHL-derivatized resin 7. To provide a control resin for protein binding studies, we also reacted Affigel-10 with ethanolamine alone to generated a ‘capped’, AHL-free resin.
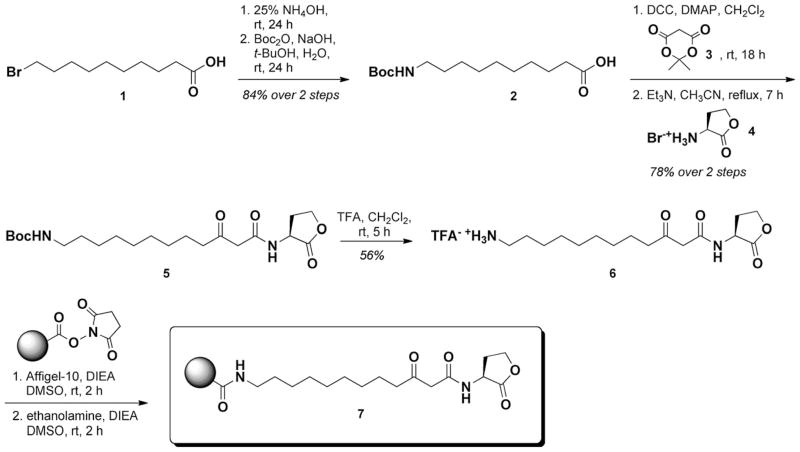
We next tested the ability of resin 7 to bind to QscR. QscR was produced in P. aeruginosa PAO-T7(pJLQhis), a QscR over-expression strain, and purified by nickel affinity chromatography using reported procedures.41 As this P. aeruginosa strain retains the synthase for OdDHL (LasI), QscR is likely isolated as a mixture of OdDHL-bound and OdDHL-free protein (and an equilibrium thereof). 43 We reasoned that this mixture would still be viable for binding experiments with resin 7. Samples of resin 7 and the control resin were incubated with purified QscR (3.75 μmol resin-bound AHL to 15 nmol QscR, or 250:1 AHL/protein) in buffer for 24 h at 4 °C,44 after which the resins were filtered and subjected to washing in order to remove any unbound/excess protein. These washings were collected, and the resin samples and washings were each diluted with gel loading buffer and heat-denatured before being loaded onto a 12% polyacrylamide gel for SDS–PAGE analysis (see Supplementary data for full details).
Figure 2 shows the SDS–PAGE data for the resin 7/purified QscR binding experiment. Resin 7 was found to cleanly bind QscR (lane 3, denoted ‘R’). Significant QscR was found in the collected supernatant (lane 4, denoted ‘S’), with reduced quantities being observed in the washing samples (lanes 5 and 6). The large quantity of residual QscR in the supernatant could be due to a high percentage of native OdDHL-bound QscR in our protein sample (see above)43 and/or the reduced ability of QscR to bind the resin-bound OdDHL analogue. Nevertheless, resin 7 bound sufficient QscR for it to be clearly observable on the gel. Further, the control resin showed no detectable binding (lane 7, denoted ‘R’), indicating that the appended OdDHL analogue is essential for binding of QscR to the resin. Collectively, these data show that resin 7 is capable of binding to a purified LuxR-type protein.
Figure 2.
SDS–PAGE analysis of purified QscR with resin 7. L: protein molecular weight ladders; P: purified QscR; R: protein denatured from resin 7 after incubation; S: flow-through solution from resin 7-QscR incubation; W1: 1st washing solution; W2: 2nd washing solution. Data for both resin 7 (lanes 3–6) and ethanolamine-capped control resin (lanes 7–10) are provided. The solid arrow indicates the presence of QscR; the dashed arrow was a GroEL contaminant.45
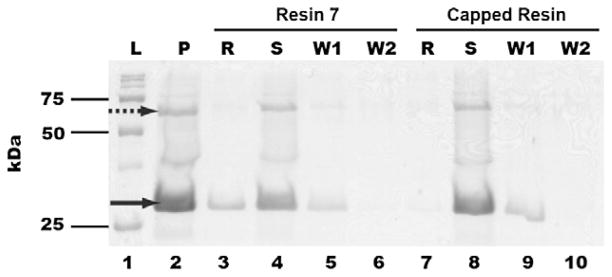
Encouraged by these results, we then evaluated the ability of resin 7 to bind and effectively remove QscR from a more complex sample, that is, cell lysates. Such a capability would be useful for the application of resin 7 to biologically relevant samples and the potential identification of new LuxR-type proteins. Co-incubation of resin 7 with crude PAO-T7(pJLQhis) lysates and analogous SDS–PAGE analysis as described above again yielded one prominent band (Fig. 3; lane 3, denoted ‘R’). This band was extracted from the gel and subjected to trypsin digest followed by ESI-MS/MS analysis, which confirmed the presence of QscR (see Supplementary data). Similar to the experiment with purified QscR, no significant binding of QscR to the ‘capped’ control resin was observed (lane 7, denoted ‘R’). We note that PAO-T7(pJLQhis) is an overexpression strain and therefore the quantity of QscR in the cell lysate is considerably higher than that in a wild-type strain.46 Even so, these initial results demonstrate the exciting potential for resin 7 to be utilized in the isolation of LuxR-type proteins from more complex biological mixtures.
Figure 3.
SDS–PAGE analysis of crude mixtures of QscR and resin 7. L: Protein molecular weight ladders; C: crude lysate; R: protein denatured from resin 7 after incubation; S: flow-through solution from resin 7-crude lysate incubation; W1: 1st washing solutions; W2: 2nd washing solutions. Data for both resin 7 (lanes 3–6) and ethanolamine-capped control resin (lanes 7–10) are provided. The solid arrow indicates the presence of QscR.
In conclusion, we have designed and synthesized a new AHL-modified affinity matrix, and demonstrated that this matrix can bind to the QscR protein from P. aeruginosa. This OdDHL-derivatized resin (7) was shown to bind QscR from both purified protein samples and crude cell lysates using SDS–PAGE. These proof-of-concept experiments illustrate how resin 7 could be used as a tool to isolate and potentially identify new AHL receptor proteins in bacteria. As mentioned above, a number of eukaryotes display sensitivity to AHLs.26 Therefore, resin 7 and related derivatives could also prove useful for the elucidation of such cross-kingdom sensing pathways. Our progress towards these goals, along with further optimization of resin 7, will be reported in due course.
Supplementary Material
Acknowledgments
We gratefully acknowledge the NIH (AI063326), Greater Milwaukee Foundation, Burroughs Welcome Fund, and Johnson & Johnson for financial support. T.P. thanks the Anandamahidol Foundation (Thailand) for a pre-doctoral scholarship. A.S.B. was supported in part by NIH Chemistry Biology Interface Training Grant (NIGMS T32 GM008505). Professor Peter Greenberg (University of Washington) is thanked for kindly providing the P. aeruginosa strain PAO-T7(pJLQhis). Dr. Matthew Lockett is acknowledged for his assistance in protein mass analyses.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2011.04.062.
References and notes
- 1.Camilli A, Bassler BL. Science. 2006;311:1113. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Platt TG, Fuqua C. Trends Microbiol. 2010;18:383. doi: 10.1016/j.tim.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galloway WRJD, Hodgkinson JT, Welch M, Spring DR. Chem Biol. 2009;16:913. doi: 10.1016/j.chembiol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Bjarnsholt T, van Gennip M, Jakobsen TH, Christensen LD, Jensen PO, Givskov M. Nat Protoc. 2010;5:282. doi: 10.1038/nprot.2009.205. [DOI] [PubMed] [Google Scholar]
- 6.Lazdunski AM, Ventre I, Sturgis JN. Nat Rev Microbiol. 2004;2:581. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua C, Greenberg EP. Nat Rev Mol Cell Biol. 2002;3:685. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 8.Welch M, Mikkelsen H, Swatton JE, Smith D, Thomas GL, Glansdorp FG, Spring DR. Mol BioSyst. 2005;1:196. doi: 10.1039/b505796p. [DOI] [PubMed] [Google Scholar]
- 9.Schuster M, Greenberg EP. In: Chemical Communication among Bacteria. Winans SC, Bassler BL, editors. ASM Press; Washington, DC: 2008. p. 133. [Google Scholar]
- 10.De Kievit TR, Iglewski BH. Methods Enzymol. 1999;310:117. doi: 10.1016/s0076-6879(99)10010-7. [DOI] [PubMed] [Google Scholar]
- 11.De Kievit TR, Iglewski BH. Infect Immun. 2000;68:4839. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nealson KH, Platt T, Hastings JW. J Bacteriol. 1970;104:313. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nealson KH, Hastings JW. Microbiol Rev. 1979;43:496. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downie JA. FEMS Microbiol Rev. 2010;34:150. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- 15.Lowery CA, Salzameda NT, Sawada D, Kaufmann GF, Janda KD. J Med Chem. 2010;53:7467. doi: 10.1021/jm901742e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattmann ME, Blackwell HE. J Org Chem. 2010;75:6737. doi: 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geske GD, O’Neill JC, Blackwell HE. Chem Soc Rev. 2008;37:1432. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Chem Rev. 2011;111:28. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 19.Amara N, Krom BP, Kaufmann GF, Meijler MM. Chem Rev. 2011;111:195. doi: 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]
- 20.Park J, Jagasia R, Kaufmann GF, Mathison JC, Ruiz DI, Moss JA, Meijler MM, Ulevitch RJ, Janda KD. Chem Biol. 2007;14:1119. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufmann GF, Sartorio R, Lee SH, Mee JM, Altobell LJ, Kujawa DP, Jeffries E, Clapham B, Meijler MM, Janda KD. J Am Chem Soc. 2006;128:2802. doi: 10.1021/ja0578698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubinsky L, Jarosz LM, Amara N, Krief P, Kravchenko VV, Krom BP, Meijler MM. Chem Commun. 2009:7378. doi: 10.1039/b917507e. [DOI] [PubMed] [Google Scholar]
- 23.Amara N, Mashiach R, Amar D, Krief P, Spieser SAH, Bottomley MJ, Aharoni A, Meijler MM. J Am Chem Soc. 2009;131:10610. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 24.Churchill MEA, Chen L. Chem Rev. 2011;111:68. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shank EA, Kolter R. Curr Opin Microbiol. 2009;12:205. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teplitski M, Mathesius U, Rumbaugh KP. Chem Rev. 2011;111:100. doi: 10.1021/cr100045m. [DOI] [PubMed] [Google Scholar]
- 27.Leslie BJ, Hergenrother PJ. Chem Soc Rev. 2008;37:1347. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 28.Taunton J, Hassig CA, Schreiber SL. Science. 1996;272:408. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 29.Harding MW, Galat A, Uehling DE, Schreiber SL. Nature. 1989;341:758. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 30.Glansdorp FG, Spandl RJ, Swatton JE, Loiseleur O, Welch M, Spring DR. Org Biomol Chem. 2008;6:4120. doi: 10.1039/b813157k. [DOI] [PubMed] [Google Scholar]
- 31.Marsden DM, Nicholson RL, Ladlow M, Spring DR. Chem Commun. 2009:7107. doi: 10.1039/b913665g. [DOI] [PubMed] [Google Scholar]
- 32.Marsden DM, Nicholson RL, Skindersoe ME, Galloway WRJD, Sore HF, Givskov M, Salmond GPC, Ladlow M, Welchd M, Spring DR. Org Biomol Chem. 2010;8:5313. doi: 10.1039/c0ob00300j. [DOI] [PubMed] [Google Scholar]
- 33.Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, Stewart GS, Bycroft BW, Pritchard DI. Infect Immun. 1998;66:36. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seabra R, Brown A, Hooi DSW, Kerkhoff C, Chhabra SR, Harty C, Williams P, Pritchard DI. Calcium Binding Proteins. 2008;3:31. [Google Scholar]
- 35.Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. Proc Natl Acad Sci USA. 2001;98:2752. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Nature. 2002;417:971. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 37.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. EMBO J. 2002;21:4393. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottomley MJ, Muraglia E, Bazzo R, Carfi A. J Biol Chem. 2007;282:13592. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 39.Zou Y, Nair SK. Chem Biol. 2009;16:961. doi: 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. J Mol Biol. 2006;355:262. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Lequette Y, Greenberg EP. Mol Microbiol. 2006;59:602. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 42.Mattmann ME, Geske GD, Worzalla GA, Chandler JR, Sappington KJ, Greenberg EP, Blackwell HE. Bioorg Med Chem Lett. 2008;18:3072. doi: 10.1016/j.bmcl.2007.11.095. [DOI] [PubMed] [Google Scholar]
- 43.Oinuma KI, Greenberg EP. J Bacteriol. 2011:193, 421. doi: 10.1128/JB.01041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Later experiments revealed that 2 h was sufficient to reach maximum resin/ protein binding.
- 45.The upper band at 60 kDa was determined to be GroEL by MS/MS, a common chaperone protein found in bacteria. The presence of this contaminant should not affect the conclusions of the experiments described herein.
- 46.LasR, which also recognizes OdDHL, was not isolated in these experiments with crude P. aeruginosa lysates. The lower abundance of LasR relative to QscR in this strain, along with the lower promiscuity of LasR relative to QscR for AHL-type ligands, are likely reasons for this. We also note that we had difficulty isolating QscR from wild-type P. aeruginosa (PAO1) using resin 7 (data not shown), which we believe is due to the low abundance of QscR in the wild-type strain.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.