Abstract
TIR (Toll/interleukin-1 receptor) domain-containing proteins play a crucial role in innate immunity in eukaryotes. Brucella is a highly infectious intracellular bacterium that encodes a TIR domain protein (TcpB) to subvert host innate immune responses to establish a beneficial niche for pathogenesis. TcpB inhibits NF-κB (nuclear factor κB) activation and pro-inflammatory cytokine secretions mediated by TLR (Toll-like receptor) 2 and TLR4. In the present study, we have demonstrated that TcpB modulates microtubule dynamics by acting as a stabilization factor. TcpB increased the rate of nucleation as well as the polymerization phases of microtubule formation in a similar manner to paclitaxel. TcpB could efficiently inhibit nocodazole- or cold-induced microtubule disassembly. Microtubule stabilization by TcpB is attributed to the BB-loop region of the TIR domain, and a point mutation affected the microtubule stabilization as well as the TLR-suppression properties of TcpB.
Keywords: BB-loop, Brucella, microtubule, Toll-like receptor (TLR), Toll/interleukin-1 receptor domain (TIR domain), Toll/interleukin-1 receptor domain-containing adaptor protein (TIRAP)
INTRODUCTION
TLRs (Toll-like receptors) are essential components of innate immunity which recognize conserved microbial components and elicit anti-microbial responses [1–3]. TLRs are characterized by extracellular leucine-rich repeats which recognize pathogen-derived molecules and an intracellular signalling domain known as the TIR (Toll/interleukin-1 receptor) domain [4]. Activated TLRs recruit TIRAPs (TIR domain-containing adaptor proteins) to their signalling domains and this molecular assembly initiates a signalling cascade [5–8].
Micro-organisms have developed various strategies to interfere with TLR signalling pathways to accomplish a less hostile environment for their survival and persistence [9–14]. Three pathogenic bacteria, namely Brucella spp., Salmonella spp. and Escherichia coli encode TIR domain-containing proteins to facilitate subversion of TLR2- and TLR4-mediated signalling pathways [9–12]. The Brucella encoded TIR domain-containing protein TcpB mimics the phosphoinositide-binding property of TIRAP [or MAL (MyD88 adaptor-like protein)] and targets the TIRAP-mediated pathway for TLR suppression [12]. TcpB exhibits cell permeability and is efficiently internalized by macrophages [15]. Recent studies have indicated that TcpB enhances ubiquitination and promotes targeted degradation of TIRAP [13].
Subcellular localization studies have indicated that TcpB co-localizes with the plasma membrane and microtubules [12]. In the present paper, we report that TcpB affects the dynamics of microtubule formation. TcpB enhances the rate of nucleation and polymerization and stabilizes the polymerized tubules. TcpB modulates the microtubule dynamics through the TIR domain which is essential for the suppression of TLRs. Changes in the dynamics and organization of microtubules contribute to the regulation of various signal transduction processes [16]. Therefore our data suggest a direct correlation between the TcpB-induced microtubule stabilization and negative regulation of TLR signalling.
MATERIALS AND METHODS
Overexpression and purification of TcpB
Expression and purification of TcpB was performed as described previously [15]. Briefly, 1 litre of E. coli BL21 cell culture harbouring the pMALTcpB or pMALTcpBG158A plasmid was induced with IPTG (isopropyl β-D-thiogalactopyranoside) to a final concentration of 0.5 mM when the D600 reached 0.6. The induced culture was then grown at 25°C for 5 h. Cells were collected by centrifugation at 5000 g for 10 min and resuspended in sonication buffer containing 50 mM Tris/HCl (pH 8.0), 1 M NaCl, 1 mM EDTA and protease inhibitor cocktail (Pierce). Cells were sonicated and then centrifuged at 16000 g for 20 min to clarify the supernatant. The supernatant was passed through a column harbouring 5 ml of amylose resin (NEB). The column was then washed with the sonication buffer followed by the same buffer containing decreasing concentrations of NaCl (750, 500, 250 or 100 mM). The bound MBP (maltose-binding protein)–TcpB was eluted with an elution buffer containing 50 mM Tris/HCl (pH 8.0) and 30 mM maltose. The eluted protein was then subjected to SP Sepharose (Sigma) ion-exchange chromatography to remove maltose followed by concentration using a Centricon protein concentrator (Millipore). The concentrated protein was dialysed in a buffer containing 50 mM Tris/HCl, 100 mM NaCl and 10% glycerol (pH 8.0) and stored at −80°C in aliquots of 50 μl.
In vitro microtubule binding assay
In vitro microtubule polymerization was performed in the presence of MBP–TcpB or MBP alone. TcpB or MBP was diluted 1:1 with G-PEM buffer [Cytoskeleton; 80 mM sodium Pipes (pH 6.9), 1 mM MgCl2 and 1 mM EGTA] at a final concentration of 5 μg/μl. To visualize microtubule binding, rhodamine-labelled porcine tubulin (Cytoskeleton) was used. Tubulin was resuspended in 6 μl of G-PEM buffer containing 10% glycerol and then mixed with 2 μl of MBP–TcpB (5 μg/μl) or MBP (5 μg/μl). Tubes were incubated at 37°C for 20 min followed by dilution of the sample with 200 μl of warm G-PEM buffer containing 30% glycerol and 200 μM paclitaxel. Polymerization products were then transferred to the wells of glass-bottomed Petri dishes coated with poly-L-lysine and incubated at 37°C for 30 min to facilitate microtubule attachment. Attached microtubules were then fixed with G-PEM/glycerol buffer containing 0.5% glutaraldehyde for 10 min followed by quenching with NaHB4 (1%) for 7 min. Wells were blocked with PBS containing 1% Triton X-100 (PBST) and 2% BSA for 10 min. Microtubules were then incubated with anti-MBP antibody (NEB; 1:5000) in blocking buffer for 20 min. Wells were washed three times with PBST followed by incubation with Alexa Fluor® 488-conjugated goat anti-mouse IgG (Invitrogen; 1:5000) for 20 min. Wells were washed three times with PBST. Excess water was removed and the labelled microtubules were preserved in Prolong Gold antifade reagent (Invitrogen). Tubules were analysed with a fluorescent microscope at ×20 magnification and images were captured using a Nikon camera.
For the microtubule-co-sedimentation assay, porcine tubulin >99% pure was used at a concentration of 5 mg/ml. Tubulin polymerization was performed with MBP–TcpB (0.5 mg/ml) or MBP (0.5 mg/ml) at room temperature (24°C) for 30 min. The polymerized microtubules were then added on the top of 100 μl of cushion buffer [80 mM sodium Pipes (pH 6.9), 1 mM MgCl2, 1 mM EGTA and 60% glycerol] supplemented with taxol in ultracentrifuge tubes. The samples were centrifuged at 50000 rev./min for 1 h in an ultracentrifuge (Beckman TL-100 rotor). After centrifugation, the supernatant was aspirated and the pellet was resuspended in G-PEM buffer. Both the supernatant and pellet fractions were analysed by SDS/PAGE.
Microtubule-polymerization assay
Porcine tubulin was resuspended in 1 ml of ice-cold G-PEM buffer containing 10% glycerol and 1 mM GTP at a final concentration of 2 mg/ml. The assay was performed in 96-well half-area plates. Controls and test samples (10 μl) were first added to the pre-warmed 96-well plates and the plates were kept at 37°C until the addition of tubulin. Ice-cold tubulin (100 μl) was then added with a multi-channel pipette and mixed two or three times by gentle pipetting. The plate was read immediately using a plate reader set at 37°C in kinetic mode (Beckman Coulter DTX 800 Multimode Detector) and the D340 was recorded for 60 min at 1 min intervals. The data were exported to Excel and analysed using Sigma Plot.
Microtubule-depolymerization assay
For nocodazole-induced depolymerization, microtubule polymerization was performed in the presence of 5 μM nocodazole as indicated above. For cold-induced depolymerization, the polymerization reaction was performed in the presence of MBP–TcpB or controls for 30 min at 37°C. The D340 was recorded, and the plate was then transferred to an ice-water bath. The D340 was recorded at 5 min intervals for 20 min.
Luciferase reporter assay
To analyse the ability of TcpBG158A to block TLR activation, HEK (human embryonic kidney)-TLR2 cells were co-transfected with various amounts of pHA-TcpB or pHA- TcpBG158A (50, 100 or 250 ng), pNF-κB-luc (50 ng) (Stratagene) and pRL-TK (10 ng) (Promega). The total amount of DNA was made constant by adding empty vector (pCMV-HA). At 24 h after transfection, cells were induced with TLR2 ligand, FSL-1 (Invivogen), and luciferase activity was assayed after 12 h using a dual-luciferase reporter assay system (Promega).
RESULTS
TcpB associates with microtubules
TcpB-induced microtubule thickening and bundling in eukaryotic cells and TcpB-decorated microtubules appeared to be resistant to nocodazole-induced depolymerization [12]. To determine the TcpB–microtubule interaction further, we performed in vitro microtubule-binding studies employing immunofluorescence as well as co-sedimentation assays. In visual microtubule-binding assays, rhodamine-labelled tubulin polymerization was performed in the presence of buffer alone or purified MBP–TcpB or MBP alone. The polymerized tubules were stained with anti-MBP antibody followed by fluorescently labelled secondary antibody. Analysis of co-polymerized microtubules by fluorescence microscopy revealed the association of MBP–TcpB along the microtubule lattice (Figure 1a, panels G–I). MBP alone did not show any affinity for microtubules, confirming that microtubule binding is contributed by the TcpB portion of the fusion protein (Figure 1a, panels D–F). The affinity of TcpB towards microtubules was demonstrated further by co-sedimentation assay. After the polymerization with MBP–TcpB or MBP alone, the microtubules were pelleted by centrifugation. Proteins present in the pellet (P) and the supernatant (S) fractions were analysed by SDS/PAGE and Western blotting. The majority of the MBP–TcpB was present in the microtubule-containing pellet fraction (Figures 1b and 1c). The result demonstrates the robust interaction between the microtubules and TcpB.
Figure 1. TcpB is associated with microtubules.
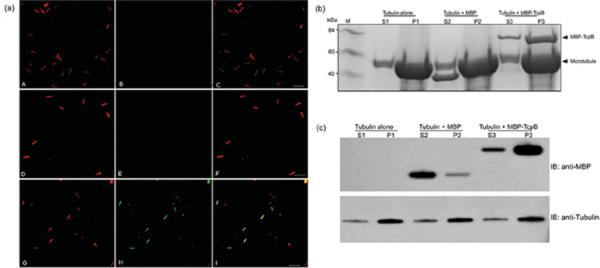
(a) TcpB binds microtubules in vitro. Rhodamine-labelled microtubules were polymerized (panel A) in the presence of MBP (panel D) or MBP–TcpB (panel G). Tubules were then fixed and stained with anti-MBP antibody followed by Alexa Fluor® 488-conjugated secondary antibody. Tubulin alone (panel B), MBP (panel E) and MBP–TcpB (panel H). Panels C, F and I represent merged images of tubulin alone, MBP and MBP–TcpB respectively. Scale bar, 20 μm. (b) Microtubule-co-sedimentation assay. Microtubule polymerization was performed with MBP or MBP–TcpB and polymerized tubules were pelleted by ultracentrifugation, followed by analysis of supernatant and pellets by SDS/PAGE. S1, S2 and S3 are S fractions of tubulin alone, MBP and MBP–TcpB respectively. P1, P2 and P3 are P fractions of tubulin alone, MBP and MBP–TcpB respectively. M, molecular mass markers (in kDa). (c) Western blot analysis of the supernatant and pellet fractions. IB, immunoblot.
TcpB stabilizes microtubules
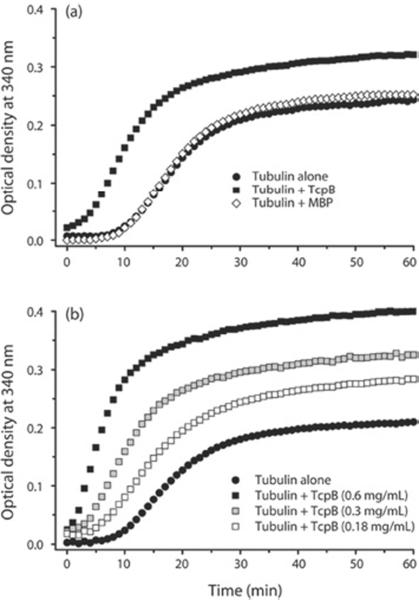
Our in vivo and in vitro analysis indicated that TcpB associated with microtubules. Next we investigated whether TcpB affected the polymerization dynamics of microtubule formation. We employed a light-scattering assay to determine the rate of microtubule polymerization in the presence of TcpB [17,18]. The microtubule-polymerization curve represents three phases of polymerization such as nucleation, elongation and steady-state equilibrium. Interestingly, MBP–TcpB enhanced the nucleation and growth phases of microtubule polymerization and the total amount of polymerized microtubules in a dose-dependent manner (Figure 2). Addition of MBP alone did not affect the polymerization dynamics of microtubule formation (Figure 2a). Paclitaxel is a microtubule-stabilizing agent that reduces the critical concentration of tubulin required for polymerization and enhances the rate of polymerization and stability of polymerized microtubules [19,20]. Our experimental data revealed that TcpB exhibited similar properties of paclitaxel (Figure 3).
Figure 2. TcpB modulates microtubule dynamics by acting as a stabilization factor.
(a) Microtubule-polymerization assay in the presence of MBP or MBP–TcpB. The assay was performed with porcine tubulin at a final concentration of 2 mg/ml in the presence of 0.5 mg/ml MBP or MBP–TcpB. (b) TcpB affects the microtubule dynamics in a dose-dependent manner. The assay was performed with increasing concentrations (0.18, 0.3 or 0.6 mg/ml) of MBP–TcpB. MBP–TcpB could efficiently enhance the nucleation and growth phases of MT polymerization and the total amount of microtubules polymerized. The light-scattering experiments were repeated at least three times in duplicate. Optical density = attenuance.
Figure 3. TcpB mimics the properties of paclitaxel.
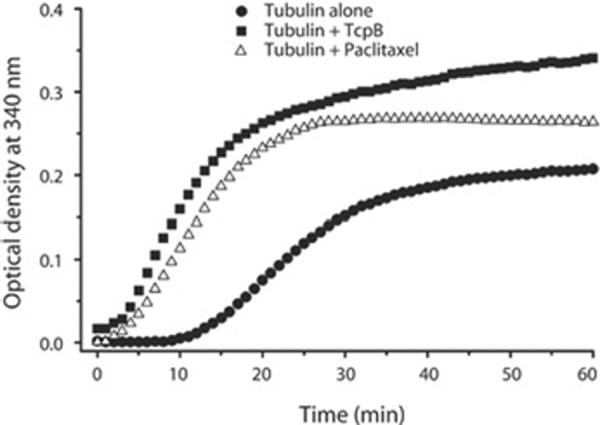
A microtubule-polymerization assay was performed with MBP–TcpB (0.5 mg/ml) or paclitaxel (5 μM). TcpB increased the rate of polymerization and accumulated polymerized tubules similarly to paclitaxel. Optical density = attenuance.
TcpB protects microtubules from depolymerization
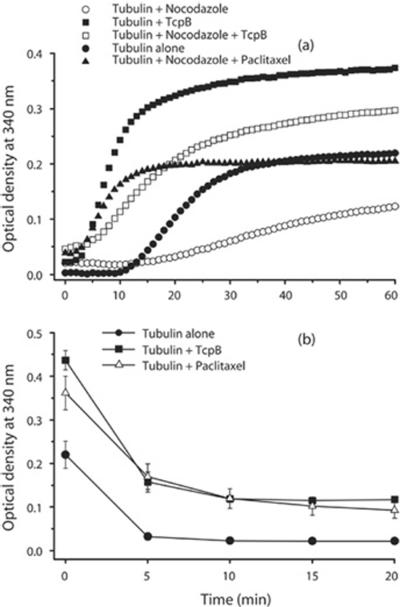
Next, we analysed whether TcpB-stabilized microtubules were resistant to depolymerization induced by nocodazole or cold. Microtubule polymerization in the presence of nocodazole affected the tubulin assembly as demonstrated by the low rate of polymerization and reduced amounts of assembled tubules (Figure 4). Remarkably, TcpB could efficiently suppress the effect of nocodazole as indicated by the enhanced reaction rate and accumulation of stabilized microtubules (Figure 4). To determine the effect of TcpB on cold-induced microtubule disassembly, we performed the polymerization in the presence of TcpB and the assembled tubules were subjected to cold. The experimental data revealed that, similar to paclitaxel, TcpB is capable of resisting cold-induced microtubule disassembly, demonstrating the efficient microtubule-stabilization property of TcpB (Figure 4b).
Figure 4. TcpB resists microtubule depolymerization.
Microtubule depolymerization was induced with 5 μM nocodazole (a) or by cold (b). TcpB could efficiently resist depolymerization of microtubules induced by nocodazole or cold. Optical density = attenuance.
Intact TIR domain is essential for TcpB-mediated microtubule stabilization
We had demonstrated previously that the intact TIR domain is essential for NF-κB (nuclear factor κB) suppression and microtubule association of TcpB [12]. A G158A mutation in the BB-loop region (Box II) (TcpBG158A) affected TLR suppression of TcpB (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/439/bj4390079add.htm). To determine whether this mutant is capable of modulating microtubule dynamics, we performed light-scattering assays with purified recombinant TcpBG158A. Interestingly, the microtubule-stabilization property of TcpBG158A was severely diminished as demonstrated by a low rate of reaction and less assembled tubulin compared with the wild-type TcpB (Figure 5a). Similarly, TcpBG158A displayed a minimum effect on nocodazole-induced microtubule disassembly (Figure 5b). Deficiency of TcpBG158A to resist nocodazole-induced microtubule depolymerization was demonstrated further by a cosedimentation assay (Figure 5c). The majority of the tubulin was present in the supernatant fraction when the reaction was performed in the presence of nocodazole and MBP–TcpBG158A mutant protein (Figure 5c). Next, we synthesized a peptide derived from amino acids 127–173 (47 amino acids) of the TIR domain of TcpB harbouring the BB-loop region. Polymerization assays revealed that the TIR peptide was capable of stabilizing microtubules in a dose-dependent manner (Figure 6a). Consistently, the BB-loop peptide could efficiently resist nocodazole- (Figure 6b) or cold- (Figure 6c) induced microtubule depolymerization, indicating the microtubule-stabilization property of the TIR peptide.
Figure 5. Intact TIR domain is essential for TcpB to stabilize microtubules.
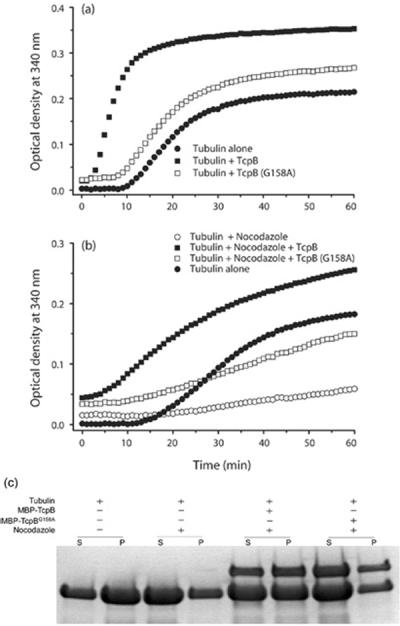
(a) Microtubule-polymerization assay performed with TcpBG158A. The mutant TcpB displayed diminished microtubule stabilization. (b) Microtubule-depolymerization assay in the presence of TcpBG158A. TcpBG158A was not as efficient in resisting nocodazole-induced depolymerization compared with wild-type TcpB. (c) Microtubule-co-sedimentation assay performed with MBP–TcpB or MBP–TcpBG158A mutant in the presence of nocodazole (5 μM). Optical density = attenuance.
Figure 6. TIR peptide is capable of stabilizing microtubules.
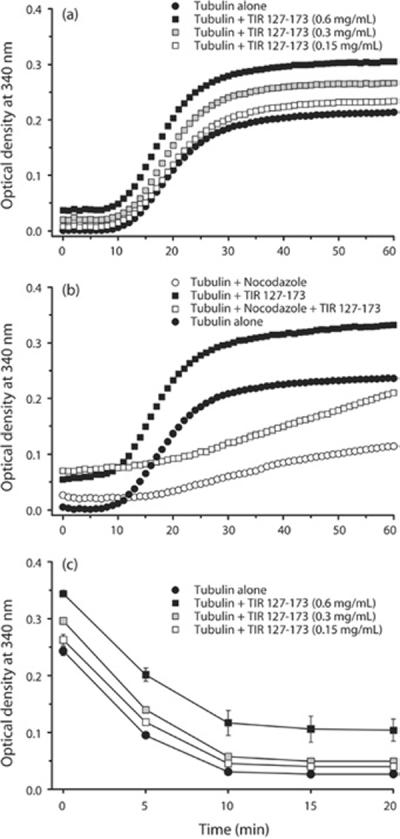
(a) The polymerization assay was performed with increasing concentrations (0.15, 0.3 or 0.6 mg/ml) of a 47-amino-acid peptide derived from amino acids 127–173 of the TIR domain of TcpB. The TIR peptide could resist nocodazole- (b) or cold- (c) induced depolymerization.
DISCUSSION
Unlike other TIR domain-containing proteins, eukaryotic cells expressing TcpB displayed an altered cell morphology, including cell rounding and shrinkage. TcpB behaved like a microtubule-associated protein and induced microtubule bundling by cross-linking the tubules. The condensed microtubule network appeared to resist the depolymerization induced by nocodazole, indicating a robust microtubule stabilization property of TcpB. Our subsequent analyses revealed that, similarly to paclitaxel, TcpB accelerated the nucleation and growth phases of microtubule formation and accumulated increased levels of stabilized tubules.
The TIR domain is a protein–protein interaction module and is vital for the biological function of all the members of the TIR family. TcpB required an intact TIR domain for microtubule modulation as well as TLR suppression as the mutant TcpB exhibited diminished activity. The BB-loop is a subregion of the TIR domain that enables interaction between the TLR and adaptor proteins [21]. The microtubule-stabilization property was located within the BB-loop region of TcpB encompassing amino acids 124–176 that is indispensable for TLR suppression of TcpB.
Microtubules are involved in many aspects of cell responses including cell division, migration and intracellular signal transduction. Microtubule networks act as a scaffold for interaction and assembly of components of signal transduction pathways [16,22]. Therefore stabilization and destabilization of microtubules directly affects various cellular signalling processes. Studies have demonstrated that microtubules play crucial roles in a number of signal transduction pathways including NF-κB, JNK (c-Jun N-terminal kinase) and MAPK (mitogen-activated protein kinase) [23–25]. Depolymerization of microtubules causes rapid and efficient activation of NK-κB [23]. Stabilization of microtubules by paclitaxel inhibits the induction of NF-κB by microtubule-depolymerization agents [23]. However, the mechanism by which depolymerization of microtubules leads to activation of NF-κB remains to be identified. NF-κB activation is driven by the ubiquitination and degradation of IκB (inhibitor of NF-κB) and several proteins of the ubiquitination machinery are associated with microtubule networks [26–29]. TcpB drives ubiquitination and degradation of TIRAP to interfere with TLR2- and TLR4-mediated signalling [13]. TcpB does not possess ubiquitin ligase activity and may recruit a cellular ubiquitin ligase for ubiquitination and subsequent degradation of TIRAP. Microtubule stabilization by TcpB may facilitate the activation and recruitment of ubiquitin machinery to TIRAP. Many additional aspects of the relationship between microtubule stabilization and negative regulation of TLR signalling pathways remain to be elucidated. Since TcpB requires an intact TIR domain for TLR inhibition and microtubule stabilization, we hypothesize that microtubule dynamics play a role in TcpB-mediated inhibition of TLR signalling.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr M.A. Beg, Department of Pathobiological Sciences, University of Wisconsin-Madison, for help with preparation of the paper.
FUNDING This work was supported by the National Institutes of Health [grant numbers 1R01AI073558 and R21 AI088038] and the Binational Agricultural Research and Development Fund (BARD) [grant number US-4378-11].
Abbreviations used
- MBP
maltose-binding protein
- NF-κB
nuclear factor κB
- P
pellet
- PBST
PBS containing 1 % Triton X-100
- S
supernatant
- TIR
Toll/interleukin-1 receptor
- TcpB
TIR domain-containing protein from Brucella
- TIRAP
TIR domain-containing adaptor protein
- TLR
Toll-like receptor.
Footnotes
AUTHOR CONTRIBUTION Girish Radhakrishnan and Gary Splitter conceived the idea. Girish Radhakrishnan performed the protein purification, microtubule-polymerization assay and immunofluorescence microscopy. Girish Radhakrishnan and Jerome Harms performed the microtubuleco-sedimentation assay. Girish Radhakrishnan and Gary Splitter analysed the data and wrote the paper.
REFERENCES
- 1.Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Adv. Exp. Med. Biol. 2005;560:11–18. doi: 10.1007/0-387-24180-9_2. [DOI] [PubMed] [Google Scholar]
- 2.Akira S. TLR signaling. Curr. Top. Microbiol. Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 4.Miggin SM, O'Neill LA. New insights into the regulation of TLR signaling. J. Leukocyte Biol. 2006;80:220–226. doi: 10.1189/jlb.1105672. [DOI] [PubMed] [Google Scholar]
- 5.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 6.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins KA, Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine. 2010;49:237–244. doi: 10.1016/j.cyto.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Kenny EF, O'Neill LA. Signalling adaptors used by Toll-like receptors: an update. Cytokine. 2008;43:342–349. doi: 10.1016/j.cyto.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, et al. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 12.Radhakrishnan GK, Yu Q, Harms JS, Splitter GA. Brucella TIR domain-containing protein mimics properties of the Toll-like receptor adaptor protein TIRAP. J. Biol. Chem. 2009;284:9892–9898. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta D, Koblansky A, Gaines J, Brown T, West AP, Zhang D, Nishikawa T, Park SG, Roop RM, 2nd, Ghosh S. Subversion of innate immune responses by Brucella through the targeted degradation of the TLR signaling adapter, MAL. J. Immunol. 2010;184:956–964. doi: 10.4049/jimmunol.0902008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao TS. Subversion of innate immune signaling through molecular mimicry. J. Clin. Immunol. 2010;30:638–642. doi: 10.1007/s10875-010-9435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radhakrishnan GK, Splitter GA. Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem. Biophys. Res. Commun. 2010;397:59–63. doi: 10.1016/j.bbrc.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gundersen GG, Cook TA. Microtubules and signal transduction. Curr. Opin. Cell Biol. 1999;11:81–94. doi: 10.1016/s0955-0674(99)80010-6. [DOI] [PubMed] [Google Scholar]
- 17.Shelanski ML, Gaskin F, Cantor CR. Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. U.S.A. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JC, Timasheff SN. In vitro reconstitution of calf brain microtubules: effects of solution variables. Biochemistry. 1977;16:1754–1764. doi: 10.1021/bi00627a037. [DOI] [PubMed] [Google Scholar]
- 19.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 20.Xiao H, Verdier-Pinard P, Fernandez-Fuentes N, Burd B, Angeletti R, Fiser A, Horwitz SB, Orr GA. Insights into the mechanism of microtubule stabilization by Taxol. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10166–10173. doi: 10.1073/pnas.0603704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toshchakov VU, Basu S, Fenton MJ, Vogel SN. Differential involvement of BB loops of Toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4- versus TLR2-mediated signal transduction. J. Immunol. 2005;175:494–500. doi: 10.4049/jimmunol.175.1.494. [DOI] [PubMed] [Google Scholar]
- 22.Etienne-Manneville S. From signaling pathways to microtubule dynamics: the key players. Curr. Opin. Cell Biol. 2010;22:104–111. doi: 10.1016/j.ceb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Rosette C, Karin M. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-κB. J. Cell Biol. 1995;128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagata K, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Best A, Ahmed S, Kozma R, Lim L. The Ras-related GTPase Rac1 binds tubulin. J. Biol. Chem. 1996;271:3756–3762. doi: 10.1074/jbc.271.7.3756. [DOI] [PubMed] [Google Scholar]
- 26.Murti KG, Smith HT, Fried VA. Ubiquitin is a component of the microtubule network. Proc. Natl. Acad. Sci. U.S.A. 1988;85:3019–3023. doi: 10.1073/pnas.85.9.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J. Ubiquitin editing enzyme UCH L1 and microtubule dynamics: implication in mitosis. Cell Cycle. 2010;9:980–994. doi: 10.4161/cc.9.5.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J. Microtubule: a common target for parkin and Parkinson's disease toxins. Neuroscientist. 2006;12:469–476. doi: 10.1177/1073858406293853. [DOI] [PubMed] [Google Scholar]
- 29.Fried VA, Smith HT. Ubiquitin: a multifunctional regulatory protein associated with the cytoskeleton. Prog. Clin. Biol. Res. 1989;317:733–744. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.