Protein quality control
Correct folding of proteins is crucial for proper protein, cell, and organ function1. Mutations and cellular stresses, some of which occur during pathology, can lead to incorrect folding and subsequent loss of protein function. If it goes uncorrected, this loss of protein function, along with the toxicity associated with the accumulation of misfolded proteins in cells, results in eventual organ failure2. Many diseases, including neurodegenerative, hepatic, endocrine, and cardiovascular disorders, are thought to be associated with, if not caused by the organ failure resulting from the accumulation of misfolded proteins3. For example, in the heart, accumulation of misfolded proteins has been associated with hypertrophic and dilated cardiomyopathies, as well as ischemic heart disease4.
Cells have evolved an elaborate protein quality control system, part of which involves organelle-initiated unfolded protein responses5–7. The unfolded protein responses that are activated upon accumulation of misfolded proteins are designed to adjust the relevant cellular machinery to enhance protein folding capacity, and/or to degrade terminally misfolded, potentially proteotoxic proteins via the ubiquitin proteasome system, or UPS. If the UPS is sufficient to remove terminally misfolded proteins, proteotoxicity can be averted; however, if the UPS is insufficient, the resulting proteotoxicity can contribute to organ dysfunction. Accordingly, the UPS aspect of protein quality control is essential for normal cell and organ function8.
Proteasomes as Mediators of Intracellular Protein Degradation
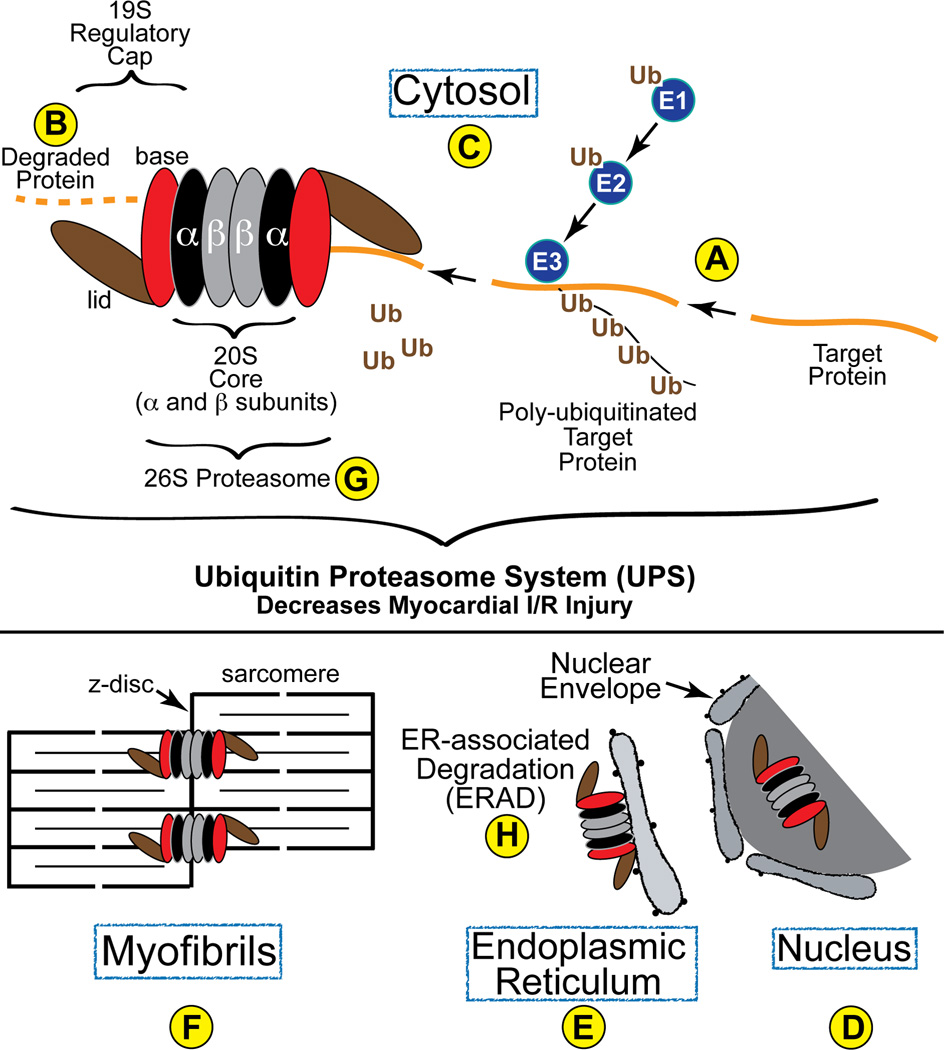
Most intracellular protein degradation via the UPS is an ATP-dependent process that involves E1, E2, and E3 ubiquitin ligases, which function in concert with chaperones to identify and ubiquitinate appropriate target proteins9 (Fig. 1A). The resulting poly-ubiquitinated proteins are then transferred to the 26S proteasome, where they are degraded (Fig. 1B). The 26S proteasome, composed of as many as 34 polypeptides in the heart10, 11, is located in the cytosol (Fig. 1C) and nucleus (Fig. 1D) of all eukaryotic cells. In the myocardium, a significant number of cytosolic proteasomes are membrane-bound, and are located in various places, such as the endoplasmic reticulum (ER) (Fig. 1E). Proteasomes also co-localize with sarcomeric z-disc proteins, such as α-actinin12 (Fig. 1F).
Figure 1. Diagram of the Ubiquitin Proteasome System in the Heart-.
Proteins are targeted for degradation and then ubiquitin is conjugated to them by the concerted transfer of ubiquitin from E1 to E2 and, finally, to E3 ubiquitin ligases, the latter of which can poly-ubiquitinate proteins, as shown (A). Poly-ubiquitinated proteins are then recognized by the 19S regulatory cap of the proteasome, which is composed of a base (red) and a lid (brown). Once within the barrel structure of the 20S portion of the proteasome, which is composed of α- (black) and β-rings (grey), proteins are degraded by the proteolytic activities resident in the β5, β2 and β1 subunits. In cardiac myocytes, proteasomes have been found in the cytosol (C), as well as associated with the z-disc region of sarcomeres (F) and intracellular membranes, such as the SR and ER (E), where they are involved in ERAD (H).
The 26S proteasome (Fig. 1G) is composed of a 20S proteolytic core made of four stacked rings of α- and β-subunits flanked by two 19S regulatory caps composed of a base and a lid. Poly-ubiquitinated proteins access the interior of the 20S core, where they are degraded into 3–20 AA long peptides by the proteolytic activities of the 20S proteasome13, 14. The protease activities of the 20S proteasome reside in the β subunits15, 16. Specifically, chyotrypsin-, trypsin-, and caspase-like protease activities, reside in the β5, β2, and β1 subunits of the 20S proteasome, respectively13. The β5 subunit is the most important for the assembly of active proteasomes17.
A Cardiac Proteasome Paradox
Numerous studies have demonstrated alterations in proteasome function in animal models of heart disease13, 14, 18; proteasome functional insufficiency has been observed most consistently in myocardial ischemia/reperfusion (I/R) injury19, 20. Such studies support the hypothesis that I/R decreases proteasome activity by reducing ATP levels, as well as oxidatively damaging and unfolding proteasome proteins13, any or all of which may contribute to myocardial injury. Studies designed to test this hypothesis have used proteasome gain- or loss-of-function in animal models of myocardial I/R injury. However, the results of these studies have generated the following paradox:
Gain-of-function using transgenic mice with increased proteasome activity showed protection from I/R injury 21.
Loss-of-function using pharmacological means to decrease proteasome activity also showed protection from I/R injury 22–24.
This paradox has clouded our view of the roles played by proteasomes in cardiac pathology. In this issue of Circulation Research, Tian et al25 have clarified this view and enhanced our understanding of UPS function in the heart by determining the effects of cardiac restricted proteasome inhibition on I/R damage in mouse hearts.
Cardiac-restricted Proteasome Inhibition in the Heart
To examine the effect of decreased cardiac myocyte proteasome activity on myocardial I/R injury, Tian et al generated a transgenic mouse line wherein a catalytically inactive, dominant interfering form of the β5 subunit of the 20S proteasome was expressed in a cardiac specific manner (T60A-β5 tg)25. Transgene expression of T60A-β5 was relatively mild as a result of using an attenuated version of the αMHC promoter, which drove cardiac myocyte expression, but at relatively low levels, compared to the αMHC promoter used in most heart transgenic mouse models. This relatively mild transgene expression was probably a critical aspect of the study, since it was shown that under basal conditions, the transgene had no deleterious effects on cardiac structure or function. Tian et al also used a transgenic mouse model they had previously generated in which the ability of proteasomes to degrade a mutant form of GFP that is susceptible to misfolding facilitated an assessment of proteasome function in the heart, in vivo. Using this mouse model, they found that, upon I/R, proteasome mediated degradation of the misfolded GFP was decreased in the area at risk and border zones, as well as the remote area. These results were confirmed using biochemical assays of proteasome activity from extracts of the same hearts. Then, using the T60A-β5 tg mice, the authors found that cardiac myocyte-restricted proteasome inhibition resulted in increased I/R injury. When combined with a previous report from the same lab, wherein a transgenic mouse model of proteasome activation showed decreased I/R damage21, the results of Tian et al contribute to clarifying the cardiac proteasome paradox, as follows:
Implications of the Study
In addition to addressing the role of proteasome inhibition on myocardial I/R injury, the study by Tian et al may help us understand other aspects of proteasome function in the healthy and diseased heart.
Opposing Effects of Global and Cardiac Myocyte-Specific Proteasome Inhibition
Tian et al found that cardiac restricted inhibition of proteasome function increased I/R injury, while other studies showed that global inhibition of proteasome function using pharmacological proteasome inhibitors, in vivo, decreased I/R injury22, 23. Caution should be exercised when comparing results from studies that used such different approaches due to a variety of caveats, including potential off-target effects of pharmalogical proteasome inhibitors, as well as the potential for differential effects of chronic proteasome inhibition by transgenesis and acute inhibition by pharmacological approaches. However, at face value, these findings support the hypothesis that inhibiting proteasomes in all cell types in the heart results in a diametrically opposed response of the heart to I/R than cardiac restricted proteasome inhibition. Additional studies in which cardiac proteasomes are inhibited in a cell-specific manner in non-cardiac myocytes, such as endothelial cells, smooth muscle cells, or fibroblasts, will be needed to address this hypothesis, which will be required to determine whether there is a need to implement cell specificity in the future design proteasome-targeted therapies for ischemic heart disease.
Potential Therapeutic Effects of Preserving Proteasome Function in the Heart
The results of Tian et al suggest that preserving proteasome function in cardiac myocytes might decrease the effects of I/R injury, which could ultimately lead to decreasing the morbidity and mortality of myocardial infarction. This conclusion correlates with the current consensus that proteasome function is impaired in animal models of I/R injury. However, there is less consensus on the effects of other models of cardiac pathology on proteasome function13, 18. Thus, while preserving proteasome function may be advantageous in the ischemic heart, it is not as clear whether it would be beneficial in hypertrophy, dilated cardiomyopathy, or heart failure. In fact, some studies suggest that pharmacological inhibition of proteasome function inhibits adaptive and maladaptive cardiac hypertrophy26, while others find that it promotes hypertrophy27. Accordingly, future studies examining the effects of genetically-modified mouse models of proteasome gain- and loss-of-function on cardiac diseases, in addition to I/R will be required to better assess the full potential of proteasome-targeted therapies.
Subcellular Localization of Proteasomes Suggests Regional-specific Functions
Using confocal immunocytofluorescence, Tian et al found that a c-Myc-tagged version of the transgene that they used to inhibit proteasome function was located to the nucleus and cytosol of cardiac myocytes isolated from transgenic mouse hearts. For the most part, c-Myc staining in the cytosol adopted a striated pattern that co-stained with α-actinin, which is localized to the z-disc region of sarcomeres (Fig. 1F). These results correlate with those of another study, which also found that some cytosolic proteasomes localize to intracellular membranous structures, such as the SR/ER of cardiac myocytes12. These results are consistent with the hypothesis that the ability of proteasomes to selectively degrade certain proteins is partly a result of their organelle-specific localization in cardiac myocytes28. For example, numerous signal transduction proteins that facilitate biomechanical sensing and signaling in cardiac myocytes are localized to the z-disc29, 30. Among these signaling molecules are components of the calcinuerin A, NFAT, PKC, ERK and p38 MAP kinase signaling systems, the activities and levels of which can be regulated by the UPS. Thus, z-disc-associated proteasomes may participate in regulating the levels of such signaling proteins. Additionally, the localization of proteasomes to intracellular membranes of myocytes, such as the SR/ER, is consistent with their involvement in ER-associated degradation (ERAD) (Fig. 1H). While ERAD has not been studied in detail the heart, in other cell types it has been shown that proteasomes located on the cytosolic face of the ER are responsible for the degradation of misfolded ER proteins that are translocated from the ER lumen to the cytosolic face of the ER, where they are ubiquitinated and degraded in a proteasome-dependent manner31.
Conculsions
In conclusion, the results of the study by Tian et al have made a significant contribution toward furthering our understanding of the roles for proteasomes in the ischemic heart. Moreover, in addition to facilitating future studies that will address important questions about the function of proteasomes in other cardiac pathologies, the T60A-β5 transgenic mouse model described by Tian et al provides proof-of-principle for a method that could be used to examine the effect of proteasome inhibition in other cell types in the heart, as well as other tissues. Such studies could extend the potential utility of proteasome-targeted therapies well beyond the heart to include numerous other tissues in which the accumulation of misfolded proteins is known to contribute to disease.
ACKNOWLEDGEMENTS
The author wishes to acknowledge Dr. Shirin Doroudgar for many insightful discussions and for critical reading of the manuscript.
Sources of Funding: National Institutes of Health PO1 HL085577, RO1 HL75573, RO1 HL104535, RO3 EB011698
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
REFERENCES
- 1.Roth DM, Balch WE. Modeling general proteostasis: proteome balance in health and disease. Curr Opin Cell Biol. 2011;23(2):126–134. doi: 10.1016/j.ceb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doroudgar S, Glembotski CC. The cardiokine story unfolds: ischemic stress-induced protein secretion in the heart. Trends Mol Med. 2011;17(4):207–214. doi: 10.1016/j.molmed.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110(10):1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44(3):453–459. doi: 10.1016/j.yjmcc.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22(1):22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shutt TE, McBride HM. Staying cool in difficult times: Mitochondrial dynamics, quality control and the stress response. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 7.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123(Pt 22):3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99(12):1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 9.Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2011;279(4):532–542. doi: 10.1111/j.1742-4658.2011.08456.x. [DOI] [PubMed] [Google Scholar]
- 10.Young GW, Wang Y, Ping P. Understanding proteasome assembly and regulation: importance to cardiovascular medicine. Trends Cardiovasc Med. 2008;18(3):93–98. doi: 10.1016/j.tcm.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, Lu H, Stefani E, Ping P. Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Mol Cell Proteomics. 2009;8(2):302–315. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, Bardag-Gorce F, Ping P. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99(4):362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301(16):H2207–H2219. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta. 2008;1782(12):749–763. doi: 10.1016/j.bbadis.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Portbury AL, Willis MS, Patterson C. Tearin' up my heart: proteolysis in the cardiac sarcomere. J Biol Chem. 2011;286(12):9929–9934. doi: 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10(5):609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 17.Jager S, Groll M, Huber R, Wolf DH, Heinemeyer W. Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J Mol Biol. 1999;291(4):997–1013. doi: 10.1006/jmbi.1999.2995. [DOI] [PubMed] [Google Scholar]
- 18.Schlossarek S, Carrier L. The ubiquitin-proteasome system in cardiomyopathies. Curr Opin Cardiol. 2011;26(3):190–195. doi: 10.1097/HCO.0b013e32834598fe. [DOI] [PubMed] [Google Scholar]
- 19.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276(32):30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 20.Powell SR, Davies KJ, Divald A. Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. J Mol Cell Cardiol. 2007;42(1):265–269. doi: 10.1016/j.yjmcc.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121(9):3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell B, Adams J, Shin YK, Lefer AM. Cardioprotective effects of a novel proteasome inhibitor following ischemia and reperfusion in the isolated perfused rat heart. J Mol Cell Cardiol. 1999;31(2):467–476. doi: 10.1006/jmcc.1998.0880. [DOI] [PubMed] [Google Scholar]
- 23.Pye J, Ardeshirpour F, McCain A, Bellinger DA, Merricks E, Adams J, Elliott PJ, Pien C, Fischer TH, Baldwin AS, Jr, Nichols TC. Proteasome inhibition ablates activation of NF-kappa B in myocardial reperfusion and reduces reperfusion injury. Am J Physiol Heart Circ Physiol. 2003;284(3):H919–H926. doi: 10.1152/ajpheart.00851.2002. [DOI] [PubMed] [Google Scholar]
- 24.Stansfield WE, Moss NC, Willis MS, Tang R, Selzman CH. Proteasome inhibition attenuates infarct size and preserves cardiac function in a murine model of myocardial ischemia-reperfusion injury. Ann Thorac Surg. 2007;84(1):120–125. doi: 10.1016/j.athoracsur.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 25.Tian Z, Zheng H, Li J, Li Y, Su H, Wang X. Genetically Induced Moderate Inhibition of the Proteasome in Cardiomyocytes Exacerbates Myocardial Ischemia-reperfusion Injury in Mice. Circ Res. 2012;111 doi: 10.1161/CIRCRESAHA.112.270983. xxx-xxx. (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedhli N, Depre C. Proteasome inhibitors and cardiac cell growth. Cardiovasc Res. 2010;85(2):321–329. doi: 10.1093/cvr/cvp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stansfield WE, Tang RH, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294(2):H645–H650. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 28.Scruggs SB, Ping P, Zong C. Heterogeneous cardiac proteasomes: mandated by diverse substrates? Physiology (Bethesda) 2010;26(2):106–114. doi: 10.1152/physiol.00039.2010. [DOI] [PubMed] [Google Scholar]
- 29.Frank D, Kuhn C, Katus HA, Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J Mol Med (Berl) 2006;84(6):446–468. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 30.Sanger JM, Sanger JW. The dynamic Z bands of striated muscle cells. Sci Signal. 2008;1(32):pe37. doi: 10.1126/scisignal.132pe37. [DOI] [PubMed] [Google Scholar]
- 31.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]