Abstract
In their natural environment, microbes organize into communities held together by an extracellular matrix composed of polysaccharides and proteins. We developed an in vivo labeling strategy to allow the extracellular matrix of developing biofilms to be visualized with conventional and super-resolution light microscopy. Vibrio cholerae biofilms displayed three distinct levels of spatial organization: cells, clusters of cells, and collections of clusters. Multiresolution imaging of living V. cholerae biofilms revealed the complementary architectural roles of the four essential matrix constituents: RbmA provided cell-cell adhesion, Bap1 allowed the developing biofilm to adhere to surfaces, and heterogeneous mixtures of Vibrio polysaccharide (VPS), RbmC, and Bap1 formed dynamic, flexible and ordered envelopes that encased the cell clusters.
Microbes within biofilms are more resistant to antibiotics, to immune clearance, and to osmotic, acid and oxidative stresses compared to planktonic cells (1–7). Despite advances in identifying the polysaccharide and proteinaceous constituents of the biofilm extracellular matrix, the mechanisms by which these factors yield a mechanically defined and spatially organized biofilm are largely unknown (8–10). The small size of most microbes has precluded multi-scale optical investigation of living biofilms. Vibrio cholerae biofilm formation involves the production of Vibrio polysaccharide (VPS) and three matrix proteins (RbmA, RbmC, and Bap1) predicted to contain carbohydrate-binding domains (fig. S1A) (11–13). To investigate the molecular mechanisms of biofilm development, we used a V. cholerae rugose variant with increased capacity to form biofilms (11). We inserted Myc, FLAG, and HA (Human influenza hemagglutinin) epitopes into its genome at the 3’ ends of the rbmA, rbmC, and bap1 genes, respectively (fig. S1B), allowing us to label these matrix proteins in vivo by supplementing the growth medium with corresponding cyanine dye-labeled primary antibodies (Fig. 1).
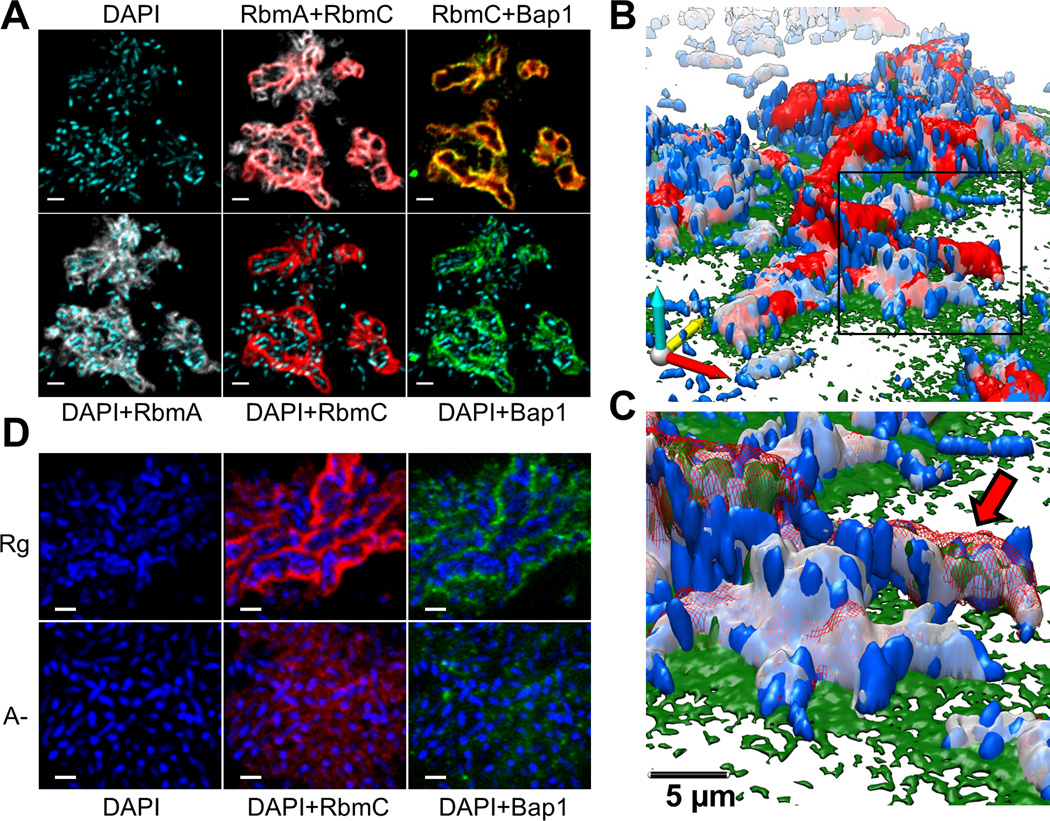
Fig. 1. V. cholerae biofilm structure.
(A) Optical section of biofilm 4 µm above coverslip. Images are pseudo-colored blue (cells), gray (RbmA), red (RbmC) and green (Bap1). RbmA localizes around and within cell clusters. RbmC and Bap1 encase cell clusters. Cells were counterstained with DAPI. Scale bars, 3 µm. (B) 3D biofilm architecture. Colors as in (B). (C) Enlargement of the boxed region in (B). Red arrow indicates one cell cluster. Red signal now rendered partially transparent to allow visualization of cells within an RbmC-containing cluster. (D) Comparison of biofilm architecture formed by rugose (Rg) and ΔrbmA (A-) strains. RbmA is required for cell cluster formation. Scale bars, 2 µm.
We used four-color confocal imaging to validate this labeling strategy and obtain a diffraction-limited overview of biofilm architecture (Fig. 1, A–C, Movie S1). Cells were mainly organized into elongated clusters whose boundaries were defined by three-dimensional envelopes of the RbmC (red) and Bap1 (green) proteins (Fig. 1C, red arrow). Within the envelope that encases the cell clusters, the relative Bap1 signal was highest in those areas with least RbmC (Fig. 1A, fig. S5 and S6). Deletion of either RbmC or Bap1 did not impair cluster formation or the resultant architecture of the envelope (11, 14) (Fig. 1D and fig. S7). The cell clusters had a regular width of 2.2±0.3 μm (N=42 clusters) while their length varied from 2 to 8 µm (fig. S8). Each cell within a cluster contacted the cluster boundary and thus the interstitial space between clusters, perhaps facilitating nutrient delivery and waste disposal.
However, although Bap1 and RbmC share 47% peptide sequence similarity (11), their spatial distribution differed notably at the interface between the coverslip and the cell clusters (fig. S9). Bap1 was concentrated at the biofilm-surface interface (14), while RbmC was absent from the interface (fig. S9 and Fig 1, B and C). Moreover, a bap1 deletion strain had a more severely altered biofilm phenotype than a rbmC deletion strain (11, 14), all pointing to Bap1 having two separable functions, namely, encasing cell clusters and attaching cells to the surface.
In contrast to RbmC and Bap1, RbmA was detected throughout the biofilm (Fig. 1, A–C) (14). Strains lacking RbmA have reduced colony corrugation and are less resistant to detergent treatment (12), but can still adhere to surfaces. Surprisingly, deletion of rbmA caused loss of cell ordering into clusters and associated RbmC-Bap1 envelopes although both of these proteins were clearly present within the biofilm (Fig. 1D. and Fig. S7). Thus Bap1 appears to help the biofilm to adhere to surfaces, RbmC and Bap1 appears to encapsulate cell clusters, and RbmA appears to participate in cell-cell adhesion (Movies S2–S6) (11, 12, 14)
To further test these hypotheses and to learn how biofilms assemble, we imaged living biofilms as they developed from a single founder cell into mature biofilms (Fig. 2A and fig. S10). We followed matrix protein secretion and organization with a continuous in situ immunostaining approach (15) in which labeled primary antibodies were added to the growth medium (Fig. 2A). At the time of initial attachment, individual founder cells did not have detectable RbmA, RbmC, and Bap1 on their surface. The first matrix protein to appear post-attachment was RbmA, which accumulated at discrete sites on the cell surface. After the first cell division, the newly formed daughter cell remained attached to the founder cell, unlike in planktonic cells, where the two cells quickly separate (Fig. 2A).
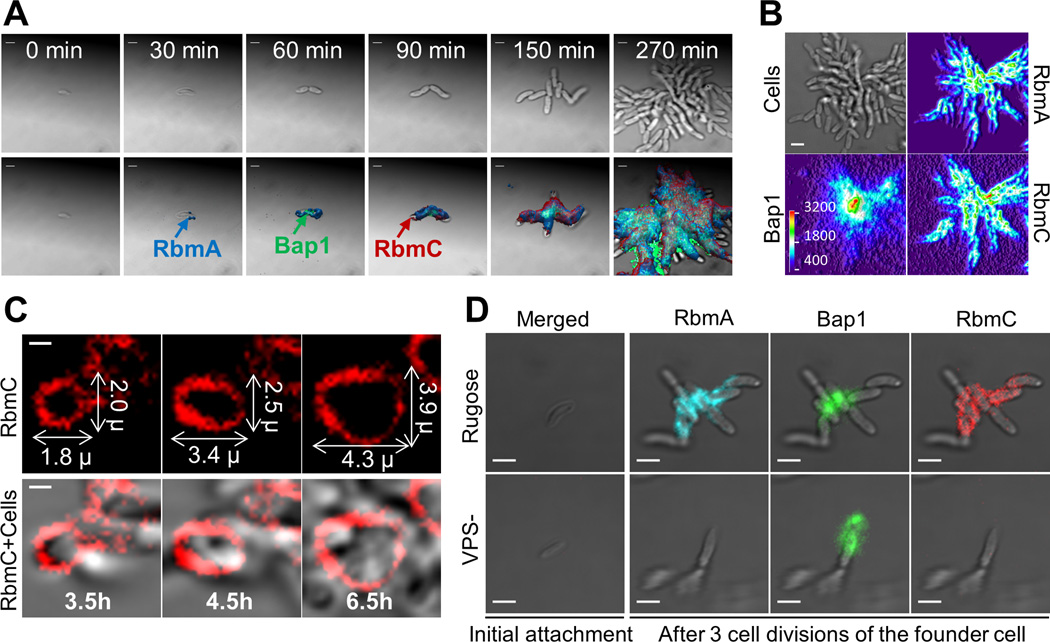
Fig. 2. Time-lapse CLSM imaging of V. cholerae biofilm development and cluster formation.
(A) Expression and subsequent distribution of matrix proteins were followed by time-lapse CLSM using continuous direct immunostaining. Cell outlines (bright field) are gray; RbmA, Bap1 and RbmC are shown in blue, green, and red, respectively. Scale bars, 2 µm. (B) Bright field biofilm image and corresponding fluorescent channel surface plots of Bap1, RbmA and RbmC obtained 4.5 hours post-inoculation. Fluorescent intensity is color-coded according to the color scale bar. Bap1 spread from a central point corresponding to the founder cell position while RbmA and RbmC were more homogenously distributed through the biofilm cells. Scale bar, 3 µm. (C) Gradual expansion of the RbmC-containing cluster tracked by time-lapse CLSM. Scale bars, 1 µm. (D) Inability to produce VPS (VPS-) prevents retention of daughter cells, accumulation of RbmA and RbmC, and blocks biofilm formation.
Bap1 then appeared at the junction between the two cells and also on the substrate near the cells (Fig. 2A). Bap1 gradually accumulated radially over distances of tens of microns from its initial location on or near the founder cell (Fig. 2, A and B). The founder cell and its immediate environment had notably more Bap1 than the rest of the biofilm for the entire 6.5-hour duration of the experiment. The radial distribution of Bap1 relative to the founder cell and the radially decaying distribution of Bap1’s concentration suggest that Bap1 is continuously secreted into solution by the founder cell and other early members of the young biofilm and then accumulates on nearby surfaces (Fig. 2B and fig. S11).
The third matrix protein, RbmC, first appeared after 90 min at discrete sites on the cell surface. Later in biofilm development, the RbmC-Bap1 envelopes formed and then grew by expansion in all directions, with the size of the RbmC-Bap1 envelope doubling within three cell divisions to accommodate the new cell mass (Fig. 2C). Biofilm formation thus involves the temporally sequenced and spatially heterogeneous secretion of matrix proteins, which may have complementary architectural functions, namely, retention of daughter cells following division by RbmA, surface functionalization by Bap1, and encapsulation of the cell clusters by RbmC/Bap1.
Next, we investigated how the RbmC and Bap1 matrix proteins interact with VPS during biofilm formation. VPS is a polysaccharide thought to form a polymeric network that gives mechanical continuity to the biofilm (8, 16–19). V. cholerae cells lacking either VPS (VPS-) or all three matrix proteins (ABC-) were unable to form 3D biofilms (fig. S12). The parent strain biofilm phenotype could be recovered by co-culturing VPS- and ABC- strains, showing that heterologous provision of these four materials is sufficient to restore normal biofilm formation (Fig. S1A and S12). VPS- cells could stick to surfaces, but subsequently produced daughter cells did not accumulate and were instead lost into the growth medium (20) (Fig. 2D and Movie S5). Although RbmA, RbmC and Bap1 proteins were synthesized (fig. S13 and S14), they did not accumulate on the surface of VPS- cells (Fig. 2D). Bap1 was detected on the substrate near the founder cell (Fig. 2D), as expected if Bap1’s main function is to adhere to diverse substrates and tether the biofilm (14). Thus, VPS is required for accumulation of the RbmA, RbmC and Bap1 on the cell surface, which in turn is needed for formation of mature biofilms (13, 14).
Because VPS was required for accumulation of matrix proteins on the cell surface, we wondered whether the opposite was also true. We directly stained the VPS with a Cy3-labeled wheat germ agglutinin (WGA) which recognizes N-acetylglucosamine sugars in the VPS (21). RbmC was essential for sustained incorporation of VPS throughout V. cholerae biofilms (fig. S15A). Without RbmC, there were occasional bright dots of VPS within the colony but at a much lower density than in the parent strain biofilm (fig. S15A). Thus, sustained retention of VPS and RbmC are codependent (Fig. 2D and fig. S15). The VPS staining also confirmed that the RbmC/Bap1 envelopes contained VPS, as expected (Fig. 1A and fig. S15B).
3D biofilm development requires a specific, mutually interdependent series of protein/VPS synthesis, secretion, capture, and crosslinking steps. The ~200 nm spatial resolution of CLSM (Confocal laser scanning microscopy) (22, 23) was however insufficient to directly visualize these developmental intermediates. We thus constructed a multicolor 3D super-resolution imaging apparatus using the stochastic optical reconstruction microscopy (STORM) (23–28) with a localization precision of 19 nm, 21 nm and 42 nm in X, Y, and Z (full width at half maximum) (fig. S16). As before, we added labels to the growth medium and imaged living biofilms. Using a Cy3-WGA reagent, VPS was first detected at several discrete sites on the cell surface at t = 15 min post attachment (Fig. 3A, white arrows). Over the next two hours, the number of VPS spots as well as their intensity increased slowly. At t = 60 min post-attachment, 3D super-resolution images of VPS organization showed that the polymer was primarily organized into 50–200 nm diameter spheroids protruding away from the cell surface (Fig. 3B, white arrow). It appears that VPS is progressively extruded from the cell as a flexible polymer that then, like all relaxed flexible polymers, adopts an isotropic, spherical configuration.
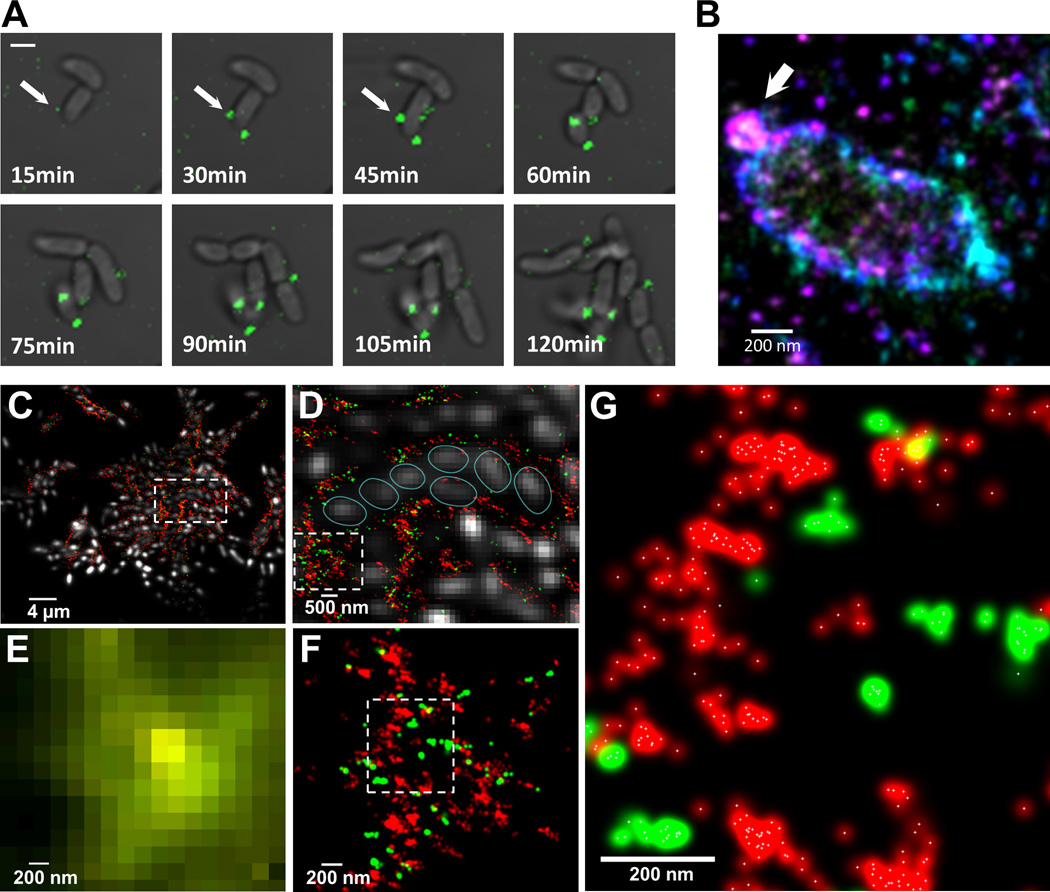
Fig. 3. Exopolysaccharide secretion, initial organization and molecular architecture of V. cholerae biofilms.
(A) Time-lapse CLSM images of VPS (green) production/secretion in V. cholerae cell during biofilm formation. Fluorescent images of VPS are merged with bright-field images of cells. Scale bars, 2 µm. (B) 3D superresolution image of a single V. cholerae cell. White arrow indicates a ball-like structure of VPS on the surface of V. cholerae cell early in biofilm formation. Color corresponds to height (−300 nm (violet) to +300nm (red)). (C) 3D two-color superresolution image (200 nm z-section) of a rugose variant biofilm showing molecular organization of VPS (red) and RbmC (green) around cell clusters. Cells were counterstained with DAPI (white). (D) Enlarged boxed region in (C) showing organization of cells within VPS/RbmC-enclosed cluster. Individual cells were outlined (cyan) for clarity. (E) Enlarged boxed region in (D) as it appears in conventional, diffraction-limited microcopy, showing unresolved VPS and RbmC signals. (F) Superresolution image of the same region in (E), showing distribution of RbmC and the VPS polymers in a biofilm matrix. (G) Enlarged boxed region in (F). White crosses indicate the center of a Gaussian-fit to each localization events.
Pseudomonas aeruginosa biofilms have been reported to self-heal within minutes after mechanical disruption beyond their yield point, implying that relatively transient interactions are responsible for maintaining the P. aeruginosa matrix (29). How could such recovery be possible if the VPS (or Psl in P. aeruginosa) were irreversibly crosslinked by matrix proteins such as RbmC? The organization of VPS and RbmC within a biofilm was visualized with two-color 3D super-resolution imaging (15). The super-resolution microscope has wide dynamic range and can detect single VPS and RbmC molecules. VPS and RbmC were not homogeneously distributed within the mature biofilm but both matrix components were confined to the envelopes encasing the cell clusters and to the interstitial space between clusters (Fig. 3, C and D). The mechanism(s) by which bacteria achieve such spatial segregation of materials within the biofilm, and thus generate a matrix architecture with sub-micron features, are unknown. Moreover, most RbmC signal was not uniformly distributed within the VPS matrix (Fig. 3, E–G). Thus RbmC and VPS may have homophilic (RbmC-RbmC or VPS-VPS) and heterophilic interactions (RbmC-VPS), where RbmC may act as a reversible crosslinker of VPS. VPS organization must also be dynamic, because otherwise the cells could not sharply re-partition RbmC and VPS into the envelopes and interstitial spaces (Fig. 3D).
We used a matrix labeling strategy to observe in real time as V. cholerae biofilms develop with single-protein and single-polymer precision, revealing assembly principles and intermediates. Cells organize into clusters within the biofilm and the mature biofilm is a composite of these clusters. An envelope composed of VPS, Bap1, and RbmC encloses these clusters. RbmA is required for these clusters formation. The VPS/Bap1/RbmC envelope is structured on the molecular level by unknown mechanism(s) and is capable of reforming, stretching, and expanding to accommodate cell growth.
Supplementary Material
Acknowledgements
We thank B. Huang for providing image processing software and to D.J. Wozniak, J.H.D. Cate, X. Nan, A. Arkin and A. Yildiz for critical evaluation of the manuscript. This work is supported by NSF (PHY-0647161 (JL)), NIH (AI055987 (FHY), GM096450 and GM068518 (XZ)). X. Z. is a Howard Hughes Medical Institute Investigator. JL acknowledges support from the DOE Office of Basic Energy Sciences (FWP SISGRKN) and an LBNL LDRD.
Footnotes
Supporting Material
Materials and Methods
fig. S1 to S17
Table S1 to S2
References (30–40)
Movies S1 to S6
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends in Microbiology. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Parsek MR, Singh PK. Bacterial biofilms: An Emerging Link to Disease Pathogenesis. Annu. Rev. Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 4.Stewart PS, William Costerton J. Antibiotic resistance of bacteria in biofilms. The Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 5.Mah T-F, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 6.Anderson GG, O’Toole GA. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 7.Reisner A, Høiby N, Tolker-Nielsen T, Molin S. Microbial pathogenesis and biofilm development. Contrib Microbiol. 2005;12:114–131. doi: 10.1159/000081692. [DOI] [PubMed] [Google Scholar]
- 8.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Micro. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 9.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010;59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 11.Fong JC, Yildiz FH. The rbmBCDEF Gene Cluster Modulates Development of Rugose Colony Morphology and Biofilm Formation in Vibrio cholerae. J. Bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. Identification and Characterization of RbmA, a Novel Protein Required for the Development of Rugose Colony Morphology and Biofilm Structure in Vibrio cholerae. J. Bacteriol. 2006;188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong JCN, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Absalon C, Van Dellen K, Watnick PI. A Communal Bacterial Adhesin Anchors Biofilm and Bystander Cells to Surfaces. PLoS Pathog. 2011;7:e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.See material on Science Online.
- 16.Sutherland IW. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 17.Karatan E, Watnick P. Signals, Regulatory Networks, and Materials That Build and Break Bacterial Biofilms. Microbiol. Mol. Biol. Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 19.Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends in Microbiology. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Moorthy S, Watnick PI. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 2004;52:573–587. doi: 10.1111/j.1365-2958.2004.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein IJ, Hayes CE. The lectins: carbohydrate-binding proteins of plants and animals. Adv Carbohydr Chem Biochem. 1978;35:127–340. doi: 10.1016/s0065-2318(08)60220-6. [DOI] [PubMed] [Google Scholar]
- 22.Pawley JB, Masters BR. Handbook of Biological Confocal Microscopy. Journal of Biomedical Optics. 2008;13:029902. [Google Scholar]
- 23.Hell SW. Far-Field Optical Nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 24.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Meth. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 26.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor Super-Resolution Imaging with Photo-Switchable Fluorescent Probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang B, Wang W, Bates M, Zhuang X. Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess ST, Girirajan TPK, Mason MD. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieleg O, Caldara M, Baumgärtel R, Ribbeck K. Mechanical robustness of Pseudomonas aeruginosa biofilms. Soft Matter. 2011;7:3307. doi: 10.1039/c0sm01467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.