Abstract
The mechanisms of mitochondrial DNA replication have been hotly debated for a decade. The strand-displacement model states that lagging-strand DNA synthesis is initiated from the origin of light-strand DNA replication (OriL), whereas the strand-coupled model implies that OriL is dispensable. Mammalian mitochondria cannot be transfected and the requirements of OriL in vivo have therefore not been addressed. We here use in vivo saturation mutagenesis to demonstrate that OriL is essential for mtDNA maintenance in the mouse. Biochemical and bioinformatic analyses show that OriL is functionally conserved in vertebrates. Our findings strongly support the strand-displacement model for mtDNA replication.
Keywords: DNA replication, mitochondrion, origin, primer, TWINKLE
INTRODUCTION
Human mitochondria contain a small double-stranded DNA genome (mtDNA) of only 16,659 base pairs (bp) that encodes 13 essential subunits of the respiratory chain. Depletion of mtDNA and different types of mtDNA mutations cause several mitochondrial diseases, and are also implicated in ageing and age-associated diseases [1]. In human cells, the mitochondrial genome is replicated by an enzymatic machinery that contains at least five different proteins: the catalytic subunit of DNA polymerase γ (POLγA) and its processivity factor (POLγB); the replicative helicase, TWINKLE; the mitochondrial single-stranded DNA-binding protein (mtSSB); and the mitochondrial RNA polymerase (POLRMT). Mutations in genes encoding these mtDNA replication factors can cause different human mitochondrial diseases [1, 2].
The exact mechanisms of mtDNA replication are still debated [3–6]. According to the strand-displacement model, DNA synthesis is continuous on both strands [7]. To ensure proper coordination of DNA synthesis, the mitochondrial genome contains two specific origins of DNA synthesis, one for each DNA strand; the heavy-strand origin of DNA replication (OriH) and the light-strand origin of DNA replication (OriL). After initiation at OriH, leading-strand DNA synthesis continues to displace the parental heavy strand (H strand) and during this first phase of replication, there is no simultaneous DNA synthesis on the lagging strand. When the replication machinery has synthesized two-thirds of leading-strand DNA, it reaches OriL; the H strand of OriL becomes single stranded and folds into a stem-loop structure, which serves as an initiation site for L-strand DNA synthesis in the opposite direction [1, 8–10]. POLRMT binds to the OriL structure and initiates primer synthesis. After about 25 nts, POLRMT is replaced by POLγ and DNA synthesis begins [11, 12]. The exact mechanisms by which POLRMT recognizes OriL and activates primer synthesis remain unresolved, but it is clear that a poly-T stretch in the loop region is required [8]. After initiation at OriL, both leading- and lagging-strand DNA synthesis proceeds continuously, until each strand is completely replicated and two daughter molecules are formed.
The relevance of the strand-displacement model for mtDNA synthesis has been disputed. Analyses with two-dimensional neutral–neutral agarose-gel electrophoresis revealed the existence of conventional duplex mtDNA replication intermediates, suggesting that leading- and lagging-strand DNA synthesis are coupled events and that the strand-displacement model might be incorrect [4]. These findings prompted proponents of the strand-displacement model to review their findings, which led to the demonstration that although OriL is the main site for initiation of lagging-strand DNA synthesis [11], initiation might also occur at other sites, but at much lower frequencies [6]. In principle, this modification of the strand-displacement model does not change the fundamentals because lagging-strand DNA synthesis, once initiated, is still continuous and not linked to the leading-strand DNA replication machinery. The data supporting the strand-displacement and strand-coupled models for mtDNA replication, respectively, are on the basis of descriptive biochemical characterization of replication intermediates and the lack of genetic data has hampered progress towards a resolution of the debate.
We have developed a reconstituted system for characterization of mtDNA replication in vitro. When combined, purified POLγ, the TWINKLE helicase and mtSSB are capable of processive leading-strand DNA synthesis on a double-stranded template [13]. Binding of POLRMT allows for primer formation and initiation of lagging-strand DNA synthesis from OriL [8]. Similar to the situation in vivo, passage of the leading-strand DNA replication machinery is essential for OriL activation and primer synthesis by POLRMT. This system allows detailed molecular analysis of the sequence requirements of OriL and other regulatory elements in the mtDNA molecule. In other systems, similar analyses are often complemented with in vivo studies, using transfected plasmid templates, in which the functional importance of specific DNA sequences can be verified by mutagenesis. Unfortunately, this coupled structure–function approach is not possible for studies of the mitochondrial DNA replication machinery, because mammalian mitochondria cannot be transfected with foreign DNA constructs [14]. In the current report, we circumvent these shortcomings of the mitochondrial system and use an in vivo saturation mutagenesis approach to establish the sequence requirements at OriL. Our data demonstrate a structural and functional conservation of OriL in vertebrates, suggesting that lagging-strand DNA synthesis is initiated by a highly conserved mechanism. Our findings give strong in vivo and in vitro support for the strand-displacement model for mtDNA replication.
RESULTS AND DISCUSSION
Saturation mutagenesis in vivo of the OriL sequence
We used a saturation mutagenesis approach in the mouse to investigate if modifications of the OriL sequence were tolerated in vivo. To increase in vivo mutation rates, we used mtDNA mutator mice carrying a proofreading-deficient form of the mitochondrial DNA polymerase γ (PolγAD257A/PolAγAD257A) that show increased overall levels of somatic mtDNA point mutations [15]. We reasoned that mutations in OriL should be underrepresented if this sequence is critically needed for mtDNA replication. Alternatively, OriL should tolerate mutations if this sequence is dispensable for mtDNA replication.
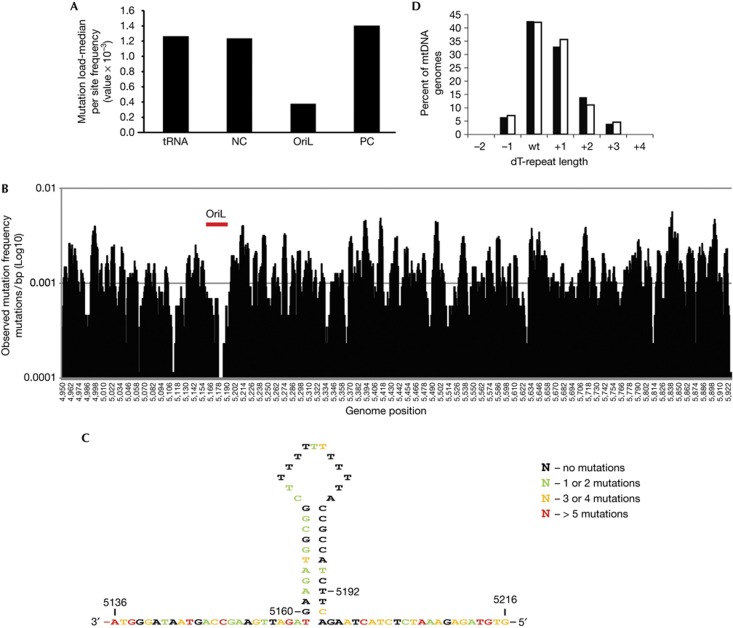
We isolated somatic tissues from mtDNA mutator mice and sequenced a 974-bp-long region covering OriL and surrounding sequences. In total we sequenced 1,747 clones and found that the tRNA genes, the cytochrome c oxidase I gene (COX I), and non-coding regions included in this mtDNA fragment showed a point mutation frequency of 1.3–1.4 mutations/1,000 bp. In contrast, the overall point mutation frequency in the OriL region was only 0.3 mutations/1,000 bp (Fig 1A). A plot of the identified mutations confirmed that there was selection against OriL-localized mutations (Fig 1B). We could only find 17 different point mutations at 13 sites in the OriL structure and all except 4 sites were located in the stem region (Fig 1C). OriL displays a very curious sequence bias, with one side of the stem loop being rich in purines (A and G) and the other side rich in pyrimidines (T and C). Strikingly, most observed mutations in the stem region were located to the purine-rich strand of OriL, which is not used as a template for primer synthesis. Just two mutations were identified in the pyrimidine-rich side of the stem that is used as template for primer synthesis. Therefore, single-site mutations in the non-template strand that lower the stability of the stem are apparently tolerated in vivo, whereas mutations in the template strand are selected against.
Figure 1.
In vivo characterization of sequence variations in the mouse OriL sequence. (A) DNA sequence analysis of proofreading-deficient (polγAD257A/polγAD257A) mice showing median mutation load in tRNA, NC and PC regions of the analysed fragment. (B) Plot of the mutations/bp for each position along the cloned regions. A 5-base sliding window starting at the number plotted. Insertions and deletions in the polyA region of OriL were excluded. (C) Schematic picture of mouse OriL. Positions where point mutations were found in the proofreading-deficient mice are indicated. (D) The percent of molecules containing poly-T stretch length variants in the single-stranded region of OriL (nts 5,172–5,182) found in the (polγAD257A/ polγAD257A) mice. mtDNA, mitochondrial DNA; NC, non-coding; nts, nucleotides; PC, protein coding; tRNA, transfer RNA.
We found a surprisingly high amount of insertion and deletion mutations in the loop region of OriL, with the length of the poly-T stretch displaying an extreme variability (Fig 1D). The wild-type (WT) poly-T length of 11 T:s was found in only 42% of the sequences, in other sequences 10 (7%), 12 (34%), 13 (12,5%) and 14 (4%) nts-long poly-T stretches were identified. The length of the T-stretch therefore deviated from the WT in most of the OriL sequences isolated in vivo. Apparently, a considerable degree of variability in the poly-T region was tolerated in vivo.
A double-stranded stem is required for OriL activity
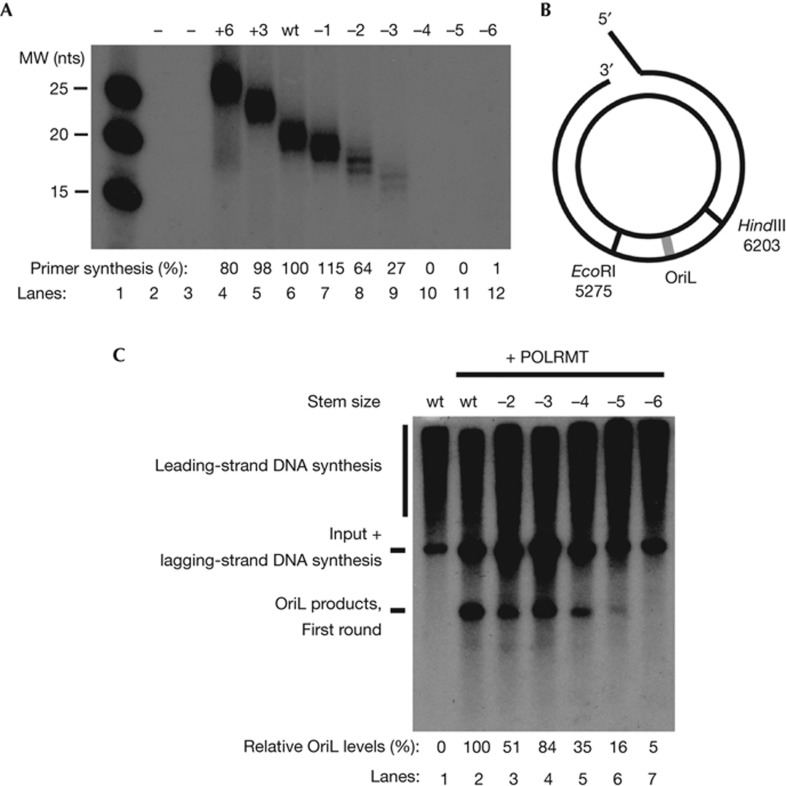
An inventory of human mtDNA sequence variation supported our findings in mice (supplementary Fig S1A online). No mutations were found in the stem region of human OriL, and observed mutations in the loop region were either transitions (purine to purine or pyrimidine to pyrimidine) or T insertions. To better understand the functional consequences of the structural variations observed in vivo, we turned to an in vitro DNA replication system, reconstituted with human mtDNA replication factors. First, we used POLRMT and investigated primer synthesis on single-stranded oligonucleotides corresponding to WT human OriL or mutant derivatives thereof. The stem size of WT human OriL is 11 bp and we could observe robust levels of primer synthesis, when this template was incubated with POLRMT and mtSSB (Fig 2A). The origin could well tolerate increases in stem size to 14 or 17 bp (Fig 2A, lanes 4 and 5). A 1-bp reduction did not affect primer synthesis, but the levels decreased when the stem was reduced with 2 or 3 bp, and primer synthesis was abolished when the length of the stem was decreased with 4 bp (Fig 2A, lanes 7–10). A shorter stem displays a lower ΔG of the stem-loop structure, leading to a destabilization of the active origin conformation at physiological temperatures (supplementary Table S1 online).
Figure 2.
Structural requirements for OriL function. (A) Primase reactions were performed with POLRMT and mtSSB on single-stranded DNA templates in the presence of [α-32P]-GTP to label newly synthesized RNA. The length of the stem size was modified as indicated and the exact sequences are provided in supplementary Table S1 online. After 30 min at 32 °C, reactions were analysed on 25% PAGE/Urea gels as described in experimental procedures. The two lanes adjacent to the marker are empty. The relative efficiency of primer synthesis compared with WT was calculated taking into consideration that the number of incorporated [α-32P]-GTP differs between the individual constructs. (B) The template used for rolling-circle DNA replication contains OriL and surrounding sequences cloned in between the HindIII and EcoRI sites in pBluescript SK (+). (C) DNA replication was performed in the presence of radioactive [α-32P] dCTP to label newly synthesized DNA. The length of the stem was reduced with −2, −3, −4, −5 or −6 bp. The products were analysed by electrophoresis on a 0.7% denaturing agarose gel. A specific product of about 2,100 nts corresponds to initiation of lagging-strand DNA synthesis at OriL. mtSSB, mitochondrial single-stranded DNA-binding protein; MW, molecular weight; nts, nucleotides; PAGE, polyacrylamide gel electrophoresis; POLRMT, mitochondrial RNA polymerase; WT, wild-type.
We also investigated if changes in stem length affected OriL-dependent initiation of lagging-strand DNA synthesis. To this end, we used a template containing a replication fork for loading the leading-strand replication machinery, which consists of a 3,886-bp double-stranded DNA region and a free 3′-overhanging terminus that acts as a primer for leading-strand DNA synthesis [8]. The template also includes an mtDNA fragment containing OriL (Fig 2B). In the presence of mtSSB, TWINKLE and POLγ, we could reconstitute leading-strand DNA synthesis on this template (Fig 2C, lane 1). Addition of POLRMT resulted in OriL-specific lagging-strand DNA synthesis (Fig 2C, lane 2). Reduction of the stem length with 2 or 3 bp still allowed a robust origin activity. Further reduction decreased origin activity, which was completely lost when the length of the stem region approached 6 bp (Fig 2C, lanes 6 and 7). For a detailed explanation of the rolling-circle replication products, please see supplementary Fig S2 online.
Our analysis demonstrates that proper origin function requires a stable stem-loop structure. Even if increased stem length is well tolerated in vitro, we do not observe any mutant versions with longer stem regions in vivo. We can of course not exclude that such structures could be identified if we performed a more extensive mutation screen, but their absence is striking given the extensive in vivo variations we observe in loop size. It is possible that the stability of the stem is a trade-off. A too long palindromic sequence might form a cruciform structure, with two opposite stem-loops extruding from double-stranded DNA, which could cause DNA stability problems. In contrast, mtSSB might denature a less stable stem-loop structure. In fact, the primary role of the OriL structure could be to prevent mtSSB binding and polymerization into the origin region, thus allowing the presentation of the single-stranded loop region to POLRMT for initiation of primer synthesis [1, 8].
A poly-pyrimidine stretch in the OriL stem region
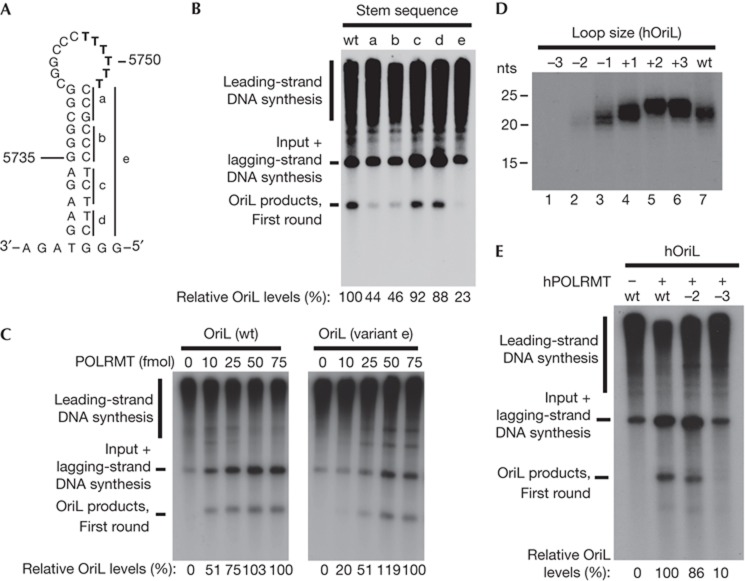
As noted above, there is a distinct sequence bias in the OriL structure, with one side of the stem loop being rich in purines and the other side rich in pyrimidines (19 pyrimidines in a stretch of 20 nts in human OriL). As the detected mutations were centred on the non-template strand, that is, the purine-rich side of the OriL stem, we speculated that this bias could affect primer synthesis activity (Fig 1C). To investigate this, we made a series of mutant constructs in the human OriL, in which parts of the stem sequence was swapped (Fig 3A). The corresponding changes were made in both strands of the stem structure to ensure that the overall stem-loop structure remained intact (supplementary Table S1 online).
Figure 3.
The OriL stem sequence and loop size affect initiation of lagging-strand DNA synthesis. (A) Mutations where done by swapping (C to G, G to C, A to T and T to A) the sets of base pairs indicated (denoted constructs a to d) or as a complete swap of the entire stem sequence (denoted construct e). (B) Activity of WT OriL and the mutant derivatives were monitored as described in Fig 2C, but in the presence of 10 fmol of POLRMT. The relative levels of first-round OriL-dependent lagging-strand DNA synthesis (the 2,100-nt band) were quantified by phopshopimaging and indicated below each lane. (C) DNA replication using wt or mutant OriL (construct e in A) in the presence of increasing concentrations of POLRMT. A specific product of about 2,100 nts corresponds to initiation of lagging-strand DNA synthesis at OriL. (D) Reduction of OriL loop size strongly reduces primase activity. Primase reactions were performed as for Fig 2A. The length of the loop size was modified as indicated and the exact sequences are provided in supplementary Table S1 online. (E) Activity of human OriL (hOriL) and the mutant derivatives with reduced loop size. Reactions were carried out as described in Fig 2C. POLRMT, mitochondrial RNA polymerase; WT, wild-type.
Initiation of lagging-strand DNA synthesis was nearly completely abolished when the entire stem sequence was swapped (Fig 3B, construct e). Changes of the region closest to the base of the stem (constructs c and d) did not have a noticeable effect on OriL activity, whereas mutations in the 6 bp of the stem region closest to the loop (5′-CCGCCC-3′; constructs a and b) reduced OriL activity, albeit not as strongly as the complete stem swap. Interestingly, by increasing the amount of POLRMT, we could overcome the reduced origin activity of the complete stem swap (Fig 3C). Therefore, a pyrimidine-rich template strand is not essential for OriL function but increases the efficiency of primer synthesis.
Our in vitro biochemistry observations were thus in excellent agreement with the in vivo saturation mutagenesis data. We had only observed mutations at two positions in the template strand of the stem and both of these sites were located outside the 6-bp stem region closest to the loop. We also used mouse POLRMT to analyse primase activity on WT and mutant OriL. All naturally occurring sequence variants of OriL were functionally active, further supporting that only active OriL variants are propagated in vivo (supplementary Fig S3A online).
Changes in poly-T region length are well tolerated
Given the extreme variability of the poly-T stretch (loop size) in vivo (Fig 1D), we also monitored if changes in the length of the loop region of OriL could affect primer synthesis in vitro. In agreement with our findings in mice, an increase in length of 1, 2 or 3 nts did not significantly change the efficiency of primer synthesis in vitro (Fig 3D, lanes 2–4). Furthermore, we could still observe near WT levels of primer synthesis when the loop was reduced with 1 nt. However, a 2-nt reduction led to a strong decrease in primer synthesis and further reduction completely abolished primer synthesis (Fig 3D, lanes 6 and 7). We also analysed if changes in the loop size affected OriL-dependent initiation of lagging-strand DNA synthesis (Fig 3E). OriL with a 2-nt reduction of the loop size could initiate lagging-strand DNA synthesis, but with reduced efficiency, whereas a 3-nt reduction inactivated the origin. We repeated these experiments with the corresponding mouse factors and observed similar effects (supplementary Fig S3B,C online). Therefore, all the naturally occurring variants in the T-stretch could efficiently initiate lagging-strand DNA synthesis in vitro.
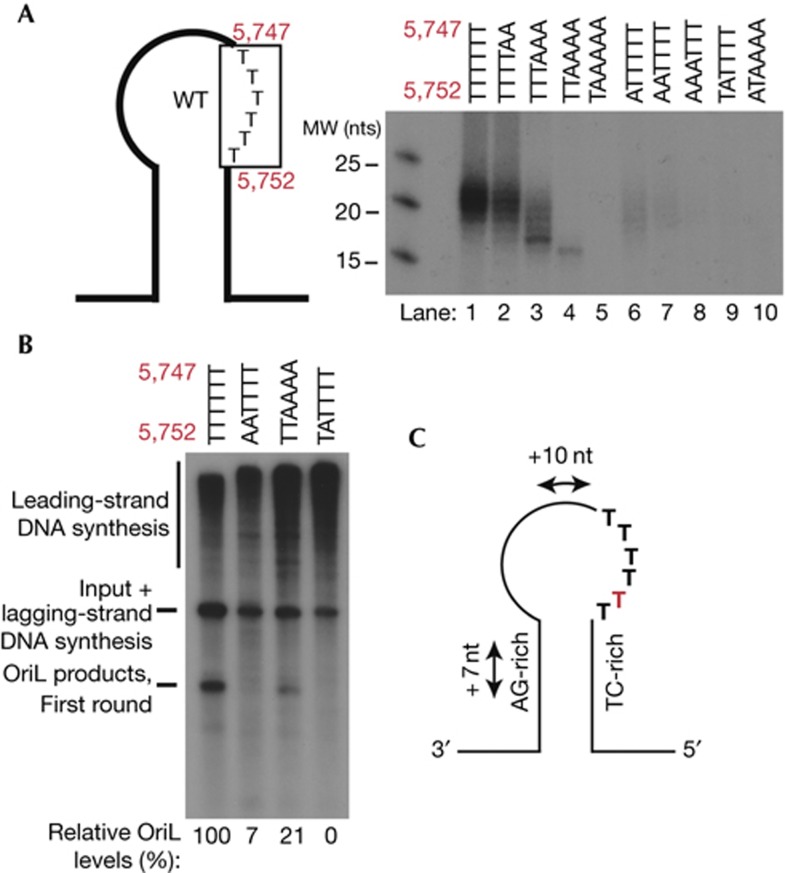
Poly-T stretch mutations affect primer synthesis
The human poly-T stretch (nts 5,747–5,752) is essential for primer synthesis, but the importance of individual positions is not known [8]. To address this, we used the primase assay on a series of mutant constructs (Fig 4A; supplementary Table S1 online). Even if all mutations in the T-stretch led to a decrease in primer synthesis, only one position was absolutely required for primer synthesis, T5751, the second T distal to the double-stranded stem (Fig 4A, lanes 5–10). In agreement with this finding, no in vivo mutations had been observed in the part of the T-stretch that was located proximal to the stem (Fig 1C). We also monitored effects of T-stretch mutations on OriL activity in vitro. In agreement with the primer synthesis assays, initiation of DNA synthesis at OriL was severely reduced, but could still be observed when the 4 T’s at position 5,747–5,750 were mutated (Fig 4B, lane 3). In contrast, mutations affecting T5,751 completely abolished OriL activity (Fig 4B, lane 4), demonstrating that this position is essential for primer synthesis and initiation of lagging-strand DNA synthesis. The sequence requirements of a functional human OriL are summarized in Fig 4C and include a loop region of at least 10 nts and a stable double-stranded stem region with pyrimidine-rich template strand. A stretch of Ts is also required, but only the T at position 5,751 is essential for primer synthesis.
Figure 4.
Primer synthesis requires specific positions in the poly-T stretch. (A) Effect of poly-T stretch mutations on POLRMT primase activity. Reactions were performed as in Fig 2A and template sequences are listed in supplementary Table S1 online. Positions of the individual Ts are indicated in the schematic representation of OriL. (B) Human mtDNA nts 5,751 and 5,752 are critical for OriL-specific initiation of lagging-strand DNA synthesis. The reactions were performed as in Fig 2C. (C) A schematic picture summarizing the structural requirements of human OriL. The essential T at position 5,751 is indicated in red. MW, molecular weight; nts, nucleotides; WT, wild-type.
OriL is functionally conserved in evolution
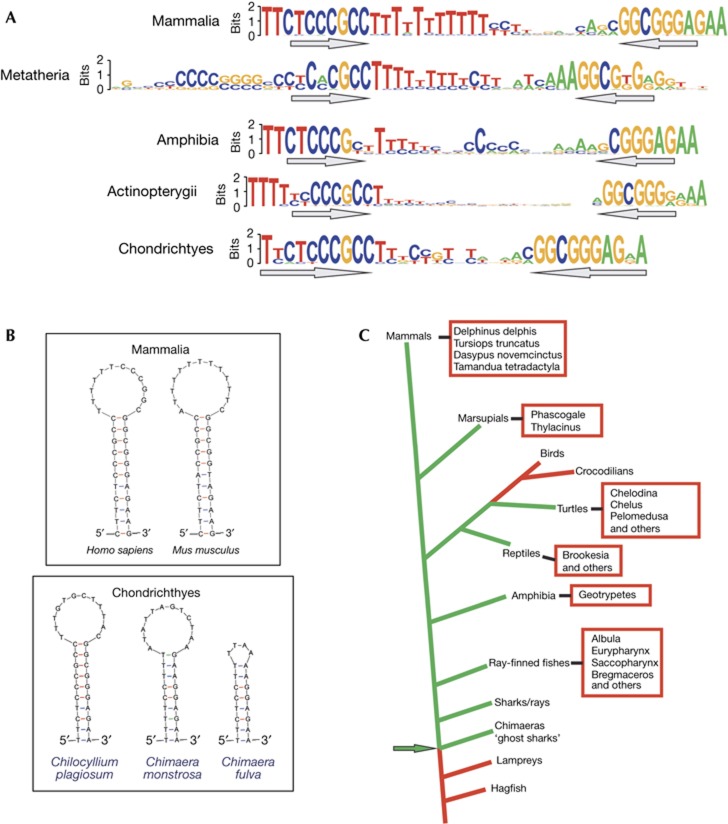
To follow-up our in vivo and in vitro analyses of OriL sequences, we analysed all available vertebrate mitochondrial genome sequences, representing 1,802 different species. Most of these genomes are carefully annotated and in many cases the OriL sequence was easily identified, either because it has been annotated as such or because of its strongly conserved location between the tRNA-Asn and tRNA-Cys genes. In certain phyla or individual species, OriL was not annotated or not present in the expected genomic location (supplementary Fig S4A online). Therefore, we also used sequence-based prediction of OriL structures. First, we employed an in-house method to identify hairpin structures with features characteristic of OriL, such as the C-rich 5′ strand, which was also important for the in vitro activity of the origin. Second, the Infernal software was used with covariance models on the basis of previously known OriL sequences [16]. Third, as our in vitro experiments had demonstrated that an efficient OriL should be thermodynamically stable, we scanned the mitochondrial genome for stable hairpin structures with the aid of UNAFold [17] (supplementary Fig S4B online).
Our analysis revealed that OriL is strongly conserved throughout vertebrates. The structure of OriL in sharks (Chondrichtyes) is very similar to that in mammals (Fig 5A,B). The deepest branching phyla where OriL seems to occur are Chimaera (ghost sharks) and Elasmobranchii (sharks, rays and skates). OriL is not present in non-vertebrates closely related to vertebrates, such as Branchiostoma (a lancelet) and Ciona (sea squirt), suggesting that OriL is an early invention in vertebrate evolution (Fig 5C).
Figure 5.
OriL bioinformatics. (A) Sequence logo of OriL from different families of vertebrates. (B) Predicted stem-loop structures for OriL sequences found in the indicated species. (C) Evolutionary conservation of OriL species marked in green, as opposed to species shown in red, harbour an identifiable OriL sequence in their mitochondrial genomes. Our complete inventory of OriL in vertebrates can be found in supplementary Table S2 online.
The bioinformatic analysis of conserved sequence elements was remarkably consistent with our findings in vitro. In addition to a stable stem-loop structure, we noted that the length of the poly-T stretch was highly variable, but there is a highly conserved T proximal to the pyrimdine-rich stretch in the stem (Fig 5A). The pyrimidine-rich sequence has an almost universally conserved CCCGCC motif, which stimulate POLRMT primer synthesis (Fig 3A,B). A poly-pyrimidine stretch on the template strand can also be observed immediately downstream of the transcription start site at LSP, suggesting that POLRMT requires a stretch of purines in the nascent transcript to efficiently initiate RNA synthesis [18]. The pyrimidine stretch could perhaps have a role in the actual primer synthesis, by stimulating the transition to transcription elongation. In support of this notion, a poly-purine stretch in nascent RNA stimulates RNA polymerase III transcription, by stimulating the formation of a more stable ternary complex [19]. Future studies are required to understand the transition between origin binding and primer synthesis by POLRMT.
METHODS
For a more detailed description of experimental details, please refer to supplementary information online.
Recombinant proteins. TWINKLE, mtSSB, POLγA, POLγB, POLRMT (Human and Mouse version) were expressed and purified as described previously [12, 13, 18].
In vitro priming on single-stranded DNA oligonucleotides. The reaction mixture contained 100 fmol single-stranded DNA oligonucleotide, 500 fmol POLRMT and 1 pmol mtSSB. After 30 min incubation at 32 °C, 12 units of RNase-free DNase I (Qiagen) was added. Reactions were processed and analysed as previously described for in vitro transcription reactions [18].
Rolling-circle mtDNA replication. Reactions were carried out as described previously [8]. The reaction mixtures (20 μl) contained 10 fmol of the double-stranded DNA template and 100 fmol TWINKLE, 250 fmol POLγA, 375 fmol POLγB (calculated as a dimer), 5 pmol mtSSB and 250 fmol POLRMT if nothing else is indicated in the figure legends. Construction of DNA templates are described in supplementary information online.
Mutation load analysis. For sequencing of mtDNA, we isolated tissue from eight genetically modified mice, carrying a proofreading-deficient form of the mitochondrial DNA polymerase γ (PolγAD257A/PolAγAD257A). The target region for the analysis was amplified by PCR and the products cloned using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen). A total of 1,747 clones (∼1,700,000 bp of sequence) were sequenced.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This study was supported by Swedish Research Council grants (to NGL, CMG, SW and MF), the Swedish Cancer Foundation (to MF and CMG), Leducq Foundation (to NGL), an European Research Council Advanced Investigator joint grant (to NGL and CMG), an European Research Council Starting Independent Investigator grant (to MF), the Swedish Society for Medical Research (SW) and a Marie Curie grant from the European Commission (SW).
Author contributions: SW, JMF, JBS, TS, NGL, CMG and MF designed research; SW, JMF, JBS, PHW, TS and MF performed research; SW, JMF, JBS, PHW, TS, CMG and MF analysed data; and SW, JMF, JBS, TS, NGL, CMG and MF wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Wanrooij S, Falkenberg M (2010) The human mitochondrial replication fork in health and disease. Biochim Biophys Acta 1797: 1378–1388 [DOI] [PubMed] [Google Scholar]
- Stumpf JD, Copeland WC (2011) Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell Mol Life Sci 68: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ (2002) Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111: 495–505 [DOI] [PubMed] [Google Scholar]
- Holt IJ, Lorimer HE, Jacobs HT (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100: 515–524 [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Yang MY, Jacobs HT, Holt IJ (2005) A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell 18: 651–662 [DOI] [PubMed] [Google Scholar]
- Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA (2005) Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev 19: 2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28: 693–705 [DOI] [PubMed] [Google Scholar]
- Fuste JM et al. (2010) Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell 37: 67–78 [DOI] [PubMed] [Google Scholar]
- Wong TW, Clayton DA (1985) In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell 42: 951–958 [DOI] [PubMed] [Google Scholar]
- Wong TW, Clayton DA (1986) DNA primase of human mitochondria is associated with structural RNA that is essential for enzymatic activity. Cell 45: 817–825 [DOI] [PubMed] [Google Scholar]
- Tapper DP, Clayton DA (1981) Mechanism of replication of human mitochondrial DNA. Localization of the 5’ ends of nascent daughter strands. J Biol Chem 256: 5109–5115 [PubMed] [Google Scholar]
- Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M (2008) Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci USA 105: 11122–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M (2004) Reconstitution of a minimal mtDNA replisome in vitro. EMBO J 23: 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileshina D, Ibrahim N, Boesch P, Lightowlers RN, Dietrich A, Weber-Lotfi F (2011) Mitochondrial transfection for studying organellar DNA repair, genome maintenance and aging. Mech Ageing Dev 132: 412–423 [DOI] [PubMed] [Google Scholar]
- Trifunovic A et al. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM (2004) The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J 23: 4606–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecherle GN, Whelen S, Hall BD (1996) Purines are required at the 5' ends of newly initiated RNAs for optimal RNA polymerase III gene expression. Mol Cell Biol 16: 5801–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.