Abstract
Taspase1 is a threonine protease responsible for cleaving MLL (Mixed-Lineage Leukemia) to achieve proper HOX gene expression. Subsequent studies identified additional Taspase1 substrates including Transcription Factor IIA (TFIIA) and Drosophila HCF. Taspase1 is essential for cell proliferation and is overexpressed in many cancer cell lines. Currently no small molecule inhibitors of this enzyme have been described. Here we report the synthesis and evaluation of vinyl sulfone, vinyl ketone, epoxy ketone, and boronic acid inhibitors designed based on the preferred Taspase1 cleavage site (Ac-Ile-Ser-Gln-Leu-Asp). Specifically, we evaluated compounds in which the reactive warhead is positioned in place of the P1 aspartic acid sidechain as well as at the C-terminus of the peptide. Interestingly, both classes of inhibitors were effective and vinyl ketones and vinyl sulfones showed the greatest potency for the target protease. These results suggest that Taspase1 has unique substrate recognition properties that could potentially be exploited in the design of potent and selective inhibitors of this enzyme.
Taspase1 is a highly conserved threonine protease that was initially purified based on its ability to cleave the MLL (mixed-lineage leukemia) protein at conserved (QXD/G) sites.1 MLL encodes a 500kD nuclear coactivator that regulates embryogenesis, cell cycle and stem cell growth.2 Deregulation of MLL by chromosome band 11q23 translocation leads to human leukemia with poor prognosis. Key MLL targets include Hox and Cyclin genes.3 Proteolysis of MLL leads to the formation of a stable heterodimer that localizes to the nucleus where it acts as a histone H3 K4 methyl transferase (HMT). Noncleavage of MLL results in a hypomorphic MLL with impairment in its HMT activity.4 Taspase1 is the only protease in mammals capable of proteolytically activating MLL, as demonstrated by the inability of Taspase1-deficent mice to cleave MLL resulting in homeotic transformations.4 In addition to MLL, we have identified MLL2, TFIIA, and Drosophila HCF as bona fide Taspase1 substrates.5 Taspase1 regulates cell cycle gene expression through cleavage-mediated substrate activation and has been shown to be essential for cell proliferation.4 Furthermore, Taspase1 is overexpressed in many cancer cell lines, and Taspase1-deficient cells are resistant to common oncogenic transformation.4 Given these findings, chemically inhibiting Taspase1 function may lead to anticancer therapeutics. However, Taspase1 has proven resistant to inhibition by general classes of serine, cysteine and metallo protease inhibitors.1b
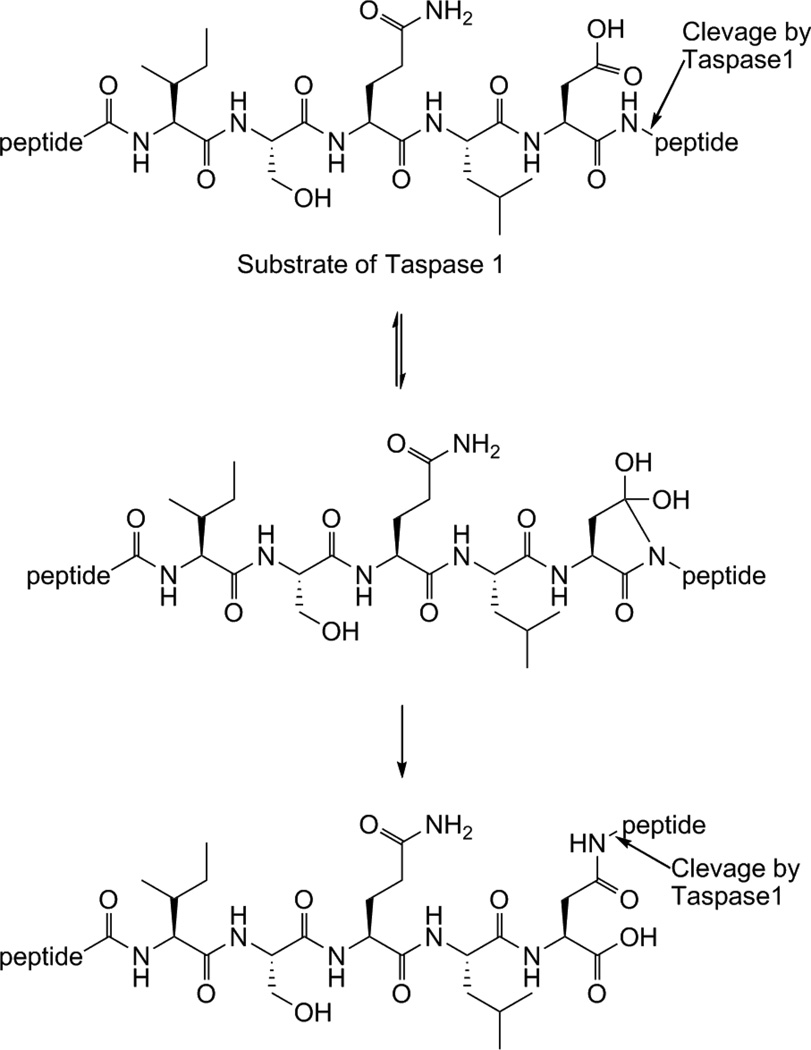
The activity of Taspase1 itself is regulated by proteolysis. It is expressed as a proenzyme that undergoes autoproteolysis to its active form.1b The crystal structure of human Taspase1 revealed significant conformational differences between the proenzyme and the active conformer. The proenzyme starts as a homodimer that is hydrolyzed into a 28kDa α and a 22kDa β subunit that produce the hetero-tetrameric active form of Taspase1.6 Interestingly, Taspase1 only shows homology to the L-asparaginase_2 family of hydrolyases. However, unlike other members of this family, it has endopeptidase activity. Taspase1 uses a threonine residue as its active site nucleophile to cleave peptide bonds C-terminal to an aspartate residue.1b In addition, Taspase1 requires a glycine residue directly C-terminal to the aspartate residue. Two Taspase1 cleavage sites have been identified on MLL (CS1 and CS2). The conserved sequence for CS2 is Ile-Ser-Gln-Leu-Asp/Gly-Val-Asp-Asp, and CS1 is Glu-Gly-Gln-Val-Asp/Gly-Ala-Asp-Asp, with the CS2 site being more optimal for cleavage.1b The fact that Taspase1 has homology to asparaginases, enzymes that hydrolyze the amide sidechain of asparagine to generate aspartic acid, suggests that it may also favor cleavage of isopeptide bonds on a substrate. Furthermore, the requirement of a glycine at the P1’ position may be explained by the need for a small residue to facilitate peptide bond transfer from the main peptide backbone amide to the aspartic acid sidechain of a substrate. A possible substrate rearrangement to produce two isoforms for cleavage by Taspase1 is illustrated (Fig. 1).
Figure 1.
A potential peptide rearrangement to yield two substrate isoforms for cleavage by Taspase1. The presence of an Asp-Gly sequence may facilitate internal transfer of the peptide bond to the sidechain of Asp. This would result in a substrate that resembles asparagine and that would require hydrolysis at the sidechain amide, similar to how asparaginases function.
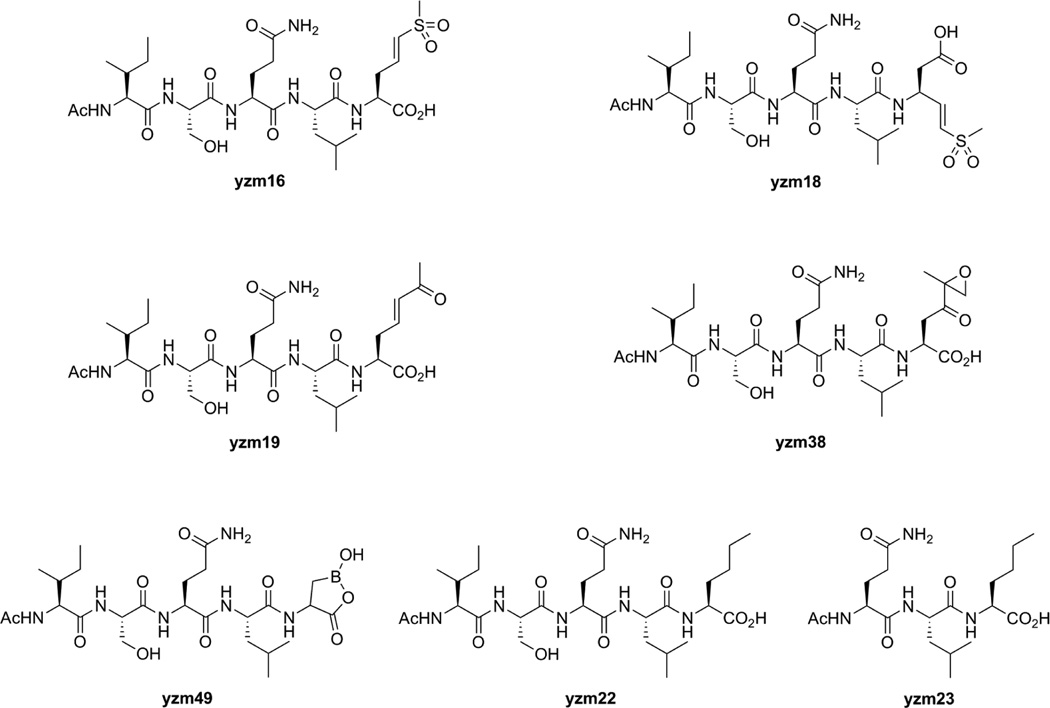
In this report, we describe the design, synthesis, and evaluation of Taspase1 inhibitors that contain a general scaffold based on the native cleavage site of MLL linked to a number of different protease-specific reactive functional groups. These inhibitors were designed to determine both the optimal “warhead” group as well to determine if placement of this group at the Asp sidechain enhanced reactivity (Fig. 2). We chose vinyl sulfones, expoxy ketones and boronates because all of these functional groups have proven to be efficient for inhibition of the catalytic threonine of the proteasome.7 Initially we synthesized a vinyl sulfone (yzm16), vinyl ketone (yzm19), epoxy ketone (yzm38) and boronic acid (yzm49) at the side chain of the P1 aspartic acid. For the vinyl sulfone warhead, we also synthesized a compound in which the reactive group was placed in the main peptide backbone, as has been described for standard protease inhibitors (yzm18).8 As control compounds, we replaced aspartic acid with norleucine to generate Ac-Ile-Ser-Gln-Leu-Nle-OH (yzm22) and Ac-Gln-Leu-Nle-OH (yzm23).
Figure 2.
Taspase1 Inhibitors (yzm16, 18, 19, 38, 49) and control compounds (yzm22, 23) used in this study.
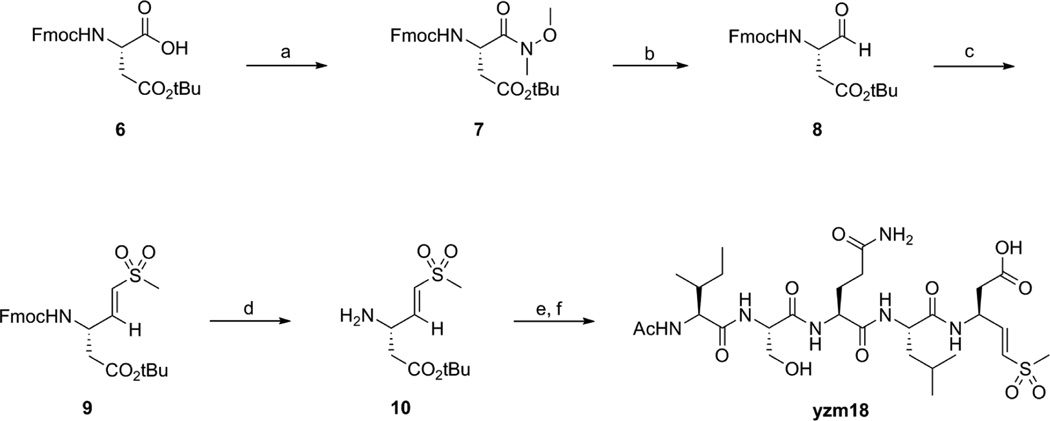
Scheme 1 and Scheme 2 outline the synthesis of yzm16 (Ac-Ile-Ser-Gln-Leu-Asp(vs)-OH) and yzm18 (Ac-Ile-Ser-Gln-Leu-Asp-vs) from Fmoc-Asp-OtBu and Fmoc-Asp(tBu)-OH respectively. The acids were converted to Weinreb amides 2 and 7 by N,O-dimethylhydroxyl-amine and then reduced to aldehydes 3 and 8 by DIBAL. Compounds 3 and 8 were coupled with diethyl methylsulfonylmethylphophonate9 to produce the vinyl sulfone compounds 4 and 9. After deprotection of the Fmoc group in a mixture of diethylamine and acetonitrile, the resulting amines were coupled with the protected peptide (Ac-Ile-Ser(tBu)-Gln(Trt)-Leu-OH) using standard peptide coupling conditions. Finally, the protecting groups were removed in TFA/TIS/H2O mixture to yield the desired products yzm16 and yzm18 that were further purified by HPLC.
Scheme 1.
Reagents and conditions: (a) N,O-Dimethylhydroxylamine hydrochloride, HBTU, DIPEA, DMF, rt; (b) DIBAL, THF, −10°C; (c) diethyl methylsulfonylmethylphosphonate, LHMDS, THF, −10°C; (d) diethylamine, acetonitrile, rt; (e) Ac-IS(tBu)Q(Trt)L-OH, HBTU, HOBt, DIPEA, DMF, rt; (f) TFA/TIS/H2O = 95/2.5/2.5.
Scheme 2.
Reagents and conditions: (a) N,O-Dimethylhydroxylamine hydrochloride, HBTU, DIPEA, DMF, rt; (b) DIBAL, THF, −10°C; (c) diethyl methylsulfonylmethylphosphonate, LHMDS, THF, −10°C; (d) diethylamine, acetonitrile, rt; (e) Ac-IS(tBu)Q(Trt)L-OH, HBTU, HOBt, DIPEA, DMF, rt; (f) TFA/TIS/H2O = 95/2.5/2.5.
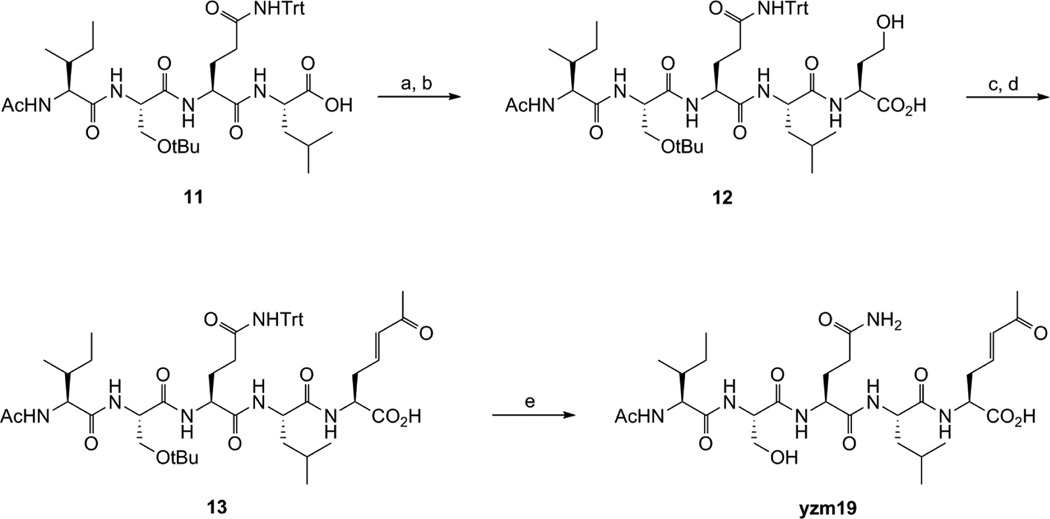
The vinyl ketone compound yzm19 was synthesized as shown in Scheme 3. Homoserine was coupled with Ac-Ile-Ser(tBu)-Gln(Trt)-Leu-OH to produce 12. The alcohol group was converted to the aldehyde by Dess-Martin Periodinane and then reacted with 1-triphenyl-phosphoranylidene-2-propanone by Wittig reaction to generate vinyl ketone compound 13. After deprotection of the protecting groups, HPLC purification afforded the desired compound yzm19.
Scheme 3.
Reagents and conditions: (a) NHS, DIC, DCM, rt; (b) Homoserine, NaHCO3, acetone/H2O, rt; (c) Dess-Martin Periodinane, DCM, rt; (d) 1-Triphenylphosphosphoranylidene-2-propanone, DCM/EtOAc, rt; (e) TFA/TIS/H2O = 95/2.5/2.5
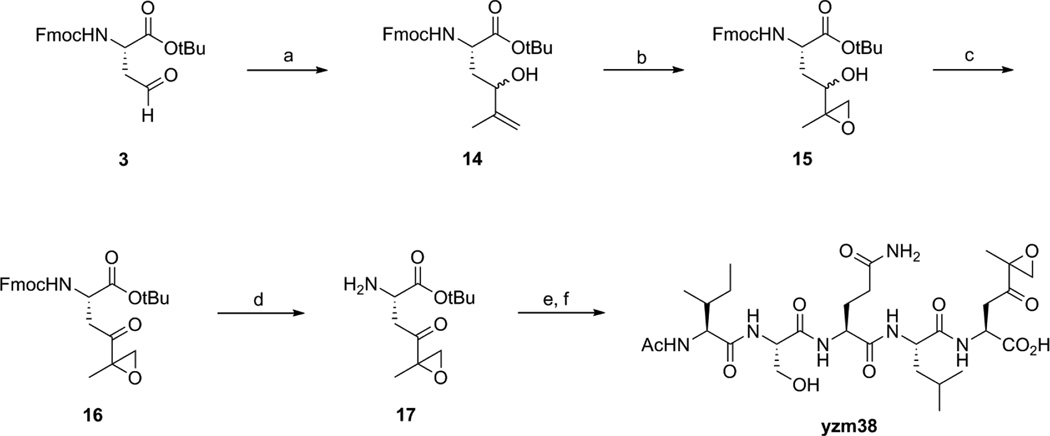
Scheme 4 shows the synthesis of epoxy ketone compound yzm38. It shares a common intermediate 3 with yzm16. Compound 14 was generated by Grignard reaction with 3 and isopropenylmagnesium bromide. The double bond was converted to an epoxide by t-butyl hydroperoxide to produce 15. The racemic alcohol group of 15 was transformed to ketone compound 16 by Dess-Martin Periodinane. The Fmoc deprotected compound 17 was coupled with Ac-Ile-Ser(tBu)-Gln(Trt)-Leu-OH. The compound was subsequently deprotected and HPLC purified to yield yzm38.
Scheme 4.
Reagents and conditions: (a) isopropenylmagnesium bromide, THF, −10°C; (b) vanadyl acetylacetonate, t-butyl hydroperoxide, DCM, 0°C; (c) Dess-Martin Periodinane, DCM, rt; (d) diethylamine, acetonitrile, rt; (e) HBTU, HOBt, DIPEA, DMF, rt; (f) TFA/TIS/H2O = 95/2.5/2.5
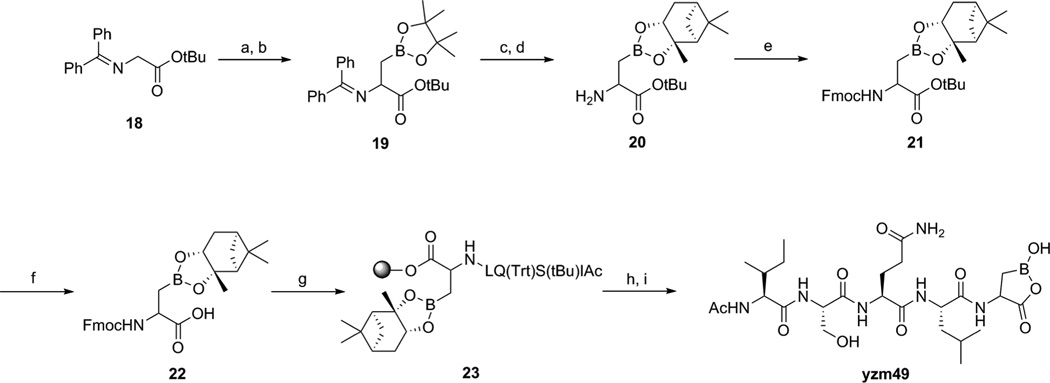
The boronic acid compound yzm49 was synthesized as shown in Scheme 5. First, commercially available N-(diphenylmethylene)glycine t-butyl ester 18 was treated with LHMDS and then 1 hour later addition of 2-(iodomethyl)-4,4,5,5-tetramethyl-1,3,2-dioxaborolane10 to this mixture led to compound 19. The deprotection of diphenylmethylene group gave rise to the deprotection of the boronic acid group. Without the protection of the boronic acid, the coupling reaction with Ac-Ile-Ser(tBu)-Gln(Trt)-Leu-OH did not occur. This problem was overcome by protection of the boronic acid with (+)-pinanediol after removal of the diphenylmethylene group generating compound 20.11 After Fmoc protection of the free amine, the t-butyl group of 21 was removed and the resulting compound 22 was loaded onto 2-chlorotrityl chloride resin. The peptide sequence 23 was then prepared on resin using standard SPPS. Cleavage from resin and deprotection of the protecting groups afforded the desired compound yzm49.
Scheme 5.
Reagents and conditions: (a) LHMDS, THF, −78°C; (b) 2-(iodomethyl)-4,4,5,5-tetramethyl-1,3,2-dioxaborolane, rt; (c) 0.2N HCl; (d) (+)-pinanediol; (e) Fmoc-OSu, acetone, rt; (f) TFA/DCM = 2/5; (g) SPPS; (h) TFA/DCM = 1/99; (i) TFA/TIS/H2O = 95/2.5/2.5
All these peptide derivatives were tested in vitro to determine their activity against Taspase1. The IC50 values presented in Table 1 were determined using a Fluorescence Resonance Energy Transfer (FRET) based reporter assay. The FRET-based assay employs a reporter substrate (MCA-KISQLDGVDD-DNP) containing a fluorogenic coumarin (MCA) group and a 2,4-dinitrophenol (DNP) quenching group. Upon Taspase1-mediated cleavage, the quencher is removed, resulting in fluorescence. These results were further confirmed by an in vitro cleavage assay utilizing a modified aa 2,500–2,800 fragment of MLL (Fig. 3). For both assays, yzm18 and yzm19 exhibited the strongest inhibitory activity (IC50 29.4 µM, 63.4 µM respectively), whereas yzm16 and yzm38 were only minimally active (IC50 >100 µM). The activity of the boronate yzm49 could not be determined because its poor solubility prevented measurement of activity at concentrations over 100 µM. We also measured the inhibition rates of taspase1 by yzm18 after different amounts of pre-incubation time and found that inhibition was not time dependent. This result suggests that yzm18 is not likely to be acting as a covalent irreversible inhibitor against this target (See Supplemental Fig. 8). The vinyl sulfone is most active when placed on the P1 side-chain whereas the vinyl ketone showed relatively strong activity when placed in the P1 side-chain position. Taken together, our study demonstrates that Taspase1 is capable of binding inhibitors that contain reactive warheads on either the main peptide backbone or in place of the P1 sidechain. This suggests that Taspase1 may be able to cleave substrates that have undergone peptide backbone rearrangement. This rather unique property may facilitate the design of highly selective inhibitors. While the compounds presented here are not highly potent, they serve as a starting point for further inhibitor design and may be highly valuable for future structural studies of the enzyme with inhibitors bound in the active site. We are currently working to validate the selectivity of the most potent compound yzm18 in cell culture models.
Table 1.
Inhibition IC50 values for yzm16, 18, 19, 22, 23, 38, 49 against recombinant taspase1 as measured using a quenched fluorescent substrate. Compounds were added simultaneously with substrate to Taspase1 (see supplemental methods).
| Compds | IC50, µMa |
|---|---|
| yzm16 | >100 |
| yzm18 | 29.40 (1.04) |
| yzm19 | 63.38 (1.65) |
| yzm38 | >100 |
| yzm49 | n.d.a. |
| yzm22 | n.d.a. |
| yzm23 | n.d.a. |
Values are means of three experiments, standard deviation is given in parentheses (n.d.a. = no detectable activity at max concentration (100µM) used).
Figure 3.
In vitro cleavage assay. A construct expressing a fragment of p45MLL from containing residues 2,500 to 2,800 with the CS1 site mutated to prevent taspase1 cleavage was used for in vitro translation. The resulting 35S-labled protein is cleaved from the p45 form to the p33 fragment upon addition of rTaspase1. For inhibition assays, compounds indicated were preincubated with rTaspase1 at the indicated concentrations for 30 min prior to addition to the in vitro translated p45MLL. Inhibition was observed as a loss of processing and accumulation of the full-length p45 fragment.
Supplementary Material
Acknowledgements
This work was supported by an NIH National Technology Center for Networks and Pathways grant U54 RR020843 (to M.B.), NIH grant R01-EB005011 (to MB).
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:XXXXXXXX
References and notes
- 1.a Hsieh JJD, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Mol. Cell. Biol. 2003;23:186. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hsieh JJD, Cheng EHY, Korsmeyer SJ. Cell. 2003;115:293. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Takeda S, Cheng EHY, Hsieh JJD. Cell Cycle. 2008;7:428. doi: 10.4161/cc.7.4.5426. [DOI] [PubMed] [Google Scholar]
- 3.a Krivtsov AV, Armstrong SA. Nat. Rev. Cancer. 2007;7:823. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]; b Liu H, Cheng EHY, Hsieh JJD. Genes Dev. 2007;21:2385. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, Kan JT, Korsmeyer SJ, Cheng EHY, Hsieh JJD. Genes Dev. 2006;20:2397. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Zhou H, Spicuglia S, Hsieh JJD, Mitsiou DJ, Hoiby T, Veenstra GJC, Korsmeyer SJ, Stunnenberg HG. Mol. Cell. Biol. 2006;26:2728. doi: 10.1128/MCB.26.7.2728-2735.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Capotosti F, Hsieh JJD, Herr W. Mol. Cell. Biol. 2007;27:7063. doi: 10.1128/MCB.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan JA, Dunn BM, Tong L. Structure. 2005;13:1443. doi: 10.1016/j.str.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 7.a Verdoes M, Florea BI, Van der Linden WA, Didier R, Van den Nieuwendijk AMCH, Van der Marel GA, Overkleeft HS. Org. Biomol. Chem. 2007;5:1416. doi: 10.1039/b702268a. [DOI] [PubMed] [Google Scholar]; b Watanabe T, Momose I, Abe M, Abe H, Sawa R, Umezawa Y, Ikeda D, Takahashi Y, Akamatsu Y. Bioorg. Med. Chem. Lett. 2009;19:2343. doi: 10.1016/j.bmcl.2009.02.117. [DOI] [PubMed] [Google Scholar]
- 8.Abdulla MH, Lim KC, Sajid M, McKerrow JH, Caffrey CR. PLoS Med. 2007;4:e14. doi: 10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikolajczyk M, Balczewski P. Synthesis. 1989:101. doi: 10.1021/ol005742b. [DOI] [PubMed] [Google Scholar]
- 10.Roy CD, Soundararajan R, Brown HC. Monatsh. Chem. 2008;139:241. [Google Scholar]
- 11.LeBeau AM, Singh P, Isaacs JT, Denmeade SR. Chem. Biol. 2008;15:665. doi: 10.1016/j.chembiol.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.