SUMMARY
The C. elegans MES proteins are key chromatin regulators of the germline. MES-2, MES-3, and MES-6 form the C. elegans Polycomb Repressive Complex 2 and generate repressive H3K27me3. MES-4 generates H3K36me3 on germline-expressed genes. Transcript profiling of dissected mutant germlines revealed that MES-2/3/6 and MES-4 cooperate to promote expression of germline genes and silence the X chromosomes and somatic genes. Based on genome-wide chromatin immunoprecipitation, H3K27me3 and H3K36me3 occupy mutually exclusive domains on the autosomes and H3K27me3 is enriched on the X. Loss of MES-4 from germline genes causes H3K27me3 to spread to germline genes, resulting in reduced H3K27me3 elsewhere on the autosomes and especially on the X. Our findings support a model in which H3K36me3 repels H3K27me3 from germline genes and concentrates it on other regions of the genome. This antagonism ensures proper patterns of gene expression for germ cells, which includes silencing somatic genes and the X chromosomes.
INTRODUCTION
A major goal for understanding how cell fates are specified and how cells can be reprogrammed to new fates is defining how chromatin states influence gene expression. Tissue-appropriate patterns of gene expression require that genes needed for tissue development reside in chromatin that can be accessed by transcription factors and transcribed by polymerases, while other genes must be kept in a repressed chromatin state. Two histone marks that are signatures of expressed and repressed chromatin are histone H3 trimethylated on Lys 36 (H3K36me3) and on Lys 27 (H3K27me3), respectively. H3K36me3 is introduced cotranscriptionally upon the passage of RNA Polymerase II through genes; this mark can also be epigenetically maintained on genes in the absence of ongoing transcription (Furuhashi et al., 2010; Krogan et al., 2003; Li et al., 2003; Rechtsteiner et al., 2010). H3K27me3 is a well-established mark of repressed chromatin that can be propagated in an epigenetic manner (Hansen et al., 2008; Lanzuolo et al., 2011; Margueron and Reinberg, 2011). H3K36me3 and H3K27me3 marks generally occupy non-overlapping regions of genomes (Ernst and Kellis, 2010; Kharchenko et al., 2011; Liu et al., 2011; this study). The notion that these marks may regulate each other's distribution is supported by two types of evidence. First, prior methylation of H3K36 prevents methylation of K27 on the same histone tails in vitro (Schmitges et al., 2011; Yuan et al., 2011). Second, Drosophila ash1 mutants, which are thought to be defective in H3K36 methylation, show spreading of H3K27me3 into and silencing of the Ultrabithorax gene (Klymenko and Muller, 2004; Papp and Muller, 2006; Tanaka et al., 2007). These in vitro and single-gene studies suggest that H3K36 methylation antagonizes H3K27 methylation, and that H3K27 methylation is otherwise a default modification. We sought to test this model in vivo on a genome-wide scale, and to examine the effects on gene expression patterns of removing H3K36me3 or H3K27me3 or both.
The C. elegans MES proteins are essential chromatin regulators in germ cells (Capowski et al., 1991). MES-2, MES-3, and MES-6 form the C. elegans version of the widely conserved Polycomb Repressive Complex 2 and generate repressive H3K27me3 (Bender et al., 2004; Ketel et al., 2005; Xu et al., 2001). MES-4, a homolog of the vertebrate NSD proteins, generates H3K36me3 on genes expressed in the germline (Furuhashi et al., 2010; Rechtsteiner et al., 2010). Loss of any of the four MES proteins causes germ cells to die. MES regulation is maternal effect: maternally provided MES(+) product promotes development of a fertile germline, while absence of maternal MES(+) product leads to death of nascent germ cells and sterile adults (Capowski et al., 1991). Previous studies focused attention on MES regulation of the X chromosomes. The X chromosomes in XX hermaphrodites and XO males are considered to be globally “silenced” during most stages of germ cell development. This is supported by the finding that histone marks associated with active gene expression decorate the autosomes but are not detected on the X chromosomes in mitotic, early meiotic, and spermatogenic germ cells, and by the low expression of X-linked genes compared to autosomal genes in dissected germlines (Kelly et al., 2002; Wang et al., 2009). Perhaps as a consequence of X silencing, germline-expressed genes are significantly under-represented on the X compared to the five autosomes (Reinke et al., 2004). The MES proteins participate in X silencing, as illustrated by the spread of marks of active chromatin to the Xs in immunostained mes-2, mes-3 and mes-6 mutant germ nuclei and the up-regulation of X-linked genes in dissected mes-4 germlines (Bender et al., 2006; Fong et al., 2002). The apparently similar involvement of the four MES proteins in X silencing is puzzling given their strongly asymmetric distributions: MES-4 and H3K36 methylation are strikingly enriched on the five autosomes and nearly absent from the X, while MES-2/3/6-generated H3K27me3 is modestly enriched on the X (Bender et al., 2004; Bender et al., 2006; Fong et al., 2002).
This paper explores how autosomally-concentrated MES-4 and X-enriched MES-2/3/6 activity contribute to silencing the Xs, identifies autosomal targets of MES regulation, and tests the model that H3K36 methylation generated by MES-4 repels H3K27me3 from germline-expressed genes. Our findings reveal how antagonistic histone modifiers can shape genome organization and tissue-appropriate gene expression patterns, and lay the foundation for understanding how loss of MES regulation and the resulting altered chromatin landscape renders germ cells susceptible to conversion to somatic cells (Patel et al., 2012).
RESULTS AND DISCUSSION
MES-4 and MES-2/3/6 cooperate to silence the X chromosomes in the germline
To better understand when and how maternal MES(+) function promotes development of a functional germline, we determined how long maternally encoded MES proteins persist in the absence of zygotically produced protein. In confocal images, MES-2 and MES-4 persisted at easily detectable levels in the primordial germ cells of newly hatched L1 larvae, but were undetectable in L2 (MES-4) and L3 (MES-2) larvae (Figure 1A, S1A,B). Their histone modifications became undetectable in L3 (H3K36me2) and L4 (H3K27me3) larvae. Quantification of immunostaining pixel intensity in L4 germ nuclei showed that in mes-4 mutants, MES-4 was reduced to 3.4% and H3K36me2 to 9.5% of wild type, and in mes-2 mutants, MES-2 was reduced to 1.3% and H3K27me3 to 1.9% of wild type (Figure S1C). These results reveal that in homozygous mes mutants, maternal MES protein and their histone modifications persist through activation of transcription in the primordial germ cells, commencement of germ cell proliferation in late L1s, and initiation of meiosis in L3s. The resulting adult germlines can produce oocytes and progeny but display compromised health (Capowski et al., 1991; Xu and Strome, 2001). These adults offered an opportunity to examine gene expression changes that follow larval loss of MES proteins and their methyl marks (Figure 1).
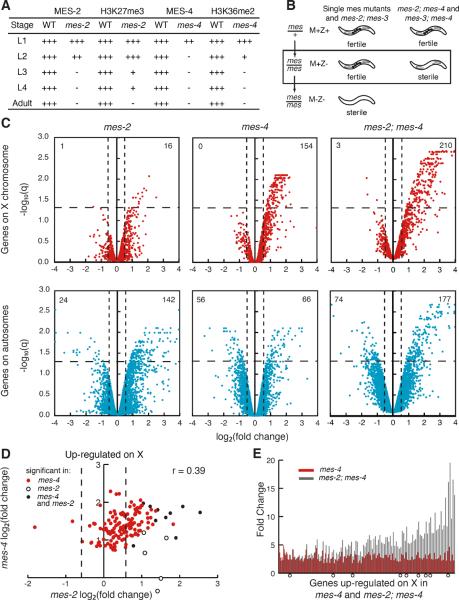
Figure 1.
Microarray analysis of germlines from mes-2, mes-4, and mes-2; mes-4 mutants compared to wild type. (A) Perdurance (+) of maternal MES proteins and histone marks in mes-2 and mes-4 mutants. See also Figure S1. (B) Summary of phenotypes of mes single and double mutants. M=maternal supply, Z=zygotic expression. See also Figure S2. Germlines were dissected from M+Z− mes mutants (box). (C) Volcano plots showing log2 of the fold change (FC) between mes and wild-type expression and the false discovery rate (−log10 q) of all genes on the microarray. Dashed lines mark the significance cut-off of q=0.05 and 1.5-fold up- or down-regulation. The numbers of genes significantly up- or down-regulated are in the top quadrants. Genes with log2(FC) >4 or <−4 are represented as 4 or −4. For validation of data by quantitative PCR, see Table S1. (D) Comparison of log2(FC) of X-linked genes significantly mis-regulated in mes-4 only (red circles), mes-2 only (white circles), and both mes-4 and mes-2 (black circles). Dashed lines show 1.5-fold up- or down-regulation. Correlation coefficient is 0.39. (E) Fold changes of genes significantly up-regulated in mes-4 (red bars) and mes-2; mes-4 (black bars). Open circles, genes significantly up-regulated in mes-2 as well.
We compared transcript accumulation in dissected germlines from mes-4, mes-2, and wild-type control adults. We previously compared mes-4 and wild-type germlines using amplicon microarrays (Bender et al., 2006). For this study we switched to long oligonucleotide microarrays, which have been reported to have greater specificity than amplicon arrays while maintaining sensitivity (Zhu et al., 2005). Expression analysis of mes-4 mutant germlines on oligonucleotide arrays identified 276 significantly mis-regulated genes in mes-4 compared to wild type: up-regulation of 154 X-linked genes and 66 autosomal genes, and down-regulation of 56 autosomal genes (Figure 1C). To verify these and subsequent microarray results, mRNA levels were measured by quantitative PCR for a subset of genes (Table S1). Expression analysis of mes-2 mutant germlines identified 183 significantly mis-regulated genes in mes-2 compared to wild type: up-regulation of 16 X-linked genes and 142 autosomal genes, and down-regulation of 1 X-linked gene and 24 autosomal genes (Figure 1C). Thus, larval decline of maternal MES-2 led to predominantly up-regulation of genes in the adult germline, consistent with the repressive role of MES-2 orthologs and H3K27 methylation in other systems (Kirmizis et al., 2004; Lee et al., 2006; Tolhuis et al., 2006). Larval decline of maternal MES-4 also led to predominantly up-regulation of genes, with a strong bias for up-regulation of genes on the X.
We compared the X-linked genes mis-regulated in mes-2 and mes-4 mutants, to assess whether MES-2 and MES-4 influence expression of the same genes. Ten of the 16 up-regulated genes in mes-2 mutants were also up-regulated in mes-4 mutants (Figure 1D). Additionally, among the X-linked genes significantly up-regulated in mes-4 mutants, most show some up-regulation in mes-2 mutants even though they were not scored as significant (i.e. FDR < 0.05). These results suggest that MES-4 and MES-2 cooperate to down-regulate expression of some of the same X-linked genes.
MES-4 and MES-2/3/6 catalyze antagonistic histone modifications, but both promote development of healthy germ cells in a maternal-effect fashion. To investigate the interplay between MES-4 and the MES-2/3/6 complex, we analyzed double mutants. If they operate in the same pathway, we expected double mutants to resemble single mutants. If they serve antagonistic roles in the same process, similar to Drosophila ASH1 and E(Z) (Klymenko and Muller, 2004), double mutants might display a less severe phenotype than single mutants. If they control a common process via parallel pathways or control different processes, we expected double mutants to display a more severe phenotype than single mutants. We saw the latter result: mes-2; mes-4 and mes-3; mes-4 double mutants display sterility a generation earlier than single mutants (Figure 1B and S2). As a control, mes-2; mes-3 double mutants resemble the single mutants. Sterile mes-2; mes-4 and mes-3; mes-4 double mutants display a range of germline phenotypes (Figure S2). 33% possess a well-proliferated and healthy-appearing germline, offering an opportunity to examine gene expression in germlines lacking both MES-2 and MES-4. This analysis identified 464 mis-regulated genes in mes-2; mes-4 compared to wild type: up-regulation of 210 X-linked genes and 177 autosomal genes, and down-regulation of 3 X-linked genes and 74 autosomal genes (Figure 1C). Compared to mes-4 single mutants, mes-2; mes-4 double mutants up-regulated more genes on the X and showed elevated up-regulation of X-linked genes (Figure 1E). Thus, even though MES-2/3/6 and MES-4 operate independently of each other (Xu et al., 2001), they cooperate at some level to repress expression of genes on the X.
MES-4 and MES-2/3/6 promote gene expression patterns appropriate for germ cells
Since mis-regulation of gene expression in mes mutant germlines is likely to contribute to sterility, an important question is whether particular classes of genes are mis-regulated. We categorized genes according to their expression in published microarray (Reinke et al., 2004) and SAGE (Serial Analysis of Gene Expression) studies (Meissner et al., 2009; Wang et al., 2009). The germline-enriched category includes genes whose expression is enriched in germline tissue based on comparing adults with and without a germline (Reinke et al., 2004). Our germline-specific category includes genes with SAGE tags in dissected germlines and not in FACS-sorted intestine, muscle, or nerve cells. Our soma-specific category includes genes with SAGE tags in at least one somatic tissue (intestine, muscle, and/or nerve) and not in the germline. Our ubiquitously expressed category includes genes with SAGE tags in germline, intestine, muscle, and nerve. We also looked at X-linked genes up-regulated in worms defective in somatic dosage compensation (Jans et al., 2009). We determined whether genes mis-regulated in mes-4, mes-2, and mes-2; mes-4 are enriched for genes in these categories.
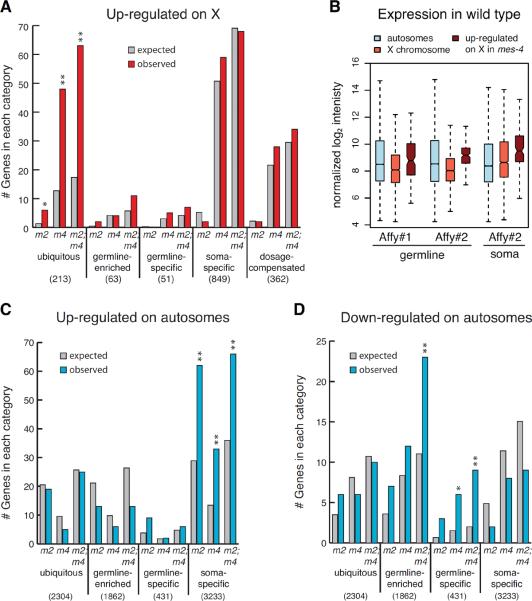
X-linked genes up-regulated in mes mutant germlines are not enriched for genes that are dosage compensated in the soma (Figure 2A), establishing that the focus of MES regulation in the germline is different than the focus of dosage compensation in somatic tissues. In all three mes genotypes analyzed, up-regulated genes on the X are enriched for genes in the ubiquitously expressed category (Figure 2A). Ubiquitously expressed genes are often among the most highly expressed (Wang et al., 2009), but the X chromosome is considered to be nearly silent in most regions of the wild-type adult germline (Kelly et al., 2002; Reinke et al., 2004). This conundrum raised the question: are X-linked genes that are up-regulated in mes mutants normally expressed at low or high levels in the germline? We found that they are among the more highly expressed X-chromosome genes in wild-type germlines (as well as in somatic tissue), based on two independent transcript profiling studies on single-color (Affymetrix) microarrays (Spencer et al., 2011; Tabuchi et al., 2011) (Figure 2B). Thus, the MES proteins dampen germline expression of ubiquitously and robustly expressed genes on the X.
Figure 2.
Genes mis-regulated in mes mutant germlines are enriched for particular expression categories. (A,C,D) Expected and observed numbers of genes in different expression categories among genes up-regulated on the X (A), genes up-regulated on autosomes (C), and genes down-regulated on autosomes (D) in mes-2 (m2), mes-4 (m4) and mes-2; mes-4 (m2; m4) compared to wild type (WT). See text and supplement for definitions of expression categories. Numbers in parentheses are the total number of genes in each category on the X or on the autosomes. Asterisks indicate significantly more genes than expected (hypergeometric test p-value < 0.01 (*) or < 0.001 (**)). See also Figure S4. (B) Comparison of autosomal and X-linked transcript levels in wild-type germlines and somatic tissue analyzed on single-color Affymetrix microarrays (Affy #1; (Tabuchi et al., 2011)) and tiling arrays (Affy #2; (Spencer et al., 2011)).
Autosomal genes up-regulated in mes mutant germlines are enriched for genes whose expression is normally restricted to somatic tissues (Figure 2C). Conversely, autosomal genes down-regulated in mes mutant germlines are enriched for genes whose expression is normally restricted to germ cells (Figure 2D). We conclude that in adult germ cells the MES proteins participate in repressing expression of genes associated with somatic development and promoting expression of genes associated with germline development.
Taken together, transcription profiling and gene class analysis reveal that the MES proteins influence gene expression in a manner appropriate for germline development, enhancing expression of certain germline genes, repressing somatic genes, and dampening expression of X-chromosome genes that are not silent in wild-type germlines, but instead are expressed at appreciable levels and in numerous tissues. These findings establish the importance of MES-4 and MES-2/3/6 in guiding gene expression patterns appropriate for germ cells, but raise the question how proteins that generate antagonistic histone modifications cooperate at a molecular level.
Methylated H3K36 and H3K27 occupy mutually exclusive domains, and methylated H3K27 is strikingly enriched on the X
To investigate how MES-4 and MES-2/3/6 contribute to regulation of gene expression in the germline, we compared the genome-wide distributions of the histone marks they generate, H3K36me3 and H3K27me3, using chromatin immunoprecipitation followed by hybridization to microarrays (ChIP-chip). We previously established that early embryos retain a germline distribution of at least some histone modifications. In particular, we showed that MES-4 maintains H3K36me3 on germline-expressed genes throughout embryogenesis and independently of ongoing transcription, and that embryo-expressed somatic genes have no to low H3K36me3 in early embryos (Furuhashi et al., 2010; Rechtsteiner et al., 2010). Figure 3 and S3 extend this analysis to H3K27me3 and confirm that early embryo chromatin retains germline signatures: germline-specific genes display elevated H3K36me3 and low H3K27me3, while soma-specific genes display low H3K36me3 and elevated H3K27me3. These findings validate performing ChIP analysis from early embryos to gain insights into germline chromatin.
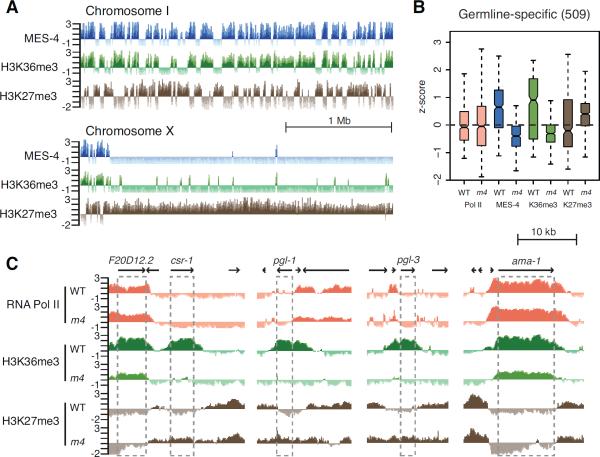
Figure 3.
Genomic distributions of MES-4, H3K36me3, and H3K27me3. (A) ChIP z-scores (standardized log2 ratios of ChIP/Input signals) of MES-4, H3K36me3, and H3K27me3 across the leftmost 3 Mb of ChrI and ChrX. (B) Levels of RNA Polymerase II, MES-4, H3K36me3, and H3K27me3 on germline-specific genes in wild type (WT) and mes-4(RNAi) (m4). Each box extends from the 25th to 75th percentile of the z-scores in the set. Whiskers extend to the 2.5th and 97.5th percentile. Wedges around the median indicate 95% confidence interval for the medians. See also Figure S3. (C) Genome browser views of germline genes (csr-1, pgl-1, and pgl-3), showing absence of Pol II in wild type, and loss of H3K36me3 and acquisition of H3K27me3 in mes-4(RNAi). F20D12.2 and ama-1 have Pol II, and do not lose H3K36me3 or acquire H3K27me3 in mes-4(RNAi).
We found that the autosomes are composed of alternating clusters of H3K36me3-bound genes and H3K27me3-bound genes (Figure 3A). These define mutually exclusive domains of these two opposing histone modifications, with a negative correlation coefficient r = −0.82 (comparing H3K36me3 and H3K27me3 on 1 kb segments across the genome). The X chromosome is strikingly different. With the exception of the leftmost 300 Mb, the X displays very few regions of H3K36me3 enrichment, and is marked by H3K27me3 along its length (Figure 3A). These ChIP-chip results in early embryos are consistent with the striking under-representation of germline-expressed genes on the X chromosome and immunostaining results showing absence of marks of active chromatin and concentration of repressive H3K27me3 on X chromosomes in the germline (Bender et al., 2004; Kelly et al., 2002; Reinke et al., 2004). The results strongly suggest that MES-2/3/6 participates directly in X repression by concentrating a repressive chromatin mark on X-linked genes. The results also raised the possibility that MES-4 and/or methylated H3K36 repel MES-2/3/6 from autosomal genes that should be expressed in the germline.
MES-4-generated H3K36 methylation antagonizes H3K27 methylation on germline-expressed genes on the autosomes and concentrates H3K27me3 on the X chromosome
To test the model that MES-4 repels MES-2/3/6 from germline-expressed genes on the autosomes, we analyzed the distribution of H3K27me3 in early embryos whose mothers were depleted of MES-4 by RNAi. Confirming that RNAi was effective, MES-4 and H3K36me3 were depleted to below detectable levels from genes with germline-specific expression, which lack transcription in early embryos (Rechtsteiner et al., 2010); ubiquitously-expressed genes with detectable transcription in early embryos, such as ama-1, retained some H3K36me3, likely catalyzed by the other H3K36 HMT MET-1, which becomes active at the ~40-cell stage of embryogenesis and which is thought to methylate H3K36 cotranscriptionally (Rechtsteiner et al., 2010). Examination of germline-specific genes revealed that loss of H3K36me3 in mes-4(RNAi) embryos was accompanied by acquisition of H3K27me3 (Figure 3B, 3C, 4C and S3). Genes that retained H3K36me3, such as ama-1, remained devoid of H3K27me3 (Figure 3C). We conclude that MES-4 activity repels MES-2/3/6 repressive activity from genes whose expression is associated with germline development.
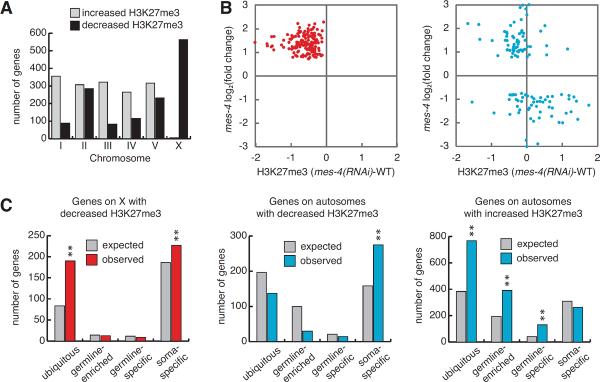
Figure 4.
Assessment of redistribution of H3K27me3 upon depletion of MES-4. (A) Number of genes on each chromosome with significantly increased or decreased H3K27me3 in mes-4(RNAi) compared to wild type. (B) For 276 genes mis-regulated in mes-4 mutant germlines, comparison of gene mis-expression (X genes, red circles; autosomal genes, blue circles) versus change in H3K27me3 levels between mes-4(RNAi) and wild-type early embryos. (C) Expected and observed numbers of genes in different expression categories among genes with significantly increased or decreased H3K27me3 in mes-4(RNAi) compared to wild type.
If acquisition of H3K27me3 is responsible for down-regulation of germline-expressed genes on the autosomes in mes-4 mutant germlines, then those genes should be restored to closer to wild-type levels when H3K27me3 is lost in mes-2; mes-4 double mutant germlines. Eight of 33 autosomal genes analyzed were restored to closer to normal levels in mes-2; mes-4 compared to mes-4 mutants (Figure S4). Two possible explanations for the remaining 25 genes showing similar or enhanced down-regulation in mes-2; mes-4 compared to mes-4 are: 1) early action of maternally supplied MES-2 and H3K27me3 in homozgyous mes mutants is sufficient to maintain repression in adults, or 2) MES-4 promotes expression of at least some germline genes independently of repelling MES-2/3/6.
We previously hypothesized that MES-4 participates in X silencing by repelling a repressor from the autosomes and focusing its repressive activity on the X (Bender et al., 2006). The above analysis suggested that MES-2/3/6 is the repressor that MES-4 repels. In support of this scenario, H3K27me3 levels were strikingly reduced on a majority of X-linked genes in mes-4(RNAi) embryos (Figure 4A and S3). Importantly, X-linked genes up-regulated in mes-4 mutant germlines displayed markedly reduced H3K27me3 in mes-4(RNAi) early embryos (Figure 4B and S3). X-linked genes with reduced H3K27me3 in mes-4 (RNAi) are particularly enriched for ubiquitously expressed genes (Figure 4C). We conclude that MES-4 activity helps concentrate MES-2/3/6 repressive activity on the Xs to dampen X gene expression.
We wondered if MES-4 repulsion of MES-2/3/6 activity helps concentrate H3K27me3 elsewhere on the autosomes. In wild-type early embryos, autosomal genes whose expression is specific to somatic cells generally lack H3K36me3 and possess H3K27me3 (Figure S3). In mes-4(RNAi) early embryos, those genes displayed reduced H3K27me3, ranging from modest to strong reduction (Figure S3). Importantly, autosomal genes up-regulated in mes-4 mutant germlines generally showed reduced H3K27me3 in mes-4(RNAi) embryos (Figure 4B), and both autosomal genes up-regulated in mes-4 mutants and autosomal genes with significantly reduced H3K27me3 in mes-4(RNAi) are enriched for soma-specific genes (Figure 2C and 3C). These findings support the view that MES-4 and MES-2/3/6 contribute not only to promoting expression of germline genes but also to repressing somatic genes in the germline.
Conclusions
Genome-wide ChIP-chip analysis and transcription profiling have advanced our understanding of antagonistic chromatin modifications and how they influence gene expression patterns during development. We show that loss of H3K36 methylation causes global redistribution of H3K27me3 and parallel changes in gene expression. This work provides mechanistic insight into how the proteins that catalyze these histone modifications cooperate to ensure germ cell survival and development in C. elegans: MES-4 function repels MES-2/3/6 repressive activity from germline genes on the autosomes and concentrates their repressive action on other autosomal regions, including somatic genes, and on the X chromosomes. This antagonism ensures proper patterns of gene expression in germ cells, which includes repression of somatic genes and the Xs. Loss of both MES-4 and MES-2/3/6 results in loss of H3K36me3 from germline genes and loss of H3K27me3 from somatic genes and the X. This likely explains the enhanced gene mis-regulation and earlier sterility of mes-2; mes 4 double mutants compared to single mutants, although both mutant and RNAi approaches to elimination of gene function suffer from gradual loss of protein and uncertainty about which effects are primary and which are secondary. Loss of MES-4 or MES-2/3/6 is not sufficient to allow expression of tested somatic proteins in the germline (Patel et al., 2012; our unpublished results). However, MES loss enables germ cells to be converted to neural or muscle fates upon ectopic expression of terminal selector transcription factors (Patel et al., 2012). Similarly, embryos lacking MES-2 display prolonged developmental plasticity and susceptibility to cell fate conversion compared to wild-type embryos (Yuzyuk et al., 2009). These studies reveal how proper chromatin states can protect cell fates and how altering chromatin context can enable reprogramming of cell fate.
EXPERIMENTAL PROCEDURES
Immunocytochemistry
Whole larvae and dissected germlines were immunostained and processed as described (Petrella et al., 2011). Antibodies and quantification of immunostaining are described in Supplemental Experimental Procedures.
Microarray Analysis
Total RNA was isolated from dissected germlines (strain details and growth conditions are in Supplemental Experimental Procedures) and made into cDNA after one round of amplification. cDNA was labeled with Cy3 or Cy5, fragmented, and hybridized to microarrays made by Washington University. Microarray normalization and analysis were performed with Bioconductor tools (www.bioconductor.org) and custom scripts using R statistical programming language. Details are in Supplemental Experimental Procedures.
Quantitative RT-PCR
PCR was done as described (Petrella et al., 2011) with total RNA from three or four biological replicates of young adult germlines as prepared for microarrays. Primer sequences are in Supplemental Experimental Procedures.
ChIP-chip Experiments
Collection of wild-type and mes-4(RNAi) early embryos, preparation of extracts, anti-H3K36me3 and anti-Pol II antibodies used, ChIP methods, and data processing and analysis are described in (Rechtsteiner et al., 2010). Anti-H3K27me3 antibody was MAb 1E7 from H. Kimura. Normalization is explained in Supplemental Experimental Procedures.
Microarray and ChIP Accession Numbers
Microarray and ChIP data were deposited to Gene Expression Omnibus. The super series accession number is GSE38160.
Supplementary Material
Highlights
MES-2/3/6 and MES-4 cooperate to regulate gene expression in the germline
MES-2/3/6 concentrates repressive H3K27me3 on somatic genes and the X
MES-4 generates H3K36me3 on germline genes and repels repressive H3K27me3
Antagonistic histone modifiers shape chromatin landscapes and gene expression
ACKNOWLEDGMENTS
We thank Teruaki Takasaki for help preparing mes-4(RNAi) embryos, Lily Shiue and Manny Ares for advice and help with microarray hybridization and use of the Agilent Bioanalyzer, Hiroshi Kimura for antibodies, and Needhi Bhalla, Oliver Hobert, and members of the Strome lab for helpful discussions. This work was supported by NIH grant GM34059 to SS and NIH NHGRI modENCODE grant U01 HG004270 to Jason Lieb (SS is a CoPI). LG was supported on NIH training grant T32 GM008646.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y, Bender L, Wang W, Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi H, Takasaki T, Rechtsteiner A, Li T, Kimura H, Checchi PM, Strome S, Kelly WG. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009;23:602–618. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee MH, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Lo Sardo F, Diamantini A, Orlando V. PcG complexes set the stage for epigenetic inheritance of gene silencing in early S phase before replication. PLoS Genet. 2011;7:e1002370. doi: 10.1371/journal.pgen.1002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B, Warner A, Wong K, Dube N, Lorch A, McKay SJ, Khattra J, Rogalski T, Somasiri A, Chaudhry I, et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development. 2011;138:1069–1079. doi: 10.1242/dev.059501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, Kimura H, Lieb JD, Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi TM, Deplancke B, Osato N, Zhu LJ, Barrasa MI, Harrison MM, Horvitz HR, Walhout AJ, Hagstrom KA. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 2011;7:e1002074. doi: 10.1371/journal.pgen.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397:161–168. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Fong Y, Strome S. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc Natl Acad Sci U S A. 2001;98:5061–5066. doi: 10.1073/pnas.081016198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics. 2001;159:1019–1029. doi: 10.1093/genetics/159.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Ping G, Shinohara Y, Zhang Y, Baba Y. Comparison of gene expression measurements from cDNA and 60-mer oligonucleotide microarrays. Genomics. 2005;85:657–665. doi: 10.1016/j.ygeno.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.