Abstract
Although the study of individual phospholipids and their synthesis began in the 1920’s first in plants and then mammals, it was not until the early 1960’s that Eugene Kennedy using Escherichia coli initiated studies of bacterial phospholipid metabolism. With the base of information already available from studies of mammalian tissue, the basic blueprint of phospholipid biosynthesis in E. coli was worked out by the late 1960’s. In 1970’s and 1980’s most of the enzymes responsible for phospholipid biosynthesis were purified and many of the genes encoding these enzymes were identified. By the late 1990’s conditional and null mutants were available along with clones of the genes for every step of phospholipid biosynthesis. Most of these genes had been sequenced before the complete E. coli genome sequence was available. Strains of E. coli were developed in which phospholipid composition could be changed in a systematic manner while maintaining cell viability. Null mutants, strains in which phospholipid metabolism was artificially regulated, and strains synthesizing foreign lipids not found in E. coli have been used to this day to define specific roles for individual phospholipid. This review will trace the findings that have led to the development of E. coli as an excellent model system to study mechanisms underlying the synthesis and function of phospholipids that are widely applicable to other prokaryotic and eukaryotic systems.
Keywords: Escherichia coli, phospholipid, phosphatidylethanolamine, phosphatidylglycerol, cardiolipin, membrane protein, enzymology, synthesis, function, genetics
1. Introduction
Analysis of phospholipids and their biosynthesis were initially carried out in plants and animals and only began to focus on bacteria in the 1960’s. Much of what we know about this area of research has resulted from a combination and interplay of studies in both eukaryotes and prokaryotes. My own interests in phospholipids began in 1969 as a postdoctoral fellow in Eugene Kennedy’s laboratory at Harvard Medical School (Fig. 1). This was an exciting time in which the basic blue print for phospholipid biosynthesis at all levels of the genetic tree had been established and awaited the application of sophisticated methods of biochemical analysis, genetics and the power of advanced molecular biology and molecular genetics. This retrospective will be largely confined to the studies in Escherichia coli and their contributions to our understanding of phospholipid metabolism and function. My personal observations at various stages of the evolution of the field will be injected. I fully recognize that major contributions have been made by studies in other biological systems for which there is insufficient space to include.
Figure 1.
The Kennedy “Clan” on the occasion of his 90th birthday. Eugene Kennedy (1919–2011) is 4th from the left in the front row. The gathering was in October 2009 at Harvard Medical School. Pictured are former graduate students, postdoctoral fellows and scientific associates of Eugene Kennedy.
2. The early studies in eukaryotic systems
2.1. Identification of phospholipids
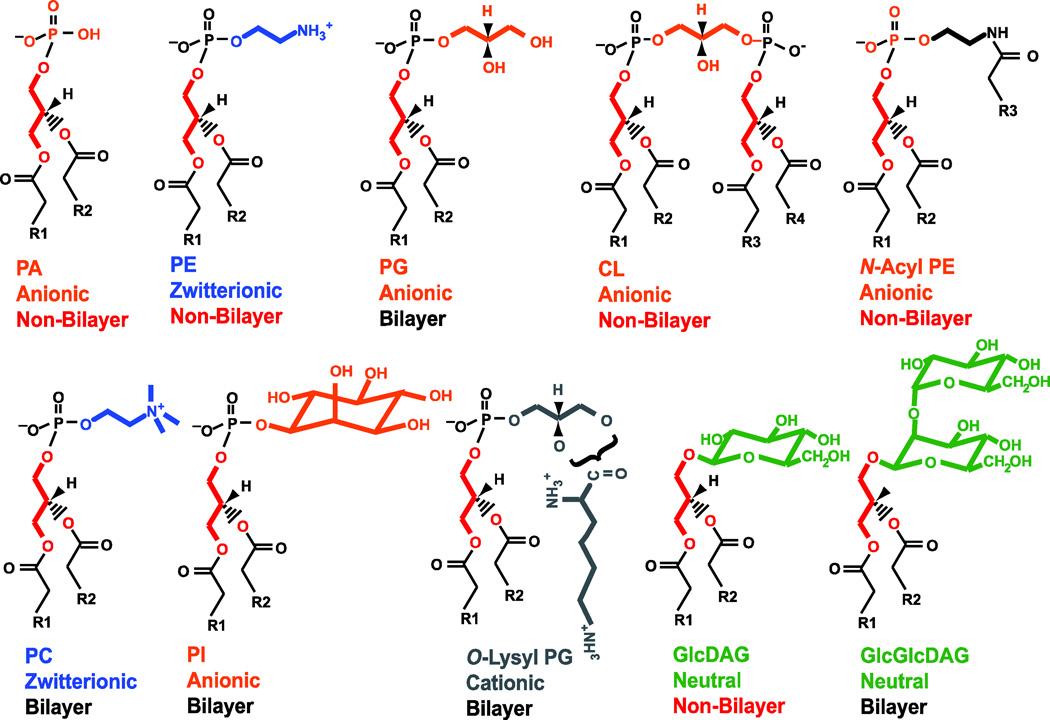
As we now know, phosphatidic acid (PA1, 1,2 diacyl-sn-glycerol-3-phosphate) is the precursor leading to the biosynthesis of the remaining phospholipids in Bacteria and Eukarya; see http://www.lipidmaps.org/ for classification and structures of glycerol-based phospholipids. Archaea phospholipids differ in that they are composed of sn-glycerol-1-phosphate in ether linkage at the 2- and 3-positions to long-chain poly isoprenoids [1]. The first report of the isolation and structure of PA was by Chibnall and Channon in 1927 [2, 3]. They began with over 20 kg of cabbage leaves from which a green colloidal “cytoplasmic fraction” free of cell wall material was first isolated. After heat treatment, the coagulated material was extracted with ether to yield about 35 g of “fatty” material, which was subjected to successive acetone treatments yielding a precipitate of which half by weight contained phosphorus. Further analysis revealed that the majority of the phosphorus-containing material was made up of divalent cation salts of PA and lyso-PA, which would later be shown to result from hydrolysis of the major phospholipid species.
In the early 1900’s lecithin (phosphatidylcholine (PC), 1,2 diacyl-sn-glycerol-3-phosphocholine) was isolated from plant and egg sources and its theoretical structure proposed. However, there were some inconsistencies in several of the reports, which resulted in some controversy in the literature. Levene and West [4, 5] questioned the thoroughness of previous analyses and demonstrated that the free amine containing lipidic material contained in samples of lecithin was cephalin (phosphatidylethanolamine (PE), 1,2 diacyl-sn-glycerol-3-phosphoethanolamine). However, by the late 1920’s the natural occurrence and structures of the phospholipid precursor (PA) and the two major amino-containing phospholipids (PE and PC) were established.
2.2. Radiolabeling of phospholipids
A major technical advancement that changed the course of characterization of biological molecules in general was the availability of man-made radionuclides. The first use of 32PO4 (32Pi) was in 1935. Chievitz and Hevesy [6] fed 32Pi to rats and traced radiolabel in tissue deposits and excretion. Thus it was now possible to follow lipidic phosphate fluxes in whole animals. Previous attempts to measure fluxes in phospholipids relied on feeding animals with elaidic acid [7] which falsely assumed that flux of this fatty acid through the lipidic phosphate pool was a measure of phospholipid metabolism. In 1937 Atrom et al. [8] noted that lipidic phosphate accumulated in largest amounts in liver, intestine and kidney of rats fed 32Pi. This ‘organification’ of phosphorus was the first proof that phospholipids were synthesized endogenously from individual building blocks. Perlman and Chaikoff [9] followed with a series of experiments that measured incorporation and turnover of 32Pi in phospholipids in various organs either from endogenous or fed supplies of fat. In the methods section the authors noted an important point, still missed today in many radioisotope experiments, to add unlabeled carrier phospholipid to samples with low levels of total lipid. In subsequent experiments the flux of 32Pi through the tissue pools of phospholipid was measured as a function of dietary supplements, but only lipidic phosphate was measured and not individual phospholipids. However, incorporation of 32Pi into phospholipid was demonstrated after incubation with homogenized liver, which set up future studies for in vitro incorporation experiments. It was not until the 1950’s with methods available for subcellular fractionation that specific lipids were identified in organelles such as the mitochondria. Swanson and Artom [10] established that the major lipidic phosphate of rat mitochondria was composed of PE and PC.
2.3. Employing cell free systems
Arthur Kornberg [11] and Eugene Kennedy [12], using soluble enzyme extracts of tissues or mitochondria, respectively, carried out the first experiments that began to delineate phospholipid biosynthetic pathways. The hallmark of Kornberg’s approach to employ enzyme preparations (not yet homogeneous as he later required) to study biosynthetic processes was evident in this early work. He used ATP, CoA and fatty acids or substituted the latter with enzymatically-synthesized palmitoyl-CoA plus enzymatically synthesized sn-glycerol-3-[32P] (GP) to synthesize radiolabeled PA using a cell free system derived from guinea pig liver. Longer fatty acid chains (16–18) were better substrates than shorter chains. Although PA had been identified in high content in previous studies, it was now clear that the conditions used previously to process the source material generated PA from PC and PE by hydrolysis. The same was true of lyso-PA, which was not observed by Kornberg but was most likely an intermediate in PA synthesis. However, whether PA is a precursor to the amine-containing phospholipids remained to be determined. Kennedy’s approach was somewhat different but the final conclusions were the same. He used isolated rat liver mitochondria incubated initially with 32Pi and then with [32P]GP with the appropriate controls to demonstrate the formation of PA, which was dependent on oxidative phosphorylation.
The literature on the synthesis of PC and PE became a bit confusing after the initial reports of Kornberg and Kennedy. Kornberg [13] demonstrated ATP-dependent incorporation by a crude enzyme preparation from liver of phosphocholine (doubly labeled in 14C and 32P) into lipidic material, which was partially characterized as PC. Kennedy [14] using a liver mitochondrial preparation appeared to demonstrate only ATP-dependent [14C]choline incorporation and not doubly labeled phosphocholine into PC. As Kennedy later showed [15] and recounted in his scientific biography [16] that he had fallen into error in the identification of PC. The then unknown compound that co-chromatographic with PC was a long-chain fatty acid ester of choline. Kornberg turned out to be correct in deducing phosphocholine as an intermediate in PC biosynthesis, but both missed that PC is made by a combination of cytosolic and endoplasmic reticulum enzymes and not mitochondria. Kennedy presented a very entertaining and informative after dinner talk at the 2003 Gordon Research Conference on the Molecular and Cellular Biology of Lipids titled the “Hits and Misses in Biochemistry.” He documented the errors and mishaps in the lipid and membrane area beginning with Otto Warburg to the date of his talk (including his own mishaps).
After the initial demonstration of PA synthesis and the apparent formation of PC in mammals, Kornberg pursued DNA replication with only occasional forays into the phospholipid field. His interests turned to membranes and phospholipids in studies wherein DnaA [17, 18], the initiator for DNA replication in E. coli, was found to require anionic phospholipids such as phosphatidylglycerol (PG) and cardiolipin (CL) for function. Kennedy on the other hand continued in the field to outline the basic design for phospholipid biosynthesis in eukaryotes and prokaryotes.
2.4. Cytidine nucleotides activate phospholipid precursors
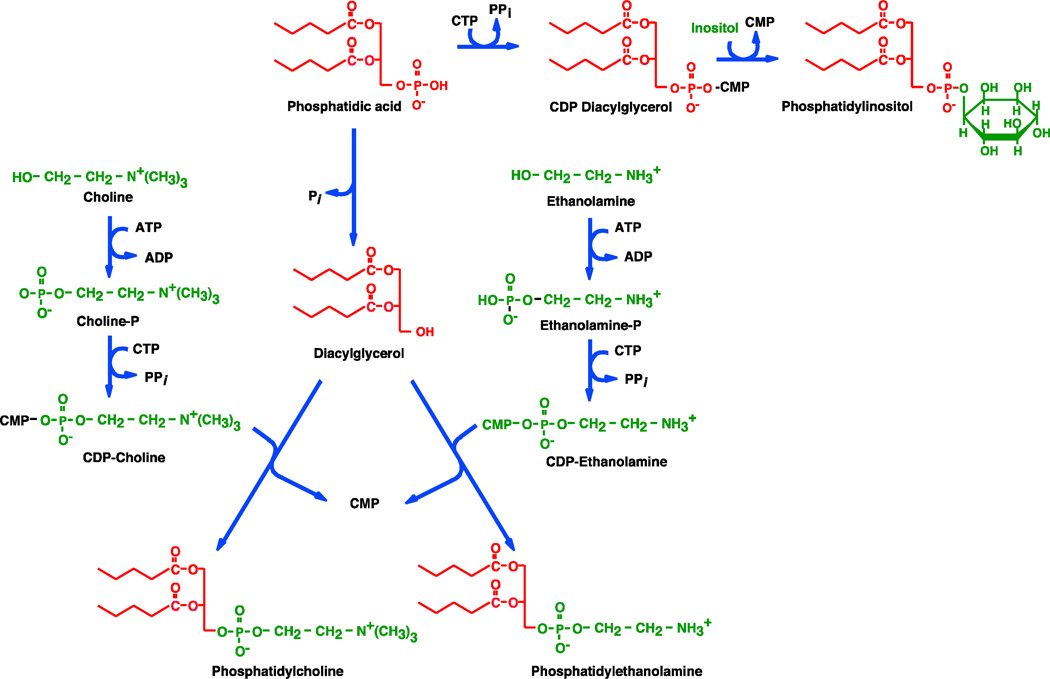
In 1956 Kennedy and Weiss [15] published the seminal paper in the phospholipid field that formed the biological foundation for future studies in all organisms thereby establishing the “Kennedy Pathways” for phospholipid synthesis. They initiated their study to resolve the conflict between Kennedy’s results and Kornberg’s results on the intermediates leading to PC biosynthesis; Rodbell and Hanahan [19] like Kornberg had also demonstrated phosphocholine incorporation into PC. Kennedy and Weiss employed a rat liver crude mitochondrial preparation that they recognized as containing other subcellular organelles and therefore referred to it as the “particulate fraction.” Using phosphocholine labeled with both 32P and 14C and 14C-labeled choline, they were able to incorporate both in an ATP dependent manner into PC, which was now more thoroughly identified. At this point the work was merely confirmatory of the findings of others. However, when they used an ATP generating system beginning with AMP, there was no incorporation into PC. Use of crystalline ATP (Pabst lot No. 122) rather than amorphous ATP (Pabst lot No. 116) also failed to support PC synthesis. The mystery was solved and the unraveling of phospholipid biosynthesis began when CTP was found as the active nucleotide in lot No. 116 supporting the pathways for both PC and PE biosynthesis shown in Fig. 2. Kennedy and Weis also showed that each of the intermediates individually supported amine-containing lipid synthesis. They also isolated CDP-choline and CDP-ethanolamine from liver and yeast and demonstrated that the amino-alcohol acceptor is sn-1,2-diacylglycerol, most likely derived from the hydrolysis of PA. As a memorial to this discovery, lot No. 116 of ATP remained in a desiccator in the rear of Kennedy’s freezer until he closed the lab.
Figure 2.
Pathways for phospholipid biosynthesis in mammalian cells. The zig-zig lines in each red structure represent the long chain fatty acids on the phosphatidyl moiety. These are mostly saturated at the sn-1 position of unsaturated at the sn-2 position of the glycerol backbone. The components of each fatty acid are color coded as to the building blocks from which they are derived. The enzymes are located in the cytoplasm and the endoplasmic reticulum. The “Kennedy Pathway” generally refers to the pathways beginning with ethanolamine and choline. Not shown are the formation of PS from PE or PC by head group exchange with serine, the decarboxylation of PS to form PE, the methylation of PE by S-adenosyl methionine to form PC, and the formation of PG and CL in the mitochondria.
Agranoff et al. [20] demonstrated the incorporation of radiolabeled inositol into phosphatidylinositol (PI) using a crude extract of rat kidneys. Interestingly, CDP-choline was the most active nucleotide species with little activity from CTP or CDP. However, addition of PA stimulated the CDP-choline-dependent reaction. They also observed incorporation of radiolabeled CMP into lipidic material, which they postulated to be CDP-diacylglycerol (-DAG) based on its breakdown products under basic conditions. Thus they proposed that CDP-choline reacted with PA to form choline and CDP-DAG, which then reacted with inositol to form PI and CMP. They got some of the players in PI synthesis correct but not the proper pathway.
Although the amounts of the putative CDP-DAG made were insufficient for complete characterization, these experiments provided important clues in establishing the pathway to PI synthesis. Paulus and Kennedy [21] investigated the effects of different buffers containing phosphate, manganese, and various cytidine nucleotides. They concluded that the previous report of inositol incorporation into PI was due to an energy independent exchange reaction that was dependent on the buffer conditions. To bypass this exchange reaction, they first used a rat kidney cell free system to generate PA from sn-glycerol-3-[32P], ATP, CoA and oleic acid. Next, formation of radiolabeled PI was found to be dependent solely on the addition of inositol and CTP to the radiolabeled PA, with the radiolabel found in the inositol phosphate after hydrolysis of PI. Radiolabeled CDP-DAG was formed from PA and CTP with radiolabel in the α-phosphate position, and synthetic CDP-DAG was also shown to be a substrate for PI synthesis. With these studies the involvement of CDP-DAG as an intermediate in phospholipid biosynthesis was established.
With this report, Kennedy had defined all cytidine intermediates necessary for phospholipid biosynthesis throughout nature. Most of the remaining work from Kennedy’s lab focused on phospholipid metabolism in E. coli, although he did return to PI metabolism in brain in the mid 1960’s when he defined the pathway for the synthesis of some of the phosphorylated derivatives of PI [22]. Work at the time suggested these PI derivatives may be of some importance as metabolic regulatory molecules, which of course we now know represent a major area of second messenger cell regulation research. Voelker and Kennedy [23] also established that the phosphocholine moiety of sphingomyelin in mammalian cells comes directly from PC and not from CDP-choline. With some exceptions, protein synthesis uses adenylate-activated amino acids, carbohydrate synthesis uses uridylate-activated sugars, and phospholipid synthesis uses cytidylate-activated precursors. From hereon this review will focus primarily on bacterial phospholipid metabolism with E. coli as the primary vehicle for study.
3. Pathway for phospholipid biosynthesis in E. coli
3.1. Synthesis of phosphatidic acid
As in eukaryotic cells, PA is the precursor to all the glycerol-based (as distinguished from Lipid A core of lipopolysaccharide (LPS)) phospholipids of E. coli. PA is synthesized in two sequential steps employing long chain acyl-CoA or acyl-ACP (acyl carrier protein) for acylation first at the sn-1 and then the sn-2 position catalyzed by the plsB [24] and plsC [25] gene products, respectively. PA [26] synthesis in bacteria is covered in detail elsewhere in this issue. Kennedy followed many of his trainees have been the central figures in defining the pathway (see Fig. 3), metabolism and function of phospholipids in E. coli. This basic framework along with the early studies of phospholipid synthesis in mammals has served as a starting point for extending the understanding of phospholipid metabolism in Archaea [1, 27], yeast [28], plants [29] and somatic cells [30, 31], which can be accessed in numerous reviews some of which are noted above.
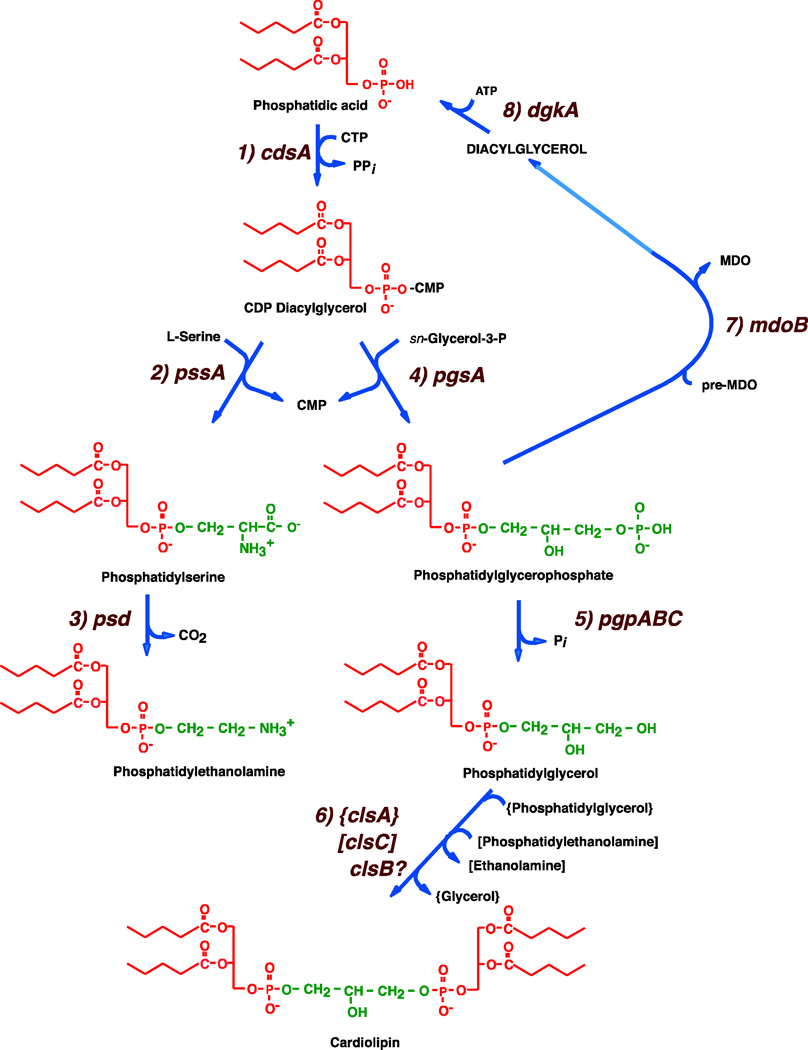
Figure 3.
Pathways for synthesis of phospholipids in E. coli. The following enzymes with their respective genes named carry out: 1. CDP-diacylglycerol synthase (CdsA); 2. phosphatidylserine synthase (PssA); 3. phosphatidylserine decarboxylase (Psd); 4. phosphatidylglycerophosphate synthase (PgsA); 5. phosphatidylglycerophosphate phosphatase (Pgp) encoded by three genes; 6. cardiolipin synthase (Cls) encoded by three genes with one substrate being PG in all three cases and the second substrate for {ClsA} and [ClsC] indicated by the brackets. Definitive identification of the second substrate for ClsB has not been established; 7. Phosphatidylglycerol:pre-membrane derived oligosaccharide (MDO) sn-glycerol-1-P transferase (MDO synthase); 8. diacylglycerol kinase (DgkA).
3.2. Formation of phosphatidylethanolamine and phosphatidylglycerol
In 1963 Kanfer and Kennedy [32] noted that rigorous characterization of bacterial phospholipids and the pathways leading to their biosynthesis were understudied relative to what was known for mammalian systems. They first labeled growing cells of E. coli with 32Pi for increasing times from 30 sec to 30 min, which was followed by a chase of label. The lipid fraction was isolated, and the incorporation of label into the mild alkaline deacylation products of phospholipids was quantified after chromatographic separation. PE, phosphatidylserine (PS), PA and phosphatidylglycerol (PG) were all detected at early time points. Label in PA and PS steadily decreased with longer labeling times consistent with being intermediates to other phospholipids. Label in PE remained stable for several generations during the chase, indicating that the phosphate moiety does not turnover to other products. Later, it will be shown that the fatty acid at the sn-1 position of PE [33, 34] is used to acylate the N-terminal amino acid of outer membrane lipoproteins followed by the reformation of PE by a specific acyltransferase [35]. An important observation was that label in PG was not stable during the chase, indicating turnover to water-soluble products due to degradation or some other phosphate labeled compound. Cardiolipin (CL) had not yet been identified in E. coli. More interesting was the fate of much of this label for the synthesis of membrane-derived oligosaccharide (MDO) of the periplasmic space, a study that Kennedy returned to in the 1970’s.
Next came the establishment of the pathway for PE and PG biosynthesis [36]. Involvement of CDP-ethanolamine was ruled out, but the conversion of PS to PE by decarboxylation was confirmed [37] to be the same as already seen in animal cells [38] while the formation of PS was quite different in E. coli. Building on the role of CDP-DAG involvement in PI synthesis, Kanfer and Kennedy tried the liponucleotide as substrate with L-serine for the synthesis of PS followed by its decarboxylation. In mammalian cells, PS is made by exchange of L-serine with the hydrophilic head group of either PE or PC by two separate enzymes [39]. Thus far CDP-DAG-dependent PS synthesis is unique to bacteria and yeast and appears to be absent in higher eukaryotes except for its presence in wheat [40]. Use of sn-glycerol-3-phosphate in place of serine resulted in the formation of PG but not PG-phosphate (PGP), which they presumed was acted on by a phosphatase as had been shown for the same reaction in chicken liver [41]. Inactivation of the PGP phosphatase in crude extracts by sulfhydryl reagents demonstrated the intermediate formation of PGP [42]. It would be nearly 50 years later before the primary PGP phosphatase is identified [43]. In 1968 Carter demonstrated the synthesis of CDPDAG from CTP and PA by a particulate fraction of E. coli [44].
3.3. Cardiolipin synthesis is different in E. coli than in eukaryotic cells
Of the major phospholipids of E. coli only the synthesis of CL remained to be established. CL was first isolated and characterized from beef heart in pursuit of the substance in alcohol extracts that reacted with sera from patients with syphilis [45]. Pangborn started with 15 beef hearts for the initial alcohol extract followed by CdCl2 precipitation. After solubilization of the precipitate with petroleum ether and about 12 more extraction/precipitation steps, 5 g of pure CL was isolated. Although Pangborn proposed a structure for CL, it was not until 1958 that Macfarlane established the correct structure [46]. Kennedy’s group [47] demonstrated the incorporation of sn-[2-3H]glycerol-3-phosphate into PG, PGP and CL by a particulate fraction of E. coli dependent on the presence of CDP-DAG. They established that PGP conversion to PG was a prerequisite for CL synthesis since use of sn-[2-3H]glycerol-3-[32P] as substrate failed to incorporate label into CL. However, the strong stimulation by CDP-DAG of the incorporation of radiolabeled PG into CL lead the investigators to make the wrong conclusion on the mechanism by which E. coli makes CL.
In virtually all simple and complex eukaryotes, CL is make by the displacement of CMP from CDP-DAG by the free hydroxyl of PG at the sn-3 position of glycerol as initially shown by van Deenen’s group [48]. However, as was later shown by Hirschberg and Kennedy [49], E. coli condenses two PG molecules to make CL with the release of glycerol. Considerable evidence had accumulated from E. coli and other bacteria in support of a non-CDP-DAG-dependent pathway for CL synthesis (see [49]). Notable among the evidence was the continued formation of CL in the absence of significant metabolic energy, the release of glycerol during CL synthesis and the incorporation of labeled PG into CL in the absence of CDP-DAG. In a series of elegant single and double label experiments, it was established by Hirschberg and Kennedy [49] that CDP-DAG stimulated the formation of CL but did not directly participate in the reaction. They also ruled out any exchange reactions between PG and existing lipids in the crude membrane preparations. With the establishment of the most common route to CL formation in eukaryotes being through CDP-DAG, a clear division between prokaryotes and eukaryotes appeared to exist. However, as will be discussed later, this line has become blurred with respect to complete restriction to either prokaryotes or eukaryotes and a third pathway for CL synthesis recently found in E. coli [50]. Again, it was not until 2012 that all three genes encoding CL synthases in E. coli were accounted for [50].
By the late 1960’s and early 1970’s, the basic outline for the synthesis of the major phospholipids in bacteria and somatic cells had been established. What followed was an era of enzyme purification, establishment of enzymological properties and the identification of genes encoding the enzymes. Around 1969 to 1974 the Kennedy lab was populated by a group of medical students and postdoctoral fellows who initiated many of the above studies and became leaders in their fields after departing. Bill Wickner initiated the purification of the first membrane-associated enzyme that carried out a step in phospholipid metabolism. He went on to successfully purify the complex E. coli replication machinery in Arthur Kornberg’s lab followed by his own work that defined how proteins are inserted into and translocated across the E. coli membrane. Chris Raetz began purification of several enzymes of phospholipid metabolism and then during his postdoc with Herb Tabor developed novel methods to isolate mutants in phospholipid metabolism. During his independent years he defined the “Raetz Pathway” for the synthesis of the membrane embedded core of LPS. Carlos Hirschberg determined how CL was made in E. coli and then in his independent career defined many of the important steps in synthesis of the carbohydrate moieties of glycoproteins. Ed Dennis studied several aspects of phospholipid metabolism in Tetrahymena and then went on to be a leader in studying the role of phospholipases in cell signaling. I was fortunate to be in the lab at the time where my interests in analysis of phospholipid biosynthetic enzymes began and was followed by pursuing the underlying genetics of these enzymes and finally studying the role of phospholipids in cell function.
4. Isolation of mutants in phospholipid metabolism
Prior to 1970 there were no mutants available in genes encoding an E. coli phospholipid biosynthetic enzyme. One of the earliest reports [51] of mutants in phospholipid metabolism was one temperature sensitive for the formation of PE. The mutation turned out to be in the pssA locus that encodes PS synthase (PssA) [52–55]. Bell [24, 56] reported a mutant (plsB) that was defective in the committed step to phospholipid synthesis, i.e. the sn-glycerol-3-phosphate acyltransferase that synthesizes 1-acyl-sn-glycerol-3-phosphate (lyso-PA). The mutant was selected as a GP auxotroph and turned to have a 10-fold higher Km for the sn-glycerol-3-phosphate substrate. Starving for sn-glycerol-3-phosphate resulted in complete cessation of phospholipid biosynthesis.
In 1975 Raetz developed a novel and rapid filter-paper based method for identifying mutants in phospholipid biosynthetic enzymes [57]. Cells were mutagenized and then grown as single colonies on agar plates. The colonies were transferred by replica plating to filter papers in duplicate for in situ assay of enzymatic activity under permissive conditions and restrictive conditions (in this case elevated temperature). Cells were lysed by incubation with lysozyme, and the filter papers, one at low temperature and one at high temperature, were incubated with radiolabeled water soluble substrates and unlabeled lipid substrates for lipid synthesis. After trichloroacetic acid precipitation of the radiolabeled lipid product on the filter papers, autoradiograms of the two filter papers were compared to see which colonies did not incorporate label at the high temperature. The method was first employed to isolate temperature sensitive mutants in the pgsA gene, which encodes PGP synthase (PgsA). Later the method was used to isolate temperature sensitivity mutants (see Fig. 3 for gene-enzyme relationships) in pssA [58], cdsA [59], and pgpAB [60].
The utility of this method lies with the fact that most fully defective null mutants in phospholipid metabolism are lethal under a variety of growth conditions. The method relied on isolating conditional defects in enzyme activities as measured in situ, which in many cases was not reflected in complete loss of function in vivo or loss in cell viability. Once a gene locus was identified, then further mutagenesis could be employed to isolate mutations that also affected growth. The procedure was also extended to somatic cells [61] and yeast [62], which opened up phospholipid metabolism to genetic analysis and provided a means to clone the respective genes in multiple organisms prior to the availability of whole genome sequences.
A temperature sensitive mutation in the psd locus was isolated by mutagenesis followed by brute force assay of individual colonies for PS decarboxylase (Psd) activity at an elevated temperature [63]. The initial mutant contained a second mutation, which rendered the cells temperature sensitive for growth but not for PS accumulation or lack of PE. The mutant PS decarboxylase in the original strain was only temperature sensitive under assay conditions in the presence of Triton X-100. However, mapping of the original locus led to a second round of mutagenesis that produced a strain in which viability and wild type lipid composition were compromised at an elevated temperature [64, 65].
As with the psd gene identification, a brute force method of screening for reduced incorporation of radiolabel into CL was used to identify the first mutant in CL synthesis now termed clsA [66]. The mutant had a 15-fold reduction of CL content and reduced turnover of PG to CL but lacked any other remarkable phenotypes. Following studies over the next 34 years would finally identify multiple genes encoding CL synthases in E. coli.
Therefore, by the early 1980’s a mutant was available in nearly every step of phospholipid metabolism in E. coli beginning with the acylation of sn-glycerol-3-phosphate. A temperature sensitive mutant of the plsC gene (responsible of acylation of lyso-PA) was reported in 1990 [25]. This set of mutants provided the basis for isolating the genes for overproduction of enzymes, construction of completely null mutants, sequencing of genes, and more sophisticated investigation of lipid metabolism and the function of individual phospholipids. However, several issues remained to be solved in the next millennium. The clsA mutant still contained residual CL. Also the pgpA and pgpB double mutants were not lethal and contained normal levels of PG and CL. These results suggested multiple enzymes responsible for these steps in the pathway.
5. Purification and characterization of phospholipid biosynthetic enzymes
5.1. Phosphatidylserine decarboxylase
The first phospholipid biosynthetic enzyme to be purified to homogeneity was Psd of E. coli [67]. Beginning in the late 1960’s purification of membrane proteins after solubilization with detergents was an increasingly popular endeavor. Fortunately, all of the enzymes of phospholipid biosynthesis in E. coli remain active after solubilization with Triton X-100 and actually require Triton X-100 for maximum in vitro activity. At the time there were only a few detergents available, gene amplification by overexpression was not yet possible, and affinity chromatography and epitope tagging were yet to be available. A young enthusiastic medical student, Bill Wickner, did a research rotation in Kennedy’s lab and began the purification of Psd. Kilogram quantities of E. coli B cells were purchased from Grain Processing, Inc. as a source of the enzyme. The term “bucket biochemistry” was not coined for the process, but Wickner’s style of attacking the problem exemplified the approach. Pilot purifications generally started with 500 g of cell paste that was lysed using a Manton-Gaulin milk homogenizer, which employed shear force through a small oriphice. Membranes were isolated by high-speed centrifugation followed by solubilization with Triton X-100 in the presence of 10% glycerol. This extract was subjected to acetone precipitation and the re-dissolved material subjected to DEAE-cellulose ion exchange chromatography. As the purification procedure was developed, this step involved a 1 L bed-volume DEAE-cellulose column developed with a 12 L linear salt gradient in Triton X-100 and glycerol. A special fraction collector was build to accommodate 200 ml fractions. Unfortunately, the collector would occasionally malfunction in the middle of the night depositing liters of effluent on the floor usually immediately behind my lab bench. Standard methods of concentrating enzyme preparations using salt precipitation or pressure dialysis were not practical due to the presence of detergents so methods of reapplying the enzyme peak to a small DEAE column followed by step elution with salt enabled the following steps. I joined the project at about this stage of development and continued it to completion after Wickner moved on to his postdoctoral position with Arthur Kornberg at Stanford. After a sucrose gradient and two sizing columns, purified Psd was obtained. The final prep used for initial characterization of the enzyme began with nearly 8 kg of E. coli cell paste and ended up with 14 mg of enzyme after 3600-fold purification.
The initial characterization of the Psd provided information that required time to explain. The dependence on Triton X-100 for activity measurements was almost absolute with a steep increase in activity with increasing detergent levels followed by an apparent inhibition at higher concentrations. Ed Dennis (former Kennedy postdoc) later analyzed the kinetics of Psd and other enzymes toward lipid substrates in detergentlipid mixed micelles using a “surface dilution” model [68] to explain enzyme substrate and detergent interactions. This model for has been extensively used [69] to analyze the kinetic behavior of many enzymes that act on lipid substrates in detergent-lipid mixed micelles. The purified protein analyzed by SDS PAGE showed a major band at 30–36 kDa (depending on the system used) and what was originally thought to be a 10% contaminant at 15 kDa. Amino acid decarboxylases general have a pyridoxal phosphate or covalently bound pyruvoyl prosthetic group. The E. coli enzyme was colorless suggesting the latter type of enzyme. Also the minor protein species was later shown to become labeled by reduction with NaB3H4 [70] or NaBH4 reduction in the presence of radiolabeled substrate [71].
An interesting coincidence is that I had done my Ph.D. training with Esmond Snell at University of California, Berkeley studying pyridoxal phosphate dependent enzymes. However, while I was in his group the first pyruvoyl-dependent amino acid decarboxylase [72] was discovered. Cloning of psd gene [73] using complementation of the psdts mutant made a further analysis of the generation of the prosthetic group of the Psd possible. Qiao-Xin Li [71, 74] in my group later showed that the enzyme was translated in a pro-enzyme form that underwent posttranslational autocatalytic cleavage at an internal serine to convert the serine to an N-terminal pyruvoyl prosthetic group of a small peptide, i.e. the apparent contaminant in the original preparation of the enzyme. S. cerevisiae and somatic cell mitochondrial Psd’s are highly homologous to the E. coli enzyme [75, 76], and the yeast gene was cloned by complementation of an E. coli psd mutant [75]. Yeast and somatic cells also contain a second pyruvoyl extra-mitochondrial Psd with little homology to the mitochondrial enzyme [77].
5.2. Phosphatidylserine synthase
Integral membrane proteins carry out the synthesis of PS in Gram-positive bacteria [78], yeast [79] and somatic cells [39]. However, Kanfer and Kennedy [37] noted that the activity was largely in the supernatant after a high-speed centrifugation of a cell lysate of E. coli. The substrates for PssA, CDP-DAG and L-serine, are the same in eubacteria and yeast while in somatic cells PS is made by head group exchange of serine with PE or PC. When Raetz initiated purification of E. coli PssA, he noted that it was tightly associated with both 30S and 50S ribosomes even in high ionic strength buffers [80]. I remember a Gordon Research Conference in 1971 where the upstart medical student Raetz went up against established researchers in defending his evidence for the lack of membrane association of the enzyme. Raetz went on to disassociate the enzyme from ribosomes using 5 M NaCl. He also demonstrated a possible covalently bound phosphatidyl intermediate based on catalysis of CMP and serine exchange with CDP-DAG and PS, respectively and a low level CDP-DAG hydrolysis activity [81]. He also isolated both CDP-DAG and dCDP-DAG from E. coli extracts [82] thus confirming their participation as lipid biosynthetic intermediates in vivo. Within his own research group Raetz always demanded such evidence in establishing a tight relationship between in vivo and in vitro experiments. He also identified [83] and partially purified [84] a membrane associated CDP-DAG-specific (not dCDP-DAG-specific) hydrolase, the function of which is still not clear. Unlike other enzymes using CDP-DAG as substrate that are phosphatidyl transferases, the hydrolase is a CMP transferase [85]. It is not clear if it is a hydrolase with water as the CMP acceptor or if it is a CMP transferase to some other molecule.
Although the PS synthases of microorganisms utilize the same substrates, there are major defenses in enzymological properties, mechanism of action and subcellular localization. The E. coli enzyme has no divalent metal ion requirement and is not membrane associated while the yeast [79] and Bacillus [78] enzymes require Mg and are integral membrane proteins. The yeast enzyme is also well characterized [28] and has many properties closer to the Gram-positive bacterial enzymes [78]. Lipid substrates with [18O] in the phosphate residue were used to demonstrate that the E. coli enzyme proceeds by a Ping-Pong reaction mechanism through a possible enzyme bound phosphatidyl intermediate, as Raetz originally postulated, while the yeast enzyme (and probably the Gram-positive enzyme) proceeds by a Bi-Bi mechanism condensing its two substrates [86] most likely not through an enzyme covalent intermediate.
After setting up my own laboratory in Texas in 1972, I initially took Kennedy’s advice and attempted to purify and characterize enzymes of phospholipid metabolism from animal cells. However, seeing no successful attempts to pursue the initial purifications started in the Kennedy group, I returned to E. coli as my enzyme source. Affinity chromatography was being used extensively to purify enzymes, and we capitalized on a “poor man’s approach” to affinity chromatography for enzymes that have substrates containing phosphate. Many such enzymes bind to cellulose phosphate-based chromatography resins, which proved to be the key step in purifying PssA by my first graduate student Tim Larson [87]. The cytoplasmic fraction of a cell lysate was subjected to several precipitation steps in the presence of Triton X-100 followed by stripping the enzyme from ribosomes with 5 M NaCl. After dilution of the salt, the enzyme bound to a phosphocellulose column even in the presence of 0.65 M NaCl. However, inclusion of 0.4 mM CDP-DAG in the elution buffer specifically eluted the enzyme in near pure form. The yield was about 2 mg of enzyme from 450 g of cell paste with 5500-fold purification. The original findings concerning the Ping-Pong mechanism were confirmed.
The rapid development of molecular genetics in the mid-1975, coupled with isolation of the first mutants in genes encoding phospholipid biosynthetic enzymes and the cloning of genes on multi-copy plasmids converged in the lipid field in a collaborate effort between Raetz’s and my lab. As noted above, Raetz had isolated a pssA temperature sensitive mutant [58]. He had acquired an early copy of the Clarke and Carbon plasmid library of the E. coli genome [88] from his old friend and former lab mate Wickner, which Raetz used to screen for plasmids that restored PssA activity and viability to his mutant. Yes the “old boys club” was beneficial in this endeavor. The plasmids increased PssA levels by an order of magnitude resulting in our purification of about 9 mg of pure enzyme from 150 g of cell paste [89]. This report may be the first example of the use of plasmid-directed overproduction of protein to increase the amount and yield of enzyme isolated from whole cells. Akinori Ohta joined my lab as a postdoctoral fellow in the late 1970’s. He had been responsible for the initial generation of pssA mutants as a student in Shibuya’s group in Japan. He placed pssA on extremely high copy number plasmids, which boosted initial specific activity of the enzyme several 100-fold so that 34 mg of pure enzyme could be obtained in 50% yield from 100 g of cell paste [90].
Investigation of the nature of the unique ribosomal location of the PS synthase indicated that this association may be an artifact of cell lysis as determined by Katherine Louie a graduate student in my group [91]. 5 M NaCl is required to dissociate the enzyme from the ribosomal fraction while physiological levels of polyamines prevent ribosomal association, which is in much less than in stoichiometric amounts. Even more interesting is that among phospholipids only CDP-DAG-detergent mixed micelles under optimal assay conditions dissociated the enzyme from ribosomes. Also the enzyme has high affinity for membranes loaded with CDP-DAG [92]. Taken together, PssA is a peripheral membrane protein that associates with membranes via its lipid substrate and may spend time as both a membrane-bound and cytoplasmic protein, i.e. an amphitropic protein. Later sequence determination revealed an enrichment of positive charges at both ends of the enzyme [93]. Reversible membrane association dependent on lipid substrate is consistent with a detailed kinetic analysis using substrate-dilution kinetics and sedimentation analyses in the presence of CDP-DAG-detergent mixed micelles carried out by George Carman while a postdoctoral fellow in my group [94]. Carman’s introduction to the enzymology of E. coli phospholipid biosynthetic enzymes carried over into to his independent career where he continues to make seminal contributions to our understanding of phospholipid metabolism and its regulation in yeast.
5.3. Phosphatidylglyerophosphate synthase
The PGP synthase (PgsA) posed a formidable problem in purification since it is an integral membrane protein. However, the introduction of affinity chromatography made initial purification from wild type cells possible. Tim Larson made a column in which periodate oxidized CDP-DAG (vicinal diol of the ribose ring converted to two aldehydes) was covalently linked to a diamine linker arm attached to Sepharose [95]. PgsA in a Triton X-100 extract of E. coli membranes bound to such a column and eluted by breaking the link between the CDP-DAG derivative and the column. A second affinity column was used [96] in which the initially bound CDP-DAG derivative was pre-treated with NaBH4, which apparently increased the specificity and allowed elution of the enzyme in high yield using CDP-DAG in the mobile phase. Enzyme (3 mg) of about 85% purity was obtained from 450 g of cell paste with an overall purification of 6000-fold.
Genetic and biochemical analyses of mutants in PG biosynthesis were initially confusing, and the cloning of the pgsA gene [97] was questioned upon submission of the work for publication to a journal not to be named. The initial pgsA mutants [57] had very low PGP synthase activity but were not temperature sensitive for growth and did not display low levels of PG and CL. A second mutagenesis was carried out by Nishijima and Raetz [98] on the original pgsA mutant to generate a strain that was temperature sensitive for growth and phospholipid synthesis. This mutant, which is temperature sensitive for growth and nearly deficient in PG and CL biosynthesis, carried a mutation in a second locus (pgsB) far removed from pgsA. Introduction of the then putative pgsA cloned gene into a pgsAB double mutant suppressed the lethal growth phenotype and resulted in overproduction of PGP synthase activity. However, introduction of the pgsB clone into the double mutant also restored normal growth but did not amplify the residual PgsA level.
To resolve the function of the pgsA gene, the DNA sequence of the gene was compared to the partial amino acid sequences of CNBr generated fragments of the amplified homogeneous enzyme [99]. The only method of DNA sequencing available at the time was the labor-intensive and time-consuming chemical method of Maxam and Gilbert [100]. There was no question now as to the identity of the pgsA gene. However, the pgsA sequence was later revised and corrected using more updated DNA sequencing technology [101]. The complete sequence of the pgsA gene was the fourth report of a phospholipid biosynthetic gene sequence, which followed that of plsB/dgkA [102] and cdsA [103].
Sorting out the function of the pgsB gene resulted in the seminal report that opened up the lifetime pursuit by Raetz to completely characterize the genetics, enzymology, structure and regulation of the enzymes responsible for the synthesis of the membrane-associated Lipid A core of LPS in E. coli as well as many related Gram-negative bacteria (see [104] for a review). Nishijima and Raetz [98] noted that at the restrictive temperature for growth of a pgsAB double mutant, two minor phosphate-positive spots (X and Y) appeared on thin layer chromatography. Appearance of the phosphate–containing compounds was dependent on the mutation in the pgsB locus that rendered the cells temperature sensitive for growth and also caused the reduction in PG and CL levels dependent on the pgsA mutation. They determined that X was a precursor to Y and both were early precursors to the Lipid A core of LPS [105], which led to defining the complete pathway for Lipid A biosynthesis in E. coli and many other Gram-negative bacteria.
The identification of minor lipid species was a fascination for Raetz up to his untimely passing in 2011. Early in his independent career Raetz labeled E. coli cells with 32Pi, extracted the lipids and subjected the extract to two-dimensional thin layer chromatography [106]. Not surprisingly after a few hours of exposure to film, he observed PE, PG, and CL. After a longer exposure additional spots appeared, which were PA, PS, some lyso-phosphates and a few unknowns. As he exposed the plate for longer times, many more spots appeared (including X and Y) until the film was almost completely black. Within the identity of these minor spots lay a wealth of clues to the origin of biosynthetic pathways leading to important lipidic biological molecules. He promoted characterizing the structure of unknown lipid molecules in mutants and clone collections to which he applied the rules of enzymology to propose pathways for their conversion to major biological constituents. The focused development of mass spectrometry applied to lipids through the LIPID MAPS consortium (http://www.lipidmaps.org) directed by Ed Dennis has been essential for such detailed identification. Based on these structural considerations, plausible substrates were synthesized and tested as substrates of new enzymes, which were purified and expression cloned to identify the genes encoding the respective enzymes. This bottom up approach served him well in identifying new pathways, enzymes and genes in lipid metabolism, which was followed by detailed enzymological and structural studies of the enzymes involved (see testimonials to Raetz’s accomplishments [107, 108]).
5.4. CDP-diacylglyerol synthase
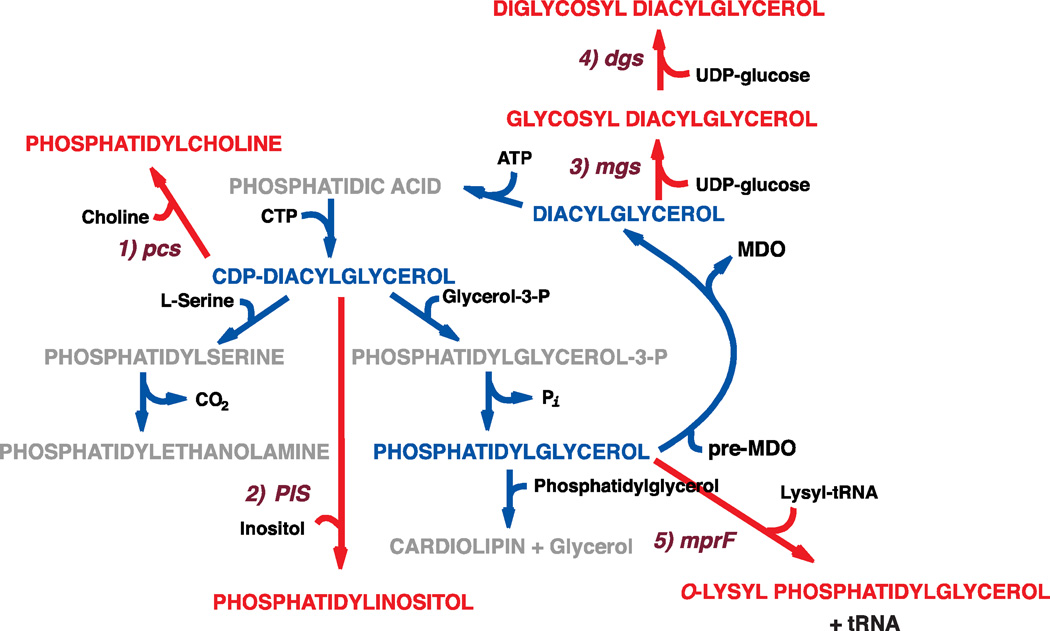
Like PA, CDP-DAG is an essential intermediate to all the phospholipids of E. coli by providing the substrate at the branch point leading (Fig. 3) to the amine-containing zwitterionic phospholipid PE and the anionic phospholipids PG and CL. Extensive focus on the CDP-DAG synthase (CdsA) and the gene (cdsA) encoding this enzyme did not occur until the 1980’s by the Raetz lab. Using the filter paper assay method, mutants were isolated with very low (d)CdsA activity with no growth rate defects [59]. However, most showed high accumulation of PA consistent with a mutation in the synthase. Once the cdsA locus was identified, further mutagenesis yielded several mutants that failed to grow at pH 8 and accumulated PA to nearly 30% of total phospholipid, which turned out to be due to a downshift in the pH optimum for the mutated synthase [109]. The accumulation was mostly at the expense of PG and CL rather than PE, which suggested that affinity for CDP-DAG may be significantly higher for the PS synthase than the PGP synthase. Coupled with the fact that withholding cytidine from cytidine auxotrophs results in high levels of PA indicated that CdsA supplied the vast majority of phospholipid precursor.
With a conditional lethal mutant in hand, cloning of the cdsA gene by complementation of the mutant was made possible followed by sequencing of the gene [103] and overproduction of the synthase for purification and characterization [110]. The plasmid amplification of the cdsA gene resulted in a 50-fold increase in starting specific activity for the synthase, which made possible the purification of about 2 mg of 90% pure enzyme from about 20 g of cell paste. Correcting for the initial overproduction, the purification from wild type cells would have been over 8000-fold. The minimum molecular mass of 27,000 Da corresponded well with the sequence predicted by the gene sequence. The enzyme followed substrate dilution kinetics with respect to PA and used both CTP and dCTP as substrates. Less than 0.5% of activity was shown using any other ribo- or deoxyribonucleotide substrate. There is considerable sequence homology with Cds’s from other organisms even though there are significant differences in molecular mass. This homology was used to identify and clone the respective synthase genes from S. cerevisiae [111], Drosophila [112], and humans [113].
5.5. Phosphatidylglycerophosphate phosphatases
The filter-paper screening technique was setup to identify mutants in PGP phosphatase (Pgp) and identified temperature sensitive mutants in two genes that encoded phosphatases (pgpA and pgpB). These phosphatases acted on several phospholipids including PA, lyso-PA and PGP [60]. However, none of the mutations conferred a temperature sensitive growth phenotype or significantly perturbed phospholipid composition except to increase PGP content from <0.1% to 1.6% of total phospholipid. PgpA appeared to be more specific for PGP while PgpB appeared to be more specific for the other two phospholipids. Overexpression of PgpA mostly increased PGP phosphatase activity [114] while overexpression of PgpB [115] increased phosphatase activity towards all three substrates with a lesser effect on PGP phosphatase activity. PgpA is localized to the inner membrane while PgpB was originally thought to be an outer membrane phosphatase, but more recent results indicate an inner membrane location [116]. Clearly either the mutations did not confer complete dysfunction in vivo at elevated temperatures for PGP phosphatase activity or additional PGP phosphatases exist.
Partial clarity was brought to the problem by construction of completely null mutations individually and together at the pgpA and pgpB loci [117] carried out by Cindy Funk a graduate student in my group. Such an approach was highly dependent on the pioneering work of Raetz in identifying genes responsible for phospholipid biosynthesis, which could be used as targets for complete gene inactivation to determine whether particular enzymes or lipids are essential or whether there are multiple enzymes for particular steps in the pathway. The null mutants behaved the same as the original temperature sensitive mutants when grown at elevated temperature so clearly there was a third pgpC gene yet to be identified. The reason Icho and Raetz [60] were able to identify PGP phosphatase mutants based on lack of significant incorporation of 32Pi into lipid at elevated temperature in the filter paper assay is that the residual PGP phosphatase activity (pgpC gene product) is rapidly inactivated at 42°C under assay conditions containing Triton X-100. However, it was left to Raetz’s group to clear up the uncertainty surrounding PGP phosphatases in E. coli by cloning the pgpC gene [43]. An E. coli genomic plasmid library was expressed in a ΔpgpAB mutant (null in both genes), and pools of individual colonies were assayed for increase PGP phosphatase activity. Using this expression cloning and direct enzyme assay approach, the third PGP phosphatase was identified. The ΔpgpABC null strain grew with wild type lipid composition provided it carried a plasmid copy of anyone of the genes. Complementing the triple mutant with anyone of the genes on a plasmid that is temperature sensitive for replication resulted in growth arrest at elevated temperatures with accompanying accumulation of PGP and loss of PG/CL synthesis.
PgpC is predicted to span the inner membrane one time with its putative haloacid dehalogenase active site facing the cytoplasm [43] where it most likely acts as the primary PGP phosphatase. PgpC homologues are found in a broad spectrum of bacteria. PgpB has been purified to homogeneity and topological organization of the enzyme in the inner membrane indicates that its putative active site faces the periplasm [116]. In addition the enzyme is among four gene products that act as undecaprenyl-PP phosphatases, which is consistent with the production of this substrate on the outer face of the inner membrane. Also PgpB is the least effective PGP phosphatase in vivo. Little is known about PgpA homologues, which are found in a narrower spectrum of Gram-negative bacteria than the PgpB family. None of the E. coli phosphatases show significant homology to either yeast [118] or somatic cell [119] PGP phosphatases, both of which appear to be the primary PGP phosphatases.
5.6. Cardiolipin synthases
CL synthases have not been purified to high degrees of homogeneity from any bacterial source due to their low amount. However, cloning of the clsA gene of E. coli [120] and overproduction of ClsA over 700-fold provided starting material for a rapid and extensive purification [121]. Although the enzyme was highly overexpressed, there was only a 30% increase in CL levels at the expense of mainly PG. Overproduction resulted in a reduced membrane potential, fragility of spheroplasts and eventual loss of viability all suggesting some compromise of membrane barrier function. However, these properties may not be due directly to increased CL since mutants in other phospholipid biosynthetic enzymes have even higher CL content without displaying these properties [93]. The only chromatography medium found suitable for purification was phosphocellulose with elution by a phosphate concentration gradient. SDS PAGE of the eluent from the final column displayed a major protein species of 45 kDa corresponding to the plasmid-amplified protein. The estimated 10,000-fold enrichment over wild type levels of ClsA approached only 80% of purity with several additional bands on the gel.
There remain important unanswered questions as to the size of mature ClsA. The gene sequence predicts a protein of mass 54–55 kDa [120], which is observed when the protein is synthesized in vitro in the absence of membranes. However, the purified protein has a mass of 45–46 kDa [121]. Post-translation cleavage removing 8 kDa from the N-terminus would explain this result but would also remove the only two predicted transmembrane domains and possibly prevent membrane insertion. Attempts to resolve this contradictory result have not been completely successful [122]. Tagging the protein at the C-terminus with a 6 × His tag results in a membrane associated inactive protein of 46 kDa that retains the C-terminal tag. Using an internal epitope tag, a full length and an N-terminal truncated (Δ2–60) ClsA were membrane associated, fully active and displayed a mass of 46 kDa and 48 kDa, respectively as had previously been shown for the truncated native protein [123]. The difference in mass indicates that the cleavage sites for the full length and truncated proteins may be different. Proper processing and activity of the full-length protein is affected by several mutations in the N-terminal domain that is removed post-translationally [122]. Most interesting are serine substitutions at positions 7 and 8 that significantly reduce in vivo activity without affecting in vitro activity. Quigley and Tropp suggested that the mutant protein is misoriented in the membrane with its active site facing the cytoplasm. Shibuya et al. [124] had suggested that the active site faces the periplasm based on the formation of phosphatidylmannitol driven by the reverse reaction when cells were grown in high concentrations of mannitol, which was not transported into E. coli in the strain they used. In this reaction mannitol would take the place of glycerol in the reverse reaction. These questions relating to ClsA posttranslational modification and orientation remain unresolved.
Since null alleles of clsA still contained low levels of CL [125], there appeared to be additional CL synthases as was found for PGP phosphatases [43]. Guo and Tropp noted two additional clsA paralog genes (ybhO and ymdC) in E. coli [123]. They were able to demonstrate CL synthase activity (using radiolabeled PG as substrate) for ybhO (now called clsB) in vitro with a crude membrane preparation from the ΔclsAB mutant only if it was overexpressed from a plasmid. However, they were not able to demonstrate increased significant CL synthesis in vivo by cells overexpressing ClsB from a plasmid in a ΔclsAB background. Guo and Tropp grew cells in rich medium containing low NaCl (0.5 g/L), which will become important later in interpreting their results. A ΔclsAB strain was reported but its phospholipid composition was not determined [126], even though it was used to establish lack of involvement of CL in polar localization of MurG responsible for peptidoglycan biosynthesis.
The ymdC gene (now called clsC) was cloned and all combinations of null mutants among the three cls genes were constructed [50]. Using both highly sensitive 32Pi labeling of cells and liquid chromatography/mass spectrometry (LC/MS) analysis, no CL was detected under a variety of growth conditions including media of high and low ionic strength or osmolarity and growth into late stationary phase. Interestingly each strain expressing only one of the three CL synthases showed the formation of CL, but the amount of CL was different for each depending on the NaCl concentration or osmolarity of the growth medium and whether the cells were analyzed during log growth or in stationary phase [50]. CL levels are know to increase at the expense of PG as cells enter stationary phase and also when cells are grown in higher ionic strength or osmolarity medium [127]. Expression of ClsA results in the highest content of CL in log or stationary phase irrespective of ionic strength or medium osmolarity. Neither ClsB nor ClsC contributes detectible CL during log growth in media of low ionic strength (i.e. 0.5 g/L NaCl) or low osmolarity. However, they do make measurable amounts of CL (ca. 2%) when cells are grown to stationary phase. This appears to be the reason Guo and Tropp did observe CL synthesis by ClsB in vivo since they used media with low NaCl content [123].
However, the story gets more interesting upon further investigation. The clsC lies within a polycistronic gene preceded by ymdB of unknown function [50]. Tan et al. [50] suggested a two-protein complex (ClsC and YmdB) transcribed from the same operon with ClsC being the catalytic subunit and YmdB being an enhancer of activity. Expression of either gene alone from plasmids in a ΔclsABC ΔymdB background only resulted in a low level of CL with a clsC-containing plasmid and no CL with a ymdB-containing plasmid. CL levels near that observed for ClsA occurred when ClsC and YmdB were expressed in tandem from the same DNA segment. ClsABC all belong to the phospholipase D superfamily and ClsA had been shown to catalyze the condensation of two PG molecules with the formation of CL and glycerol [49]. YmdB, consistent with the inability to catalyzed CL synthesis, does not contain the phospholipase D motifs. Mutation of the putative catalytic domains of ClsC inactivated the enzyme. However, ClsC carries out CL synthesis by a novel mechanism involving a phosphatidyl transfer from one PE molecule to PG rather than a phosphatidyl transfer from one PG molecule to a second PG molecule [50].
Establishment of the novel mechanism for ClsC relied completely on employing mass spectrometry to analyze the products. First, a PG containing a non-native 17:0/14:1 fatty acid composition was used as substrate with a crude membrane preparation from the following strain. A ΔpgsA strain (discussed latter and completely lacking PG and CL) was made null in clsABC and ymdB. This mutant was then used to express either ClsA or YmdB-ClsC from a plasmid. When membranes containing ClsA were incubated with the non-native PG, both PA moieties of CL displayed the non-native fatty acid composition. This is consistent with using two PG molecules to make CL with the release of glycerol. However, YmdB-ClsC-containing membranes only incorporated the non-native fatty acids into one of the two PA’s of CL with the other having a fatty acid composition similar to that of endogenous phospholipids of E. coli. Use of CDP-DAG containing non-native fatty acids ruled out its involvement as a second substrate. Next, the same experiment was carried out using both PG and PE with different but non-native fatty acid compositions. The resulting CL using the YmdB-ClsC membranes contained one PA derived from the PG and one from the PE. Therefore, ClsC catalyzes a condensation between PE and PG to yield CL with the apparent release of ethanolamine. In light of this result, a further investigation as to the mechanism of ClsB is needed. The phospholipid pathway outlined in Fig. 3 has been modified from earlier versions to show the new information with respect of CL synthesis (step 6). The same might apply to the CL synthases from other bacteria, which have all been assumed to catalyze the condensation of two PG molecules. A recent report [128] suggested a “bacterial-type” CL synthase in Trypanosoma brucei, which would be the first in a eukaryote. This identification of mechanism was based solely on informatics identification of the phospholipase D catalytic motif without reporting evidence for in vitro CL synthesis dependent on gene expression. As we now known, more rigorous verification of substrates and products are required before a mechanism for CL synthesis can be suggested in any cell type.
ClsA is expressed under all growth conditions and contributes the majority of CL [50]. ClsB and YmdB-ClsC contribute CL mostly during late log and stationary phases with only minor contribution during log phase. In low osmolarity growth medium all three Cls’s make less CL with ClsB and YmdB-ClsC contributing barely detectible amounts, which increase with increasing osmolarity/ionic strength of the growth medium. Whether these media effects reside at the enzyme catalytic level or are due to differences in gene expression remains to be determined. There are no dramatic phenotypes due to the loss of CL, although null mutants in clsA, which fail to induce high levels of CL in stationary phase, lose viability more rapidly during long-term incubation in stationary phase [127]. The existence of multiple genes encoding CL synthases with homology to the E. coli enzymes is wide spread among bacteria as is summarized in this report [50]. However, eukaryotic CL synthases show no homology to the bacterial enzymes, which is not surprising due the differences in reaction mechanism.
Conclusions about how and under what conditions CL is made in E. coli (and probably in other organizes) needs careful reinvestigation. Shibuya et al. [129] suggested that the low level of CL observed in a clsA null mutant might be due to a side reaction of the PS synthase. This was based on the observation that in a pssA temperature sensitive mutant also null in clsA, the level of CL increased with the increase in catalytic activity of the PS synthase. The molecular basis for this observation probably lies in the fact that the then unknown ClsC utilizes increasing amounts of PE made by increased PS synthase activity. Guo and Tropp [123] concluded that ClsB did not make CL in vivo because overexpression of ClsB could not rescue the lethality of the pssAts ΔclsA strain at elevated temperatures. However, they used low salt media in their experiment where ClsBC do not contribute significant levels of CL. Finally, what are the functions of ClsB and YmdB-ClsC, if ClsA contributes the majority of CL under all growth conditions.
6. Diglyceride kinase and membrane derived oligosaccharide biosynthesis
As noted earlier, Kanfer and Kennedy [32] reported stable incorporation of 32Pi into PE but rapid turnover of the label incorporated into PG. Schulman, a graduate student in the Kennedy lab, and van Golde, who was on sabbatical leave from the Utrecht lipid powerhouse, returned to this problem in the mid 1970’s and discovered a group of glucose-containing oligosaccharides decorated with moieties derived from the head groups of phospholipids [130]. These were termed “membrane-derived oligosaccharides” (MDO) and were localized to the periplasmic space. In an update of the earlier labeling and stability experiment, E. coli was label with sn-[2-3H]glycerol-3- 32P. It should be noted that use of sn-[2-3H]glycerol-3-P to label cells is very specific for phospholipids because any other use of this compound must go through the formation of dihydroxyacetone phosphate with loss of tritium label. Again label incorporated into PE was stable during the chase while significant amounts of both labels were lost from PG and CL to water soluble, chloroform insoluble compounds containing glucose as the only sugar. The heterogeneous mixture of compounds all contained glycerophosphate with some containing succinic acid in ester linkage. The material made up about 1% of the dry wet of cells. Subsequent work showed that the glycerophosphate as the sn-1 phosphate was derived from PG and was attached via a phosphate ester at the 6 position of one of 10–12 glucose molecules of the highly branched oligosaccharide [131, 132]. The chirality at the 2-position of glycerophosphate was an important clue to its source. The PGP precursor to PG has a phosphate at both the 1- and 3-position of the unesterified glycerol with the 1-phosphate remaining after conversion to PG. Some forms of MDO also contained phosphoethanolamine, derived from PE, at the same position. Lack of significant turnover of PE in previous experiments is probably due to the relative amount of PE versus PG. The oligosaccharide is synthesized from UDP-glucose [133], and the phosphate-containing decorations are derived from membrane phospholipids by enzymes located in the periplasmic space [131, 134].
The expected lipid byproduct of such a head group transfer would be sn-1,2-diacylglyerol. Mutants in diglyceride kinase (dgkA gene) accumulate its lipid substrate as high as 8% of total glycerol-based lipids [135]. Blocking MDO synthesis at the level of UDP-glucose utilization or blocking glucose synthesis greatly reduced diglyceride formation. Overall the data supported a “diglyceride cycle” in E. coli [135] where MDO synthesis generates diglyceride from phospholipid head group utilization (Fig. 3). The diglyceride is then fed back into the phospholipid biosynthetic pathway with the formation of PA via the diglyceride kinase. The level of diglyceride formation and the rate of flux through the cycle is inversely proportional to the osmolarity of the growth medium because MDO’s maintain the osmotic pressure of the periplasmic space of not only E. coli but Gram-negative bacteria in general [136]. The negative effect on growth of a dgkA mutant in medium of low osmolarity was effectively used to isolate mutants in the biosynthesis of MDO [137].
Therefore, a major route for turnover of PG is dependent on the demand for MDO while in stationary phase PG turns over to CL. However, nothing is known about the turnover of CL. Contrary to earlier findings PE does turnover again dependent on the demand for MDO. Although the 1-acyl group of PE is the major source for acylation of the N-terminal cysteine of the major outer membrane lipoprotein (encoded by lpp) plus probably all of these types of lipoproteins [33, 34], strains completely lacking PE still acylate lipoproteins using other phospholipids [138]. Another route for PE turnover is transfer of phosphoethanolamine to the Lipid A core of LPS [139], which also generates diglyceride. Interesting all of these turnover steps use phospholipid parts as precursors for the synthesis of other important biomolecules without the complete degradation of any one phospholipid species.
7. Construction of null mutants
7.1. Overview
The initial isolation of conditional mutations in many of the genes encoding phospholipid biosynthetic enzymes was a significant development that led to cloning of the genes, overproduction of the gene products, identification of orthologs in other species and purification of many enzymes of phospholipid metabolism. However, these mutants did not address the issue of the function of or necessity for specific phospholipids. The set of cloned genes now made it possible to construct null mutants at several points in the pathway. The importance of this approach was illustrated above in uncovering multiple genes such as pgpABC and clsABC. However, many of the genes of interest appear to be essential especially if there is only a single gene encoding a particular function. In fact the most recent update of the Keio library of single null gene constructs of the entire E. coli genome (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp [140]) only lists clsA, pgpAB, dgkA, and cdh as non-essential genes. It is understandable that plsBC and cdsA are essential genes because of their early position in the pathway (Fig. 3) and the lack of additional genes encoding these activities. It turns out that the remaining “essential” genes can be made null producing strains that can be propagated under defined culture conditions or if combined with second site suppressor mutations.
My interest in constructing null mutants in genes related to phospholipid metabolism started during a sabbatical with Jeff Schatz in Switzerland in 1984 where I also initiated my second research focus on the role of specific phospholipids in yeast mitochondrial function. Yeast geneticists had developed easy recombinant methods to generate gene knockouts and could utilize diploid strains to isolate disrupted essential genes. Upon returning from sabbatical, my lab set out to determine which if any of the major phospholipids of E. coli were essential for viability or if cells lacking of these lipids displayed phenotypes that would provide clues to functions of each lipid. Unfortunately, no easily usable systems for gene disruption, as in yeast, were available for E. coli at the time. Direct transformation with linear DNA followed by recombination was not very efficient due to highly active nucleases. Systems using the λ Red recombinase now make construction of null mutants very easy [141], and null mutants are available in all non-essential genes from the Keio library [140]. The laborious method initially used to construct strains with selectable drug-resistance markers inserted into specific genes is described in [142]. Phil Heacock joined my group as a technician shortly after my return from sabbatical. He has carried out much of the complex molecular genetic manipulations of E. coli strains that have been used in my group up to today.
7.2. Mutants lacking phosphatidylglycerol and cardiolipin
The first gene in phospholipid biosynthesis to be disrupted was pgsA [142] followed the next year by clsA [125]. Since we assumed the gene is essential, disruption (pgsA30::kan) was carried out in the presence of a copy of the wild type gene carried on a low copy number plasmid that was temperature sensitive for replication, which facilitated loss of the plasmid during growth at 43°C. Various parameters were followed during growth of the final construct at 43°C. PgsA activity dropped from the overproduced level due to the plasmid to about wild type levels within the first few generations and then continued to undetectable levels at the point of cell arrest of the culture at 4.5 h. Culture content of PG began to drop at about the time PgsA activity levels were 25% of wild type while the viability judged by plating for single colonies at 30°C began to drop about the time PG and PgsA activity levels fell below wild type levels presumably due to those cells that had lost the plasmid. At the point of culture arrest, the total of PG, CL, and PA was about 5% of total phospholipid. Therefore, there was a good correlation between loss of the covering plasmid, cell viability, PgsA activity and PG plus CL levels. Cells continued to grow until PgsA levels dropped below a level to sustain PG synthesis necessary for continued growth. Even though viability was compromised as shown by the inability to form single colonies after loss of the plasmid, there was no apparent lysis of cells. The conclusion was that PG was essential for cell viability but why it is important still remained to be determined. That is dead cells tell few stories. We needed to have viable cells compromised in specific functions.
We next made one of the first inducible systems for regulating phospholipid composition by placing the pgsA gene under control of the PlacOP (promoter of the lac operon). In order spread out the induction curve as a function of isopropyl-ß-Dthiogalactoside (IPTG) inducer concentration, it was necessary to have a strain lacking the lactose permease (LacY) [143]. With this system we were able to vary anionic phospholipid (PG + CL + PA) content in an IPTG-dose responsive manner from the minimal level of about 5% to wild type levels of about 25%. Below about 15% of PgsA wild type levels the level of PG and CL began to decrease and cell growth rate slowed and finally arrested at zero inducer levels. An important observation was that cells were still viable and rapidly resumed growth with addition of IPTG. Therefore, cell membrane integrity was not compromised. Cell growth was limited by the capacity of the residual PgsA activity to supply sufficient PG either as a precursor for some other macromolecule (MDO or lipoproteins), a regulator of an essential metabolic process, or a necessary constituent of the membrane bilayer. What we did not appreciate at the time was that PlacOP is a stochastic promoter that behaves in an all or none fashion [144]. Therefore, the level of enzyme and PG in the culture as a whole was dose dependent on IPTG because IPTG concentration determined the number of cells that were fully induced and not the level of PG in each cell of the culture. Later we would use promoters that gave homogenous induction over all cells of the culture.
7.2.1. Suppressors of the ΔpgsA allele
Upon further investigation things got very complicated and in the end our conclusion was correct that PG is essential in wild type cells, but cells could be constructed that were robustly viable without PG and CL. Miyazaki et al. [145] had previously generated a very tight temperature sensitive mutant of pgsA in a strain (SD12) that carried several other mutations. Among them was a mutation in the synthesis of the major outer membrane protein (lpp gene) [146]. The mutant grew well at 30°C with barely detectible PG, CL or PgsA activity. Aware of this observation, we introduced the ΔpgsA allele into SD12 and again concluded that pgsA gene is essential [142]. Later it was shown that lethality of strains lacking the capacity to make PG was due to several reasons. First, lack of transfer of a diacylglycerol moiety from PG in thioether linkage to the N-terminal cysteine [147] of the lipoprotein results in its accumulation in the inner membrane instead of its transport to the outer membrane [148]. Since this protein, although small, has the highest copy number of any protein in E. coli, this accumulation led to cell growth arrest and eventually cell death. Cells lacking the lipoprotein have some compromised barrier function of the outer membrane but otherwise are viable. Therefore, SD12 should have been viable with a ΔpgsA allele as was eventually shown [146]. However, it turns out that the ΔpgsA allele confers does not prevent growth at 30°C but results in poor growth at 37°C with loss of viability above 40°C even in the absence of lipoprotein synthesis [149, 150]; there is also lack of growth in minimal medium and medium of low osmolarity, the latter of which maybe due to the lack of sufficient MDO synthesis. Therefore, during the curing of the temperature sensitive covering plasmid in our original construction of the ΔpgsA allele, the cells became temperature sensitive and did not survive. It turned out that viable ΔpgsA strain [149] contained an additional mutation rcsA gene which suppressed activation of RcsCDB phosphorelay signaling system allowing the cells to grow at 37°C but not at 42°C where cell lysis still occurred [150, 151]. Therefore, by introducing mutations in the phosphorelay system, lpp and pgsA, a robust viable strain completely lacking PG and CL is available [149, 152]. As a side note, it is not clear whether PGP phosphatase activity is absolutely required. The evidence for lethality is based on curing a covering plasmid at 42°C in a largely wild type background except for ΔpgpABC [43], which is the same evidence used for establishing essentiality for the pgsA gene. Whether the same suppressors that rescue the ΔpgsA strain would rescue the ΔpgpABC strain has not been tested.
7.2.2. Other functions of anionic phospholipids
The initial observation that PG is essential in wild type cells was correct and with the efforts begun by the Shibuya lab followed through by Matsumoto and Hara labs several important functions of PG have been established. Although the major outer membrane lipoprotein (and possibly other lipoproteins) and MDO are not absolutely required for E. coli survival, they are important under certain growth conditions to stabilize the membrane envelope. Also ΔpgsA strains carrying the above suppressors are viable at 37°C but not at 42°C, which has not been explained. Are there other functions that require PG or CL? Both in vivo and in vitro experiments support a requirement for anionic phospholipids in translocation of proteins across the inner membrane, initiation of DNA replication and proper location of the division septum at mid-cell. Components required for all of these functions are amphitropic proteins that reversibly interact with anionic phospholipids at the membrane surface. Since PA and CDP-DAG are absolutely required, it is not possible to make a strain completely lacking anionic phospholipids, which may partially substitute for PG and CL.
A strain carrying the pgsA3 allele (not a null allele but contains very low levels of PG or CL [145]) and an lpp mutation is compromised in SecA-dependent translocation of outer membrane across the inner membrane [153]. Inverted membrane vesicles made from the anionic phospholipid depleted strain also displayed a significantly slower rate and steady state level of protein translocation than control membranes including those in which only CL was depleted. In a ΔpgsA strain (also with a lpp mutation) in which PG/CL levels were regulated using IPTG, the rate of protein translocation was proportional to the level of PgsA induction [154, 155]. For membranes isolated at different levels of induction, there was an IPTG-dependent dose response to SecA (cytoplasmic protein required for pre-protein translocation across the inner membrane) membrane binding, stimulation of latent required SecA ATPase activity, and effectiveness of protein translocation. However, this dose response relationship probably reflected the proportion of fully induced cells. Finally, addition of PG to the anionic phospholipid depleted inverted membrane vesicles restored SecA-dependent protein translocation [154].
Kornberg had reconstituted DNA replication in vitro and observed that DnaA initiates DNA replication in an ATPase dependent manner but fails to continue new rounds of DNA replication because the resulting ADP is tightly bound to DnaA. Initially, CL but not PG (dipalmitoyl) when added to the in vitro system reactivated DnaA for additional rounds of initiation by inducing ATP exchange for ADP [18]. Later it became apparent that PG was equally effective when containing unsaturated fatty acids [17, 156]. Choice of fatty acid composition is also of importance in in vitro experiments with avoidance of phospholipids with both chains saturated or both chains unsaturated. The former does not disperse as well as the latter and neither is highly represented in E. coli phospholipid species. Using the ΔpgsA strain in which PG levels could be regulated by IPTG, Weiming Xia in my group demonstrated that growth arrest of this strain at limiting PG levels could be suppressed by activating a DNA initiation system that bypasses the involvement of DnaA [157]. DnaA, which is isolated as a soluble protein, has high affinity for anionic phospholipid-containing liposomes [158, 159] and a photoactivated anionic phospholipid crosslinks to DnaA at a positively charged amphipathic helix in DnaA [158]. A leucine to lysine mutation in this helix renders DnaA function independent of anionic phospholipids and restores growth to cells limited in anionic phospholipid content [160].
MinD is an ATPase and is part of the Min system that assures organization of the division site at mid-cell [161]. It oscillates from cell pole to pole where it binds in association with MinC to the membrane through a C-terminal positively charged amphipathic helix when in the ATP-bound form. The MinCD complex shields the poles from components that build the division septum at mid-cell. Binding by MinE stimulates ATPase activity followed by the depolymerization of MinD and release from the membrane. In a mutant completely lacking PE and now containing only anionic phospholipids, the cycling of MinD, labeled by Green Fluorescent Protein, between the cell poles is disrupted. The dwell time for MinD in association with the membrane surface through CL-enriched membrane domains is now significantly increased [162]. We had previously demonstrated that similar but stable domains visualized by the CL specific dye Nonyl Acridine Orange (NAO) exist in this PE-lacking mutant [163]. NAO also labeled the cell poles and the division site in wild type cells consistent with the enrichment of CL at the cells poles [164]. Although not directly demonstrated, the pattern of MinD oscillation in cells lacking PE matches the distribution of CL domains along the membrane. MinD binding to liposomes is dependent on ATP and the presence of anionic lipids in a protein cooperative manner [162] mimicking the oligomerizatoin of MinD on the membrane surface in vivo. MinD binding to liposomes containing anionic phospholipids perturbs lipid organization [165] and is enhanced when anionic lipid domains are induced in the presence of neutral zwitterionic phospholipids [166].
The combined in vivo and in vitro results are strong support for involvement of PG and/or CL in the above function of amphitropic proteins. On the other hand ΔpgsA strains (completely lacking PG and CL [151]) display normal growth, DNA replication, and pole-to-pole oscillation of MinD [167] although they are shorter than the parent wild type strain. The most likely explanation is that E. coli is less attuned to the structural properties of its membrane lipids than eukaryotic cells and relies mostly on their less constraining physical and chemical properties. ΔPgsA strains, such as UE54, still contain nearly 10% of the phospholipid as anionic. In the complete absence of PG and CL and with 90% of the phospholipid as PE, NAO still bound to something at the cell poles but only elicited green fluorescence, normally seen with any anionic phospholipid, and not a red shift in fluorescence normally observed with CL [167]. Analysis of the lipids that localized to the poles (which are derived from the septum area) in UE54 showed a high enrichment in PA as well as a newly discovered phospholipid in E. coli, N-acyl PE. The localization of CL normally to the cell poles is believed to be due to its ability to induce negative curvature in the lipid bilayer because of its small head group and large hydrophobic domain. Both PA and N-acyl PE are also anionic lipids and have similar non-bilayer prone properties as CL (see Fig. 5). It is not possible to completely eliminate anionic phospholipids from E. coli since PA and CDP-DAG are essential intermediates to all phospholipids. E. coli shows greater promiscuity than mammalian cells with respect to the structure of supporting lipids so that most anionic phospholipids will support enzyme function in vitro. Therefore, the best evidence for specific roles for these lipids in the above process in vivo stems from studies of the interaction of specific proteins with lipids in vitro correlated with modification of cellular processes in mutants with reduced anionic phospholipids.
Figure 5.
Structure, physical and chemical properties of lipids native to E. coli and those introduced from other sources. See Fig. 4 for pathways of synthesis and genes encoding enzymes responsible for the synthesis of each lipid except N-acyl PE, which is discussed under section 7.2.2. The glycerol backbone (red) shown in ester linkage to fatty acids (aliphatic chains R1-R4) at the sn-1 and sn-2 positions and in either phosphodiester linkage for phospholipids or in a glycosidic linked for glycolipids at the sn-3 position. Head groups are color coded to indicate the charge nature of each head group.
7.3. Phosphatidylethanolamine and phosphatidylserine are not absolutely essential
With the success in making null mutants in pgsA, we decided to determine if the amine-containing phospholipids of E. coli were essential. The existing temperature sensitive mutants in pssA and psd become filamentous as PE levels decrease [58] or PS accumulates due to lack of conversion to PE [65], respectively. In the case of the psd mutant growth arrest could be suppressed by supplementing rich medium with Mg2+. PS levels rose from 48% in low Mg2+ to 76% in high Mg2+ with PE levels reduced to 5% of total phospholipid. With the pssA mutant grow arrest occurred when PE levels dropped from 85% to 32% of total phospholipid; CL levels also rose from 17% to 58%. Addition of Mg2+ to the growth medium also suppressed the growth arrest phenotype without correcting phospholipid composition [53, 168]. Since Mg2+ appeared to suppress the growth phenotype of the pssA and psd mutants, we set out to make null alleles of these genes.
Although we were able to make several mutants in the psd locus to study the formation of the Psd pyruvoyl prosthetic group [71, 74], interruption of the gene by a drug marker was unsuccessful [169]. Covering a psd::kan allele with the plasmid copy of the psd gene alone did not restore viability even in the presence of divalent cations, but including the neighboring gene (yjeP) in tandem on the plasmid restored viability and normal phospholipid composition. We concluded that the two genes are in an operon and that interruption of psd had a polar effect on yjeP transcription. YjeP is listed as a non-essential gene in the Keio Collection and is predicted to encode a mechanosensitive channel. Possibly the membrane related function of this protein is essential in a background in which membrane lipid composition is altered. Further investigation into the relationship between Psd and YjeP function has not been initiated.
A ΔpssA strain [93] was constructed in a similar manner to the ΔpgsA strain [142] using a temperature sensitive covering plasmid carrying a functional copy of the gene. After a shift in growth temperature to 43°C, the mutant strain followed a similar pattern to the ΔpgsA strain in that PssA activity, PE levels (reduced to 30% of total phospholipid) and cell viability declined after the plasmid was lost with growth arrest after about 13 generations. The unexpected result was that supplementation of the growth medium with divalent cations (Ca2+, Mg2+, Sr2+, but not Ba2+) suppressed the growth arrest phenotype but not filamentous growth, loss of PssA activity or elimination of PE. Cells did grow about half as fast as wild type cells and did not reach as high a cell density before entering stationary phase. A concern in studying the mutant is that newly prepared null mutants grow slowest and with multiple culturing grow better suggesting that suppressor mutations may be generated with time. Maximum growth rates are obtained in 15 mM Ca2+, 50 mM Mg2+, or 15 mM Sr2+ with no grow in Ba2+. Several other divalent cations, monovalent cations, various osmotic stabilizers, or trivalent cations (Heacock and Dowhan, personal observation) did not support growth. The dependence of growth rate on Sr2+ concentration is bell-shaped with no growth at 5 or 20 mM. These results suggested a specific requirement for a set of divalent cations to stabilize a physical property of the lipid bilayer (discussed below) rather than stabilization due to ionic strength or osmolarity. However, rapid dilution of the culture without divalent cations or into EDTA results in cell lysis. The mutant has a general requirement for various mixtures of amino acids including tryptophan (Heacock and Dowhan, unpublished result) and therefore requires rich medium for growth. There are only minor differences in the fatty acid composition among the wild type and mutant strain grown in the different divalent cations [170]. Radiolabeling with 32Pi showed no detectible PE, lyso-PE or PS after long exposure to film of extracted lipids separated by two-dimensional thin layer chromatography [93]. No new phospholipid species were apparent, and the phospholipid to protein ratio was maintained by an increase in CL over PG and increases in PA and CDP-DAG. However, an exhaustive search for PE using very high specific activity 32Pi and isolation of the neutral lipid fraction that did not adhere to a DEAE column revealed a labeled component that behaved as PE in three different chromatography systems. This material represented about 0.007% of total phospholipid or about 1500 molecules per cell. Whether or not his material is PE has never been further investigated; analysis of the extracted lipids from the strain using liquid chromatography/mass spectrometry (LC/MS) did not detect PE (Raetz, personal observation). Therefore, a strain of E. coli was now available which has been used to probe the possible functions of PE.
7.3.1. Divalent cation requirement in the absence of phosphatidylethanolamine
There appears to be a synergistic relationship between PE and CL in E. coli. Conditions can be found to support viability of strains lacking either lipid [50, 93]. However, as originally reported [93] and with numerous attempts over the years, a ΔpssA ΔclsA strain is not viable. Both phospholipids have the unique ability to introduce stress in the bilayer through the capacity to form non-bilayer (hexagonal II or HII phase) structures due to the negative curvature induced by the relatively larger hydrophobic domains as compared to the hydrophilic head group domains [171, 172]. However, CL only imparts negative curvature in the presence of three divalent cations (Ca2+ > Mg2+ > Sr2+ ≠ Ba2+) [172, 173] with the same efficiency as these ions support growth of a ΔpssA strain [93, 170, 174]. Mg2+ concentrations inside E. coli are on the order of 100 mM [175] and are uniform over the membrane and cytoplasm. Ca2+ and Sr2+ are actively pumped out of E. coli to maintain µmolar levels [176] with localization at the cell envelope [175] suggesting a stabilizing role for the highly negative LPS layer, which is disrupted by chelating agents. Therefore, the site of the divalent cation requirement in ΔpssA strains is most likely somewhere amongst the outer leaflet of the inner membrane and the inner and outer leaflet of the outer membrane.
Further evidence supports a specific role for CL and/or PE based on a physical property of these phospholipids that impart negative curvature and therefore lateral stress to the membrane bilayer. First, the ion requirement is isolated to a subset of divalent cations specifically affecting the physical organization of CL. Ionic strength and osmotic effects have been ruled out by monovalent cations, trivalent cations and high osmolyte concentration. Support of growth and cellular CL levels correlated closely with divalent ion-dependent HII phase induction of CL [170, 174]. CL levels were lowest (27% of total phospholipid) in cells grown in Ca2+ (the strongest inducer of the HII phase) and highest (40–45%) when grown in the weaker inducers (Mg2+ and Sr2+) of the HII phase; Ba2+ did not induce the HII phase and did not support growth. The phase transition from bilayer to non-bilayer phase for phospholipids extracted from wild-type E. coli begins about 10°C above the growth temperature of 37°C and has a mid-point at about 55°C. Phospholipids extracted from the ΔpssA strain grown in 50 mM Ca2+ and analyzed at the same Ca2+ concentration also displayed a wild type phase transition as a function temperature. However, analysis in vitro at 50 mM Mg2+ resulted in a mid-point transition above 75°C. Similarly phospholipids extracted from the mutant grown in 50 mM Mg2+ displayed a wild type transition profile in 50 mM Mg2+ but a transition mid-point at about 35°C in 50 mM Ca2+. Furthermore, lipids extracted from cells grown in 15 mM Sr2+ (maximum for growth) displayed no transition in either the absence of Sr2+ or at 20 mM Sr2+ (neither of which supported growth) and a decreasing mid-point transition from 65°C to 60°C to 55°C at 10, 15 and 12 mM Sr2+, respectively. For wild type cells growth, their phospholipid composition, and the phase transition profiles of extracted lipids were not influenced by the divalent cations in the growth medium or during in vitro analysis. Charge shielding of the high anionic phospholipid composition of the mutant lipid bilayer was equal with Ba2+ and the other divalent ions yet Ba2+ did not support growth. Ba2+ also raised the mid-point of the phase transition when added to lipids analyzed in the presence of Mg2+ [174]. Finally, fatty acid composition can affect the phase transition profiles, but the fatty acid composition was not affected by the divalent cation in which cells were grown. These results strongly support a role for PE and/or CL in providing bilayer stress, which appears to be required for cell viability. However, pssA mutants are more sensitive to antibiotics [177], the outer membrane of ΔpssA cells are leaky to macromolecules as evidenced by the presence of the periplasmic RNase in the growth medium [178], and LPS is not decorated with phosphoethanolamine [179]. All of these latter changes in the cell envelope also affect the E. coli barrier to external assault.
7.3.2. Systematic regulation of membrane phospholipid content
As noted above, placing the pgsA gene under regulation of the PlacOP allowed control of steady-state anionic phospholipid content at the two extremes of wild type and about 10% of total phospholipid. Due to the stochastic nature of the promoter [144], apparent intermediate levels of induction result in a change in the ratio of fully induced to uninduced cells and not an intermediate level of lipid composition in each cell. The pssA gene was placed under control of ParaB with gene expression controlled by arabinose [180] resulting in PE levels between 2 and 75% in the culture as a whole. This promoter is also not optimal for regulating lipid composition because it is induced in stationary phase in the absence of arabinose and behaves in a stochastic manner in the presence of varying concentrations of arabinose [181]. However, the PLtetO-1 (tet promoter regulated by tetR (repressor) and induced by anhydrotetracycline) acts in a dose-responsive manner with respect to each cell in the culture [182] so that the effect of intermediate levels of PE can be studied [183] as well as the extremes of PE [184]. The potential exists to utilize this promoter in regulating other genes of phospholipid metabolism.
Heterologous expression of foreign genes in E. coli is not a new idea and has been extensively used to establish the identity of foreign gene products. We constructed a set of E. coli strains (Fig. 4) in which primarily PE was replaced by a number of lipids not found in E. coli in order to determine which properties of PE suppress various phenotypes of PE-lacking cells. Expression of the PC synthase from Legionella pneumophila in an E. coli cell lacking PE (ΔpssA) resulted in 70% of total phospholipid as PC [185]. The yeast PI synthase produced about 10% PI in a wild type E. coli [186]. The two glycolipids glucosyl diacylglycerol (GlcDAG) and diglucosyl diacylglycerol (GlcGlcDAG) were produced at about 30–40% of total glycerol-based lipids in ΔpssA mutants of E. coli expressing the Acholeplasma laidlawii GlcDAG synthase [178] either alone or with the GlcGlcDAG synthase [178], respectively. The lysyl t-RNA:PG lysine transferase from Staphococcus aureus [187] has been expressed in wild type E. coli and the ΔpssA strain at 40% and 70%, respectively of total phospholipid (Heacock and Dowhan, unpublished data). The cells are viable except in the latter case tend to mutant the mrpF gene (encoding the transferase) or lose the plasmid suggesting the cells are compromised with the net positively charged phospholipid. This set of mutants has been reviewed [188], and the foreign genes introduced into E. coli with the respective lipid synthesized are summarized in Fig. 4. The physical and chemical properties of each of the natural and foreign lipids are summarized in Fig. 5. A similar approach can be used to replace other natural lipids with a spectrum of foreign lipids. However, once a phenotype related to the loss or replacement of a given lipid is established in vivo, it is critical to reconstitute the system in vitro and demonstrate a similar effect of lipid on the process under study. The use of this approach is exemplified by the following characterization of the role of PE is determining the structure of LacY.
Figure 4.
Synthesis of foreign lipids in E. coli. See Fig. 3 for the native pathways (blue arrows) and native lipids (blue and grey). The following enzymes with their respective genes named carry out (red arrows): 1. phosphatidylcholine synthase (Legionella pneumophila [185, 248]); 2. phosphatidylinositol synthase (Saccharomyces cerevisiae [186]); 3. glucosyl diacylglycerol synthase (Acholeplasma laidlawii [212]); 4. diglucosyl diacylglycerol synthase (Acholeplasma laidlawii [211]); 5) lysyl t-RNA:phosphatidylglyerceol lysine transferase (Staphococcus aureus [187]).
7.3.3. Role of phosphatidylethanolamine in topological organization and function of solute transport proteins
7.3.3.1. Early work on lactose permease
I have had a love-hate relationship with LacY of E. coli since my postdoctoral years with Eugene Kennedy. I joined Kennedy’s group in 1969 because I was fascinated with the emerging emphasis on studying the structure and function of membrane proteins, which I though would build on my expertise in similar studies of soluble proteins. Fox and Kennedy [189–192] had just published their series of seminal papers that identified a membrane component (M Protein) encoded by the lac operon and showed it was the lacY gene product. They specifically labeled and solubilized the M Protein as well as studied and postulated mechanisms for energy dependent uphill accumulation and energy independent downhill equilibration of ß-galactosides by this transport system. I spent my first year trying to purify LacY to homogeneity and failed miserably hence my initial hatred for LacY. Cholate-based detergents do not solubilize LacY and exposure to Triton X-100 results in irreversible aggregation of LacY. It was not until the early 1980’s that a joint venture between the Tom Wilson and Ron Kaback labs, aided by the availability of the new detergent octylglucoside, resulted in purification of LacY to homogeneity [193], reconstitution of the protein into proteoliposomes [193, 194], and characterization of its transport properties as a function of liposome composition [195, 196]. LacY reconstituted into E. coli total phospholipids carried out both uphill and downhill transport of substrate. However, liposomes containing only PG and CL or dioleoyl-PC in place of E. coli PE only carried out downhill transport. Use of progressively methylated PE showed a decline in uphill transport with PC again being inactive. They concluded that LacY required the free amino group of either PE or PS for uphill but not downhill transport. These previous observations on the dependence of LacY function on lipids began our use of LacY as a model system to study the role of specifically PE in membrane protein folding and function, which redeemed my love for LacY.
7.3.3.2. Lactose permease properties in cells lacking phosphatidylethanolamine
We know a lot about the physical and chemical properties of individual lipids and lipid mixtures organized into non-covalent structures in solution such as bilayer or nonbilayer prone, inducement of negative or positive curvature, fluidity and the charge density of the lipid-aqueous interface [197]. Due to the polymorphic nature of lipids in solution as a function of solvent conditions and lipid mixtures coupled with a lack of understanding of how the above variation of lipid organization translates into biological function, many artifacts can arise in in vitro function studied that result in conclusions with no biological significance. On the other hand lipids have multiple functions and changes in membrane lipid composition results in pleiotropic effects on cell processes that cannot be sorted out by in vivo studies alone. Therefore, it is imperative to couple in vitro studies with in vivo studies in order to arrive at precise understanding of the role of specific lipids. With a mutant in hand completely lacking PE, we could now establish whether the apparent requirement for PE in supporting uphill transport is of biological significance and determine the molecular basis for the lack of uphill transport in the absence of PE in proteoliposomes.
I was fortunate that Dr. Mikhail Bogdanov moved from Russia to join my laboratory shortly after our isolation of the PE-lacking mutant. He came with an enthusiastic and intense interest in lipid-protein interactions, and we have worked together ever since in defining roles for PE in cell function. Bogdanov measured LacY transport function in PE-containing and PE-lacking cells and was able to replicate the Wilson in vitro observation that only PE-containing cells supported uphill transport and that PE-lacking cells carried out only downhill transport but at the same rate as the PE-containing cells [198]. Lack of uphill transport was not due to a change in membrane permeability, a reduced proton electrochemical gradient or a reduced level of LacY in membranes. LacY carries out uphill transport by a proton-substrate one-for-one symport mechanism driven by coupling uphill movement of substrate directly with downhill movement of a proton. We concluded that this coupling was severed in PE-lacking cells. The question of why PC does not support uphill transport had to wait for another day. The following 17 years have been spent establishing the molecular basis for the PE requirement using both in vitro and in vivo approaches.
We suspected that LacY was not in a completely native structural organization when assembled in PE-lacking cells. A fortuitous finding in the Kaback laboratory allowed us to begin structural studies on LacY prior to solution of its more detailed crystal structure [199]. None of our progress would have been possible without the generous assistance of Ron Kaback and the knowledge and resource base available from his lab. The Kaback lab had isolated the conformation-specific monoclonal antibody 4B1 (mAb 4B1) [200] directed at the extramembrane domain P7 of LacY (Fig. 6). Reaction of LacY with mAb 4B1 in spheroplasts of E. coli blocked uphill transport [201, 202]. We found that mAb 4B1 did not react with LacY assembled in spheroplasts of PE-lacking cells and unlike LacY derived from PE-containing cells did not recognize LacY after SDS PAGE by Western blotting; polyclonal Ab recognized LacY from either cell type. Binding of mAb 4B1 to the P7 domain lowers the abnormally high pKa of the glutamate residue at position 325, which is part of the proton wire associated with uphill transport [201, 202]. Therefore, we concluded that LacY assembled in the absence of PE lacked the proper structural organization of an epitope in domain P7 that is associated with uphill transport. Lack of recognition of LacY by mAb 4B1 has been a useful marker of mis-folding of LacY in all of our studies of lipid-LacY interactions.
Figure 6.
Topological organization of LacY as a function of the presence or absence of PE. Cytoplasm is at the top of each figure, TMs are noted by rectangles, NT and CT refer to the N and C terminus respectively, and extramembrane domains oriented to the cytoplasm (C) or periplasm (P) in PE-containing cells are indicated. (A) LacY topology as determined in PE-containing (+PE) cells is depicted. The approximate position of D325 in TM X is indicated. (B) LacY topology after assembly in PE-lacking cells (−PE) cells. The exposure of TM VII (red) to the periplasm results in the loss of salt bridges of TM VII with TM X and TM XI as noted by the charges. (C) LacY topology determined after induction of PE synthesis in cells where assembly of LacY initially occurred in the absence of PE.
Bogdanov developed a very useful procedure termed an “Eastern-Western” blot in which proteins after separation by SDS PAGE are transferred to a solid support coated with specific phospholipids (Eastern blot). Proteins from the gels are electrophoretically transferred to the area of the solid support coated with lipid (Western blot), SDS is removed during electrophoresis, and the proteins refold in the presence of lipid. Using this technique he demonstrated that LacY from PE-lacking cells could be recognized by mAb 4B1 if reconstitute with PE, but not PG/CL or dioleoyl-PC [203], consistent with the lack of function of LacY both in vivo and in vitro. A series of experiments followed indicating that LacY requires PE in a late folding event to organize into a proper functional conformation after exit from the translocon. PE acts much as a molecular chaperone in that PE is required during folding but not once LacY is properly folded [204–206]. Thus we coined the term “lipochaperone.”
7.3.3.3. Phospholipids as membrane protein topological determinants
What is the structure of LacY assembled in cells lacking PE? We suspected some type of major structural rearrangement of LacY in the membrane. Fortunately again, Kaback had generated a library of single and multiple cysteine replacement mutants in LacY lacking its native cysteine residues. These were used to map the topological organization and packing of the 12 transmembrane domains (TMs) and the extramembrane domains connecting neighboring TMs. The basic structure of LacY based on accessibility to these cysteines by sulfhydryl reagents [207] was later confirmed by X-ray crystallography [199, 208]. We used a similar approach, substitutedcysteine accessibility method as applied to TMs (SCAM™), to defining the topological of LacY as a function of membrane lipid composition. This method uses a membrane impermeable biotinylated sulfhydryl reagent to probe the accessibility of single cysteine replacements of LacY expressed in cells either before or after disruption of the cell membrane [209, 210]. The orientation of TMs is determined by the pattern of exposed and protected cysteines engineered into putative adjacent periplasmic or cytoplasmic extramembrane domains, respectively. All extramembrane domains are exposed after cell disruption while cysteines in TMs are protected.
To our surprise we found that LacY assembled in membranes lacking PE displayed a complete inversion of the N-terminal six TM α-helical bundle with respect to the plane of the bilayer and the C-terminal five TM α-helical bundle. TM VII was no longer a TM but was exposed to the periplasmic space [180, 184, 188, 197]. This structural rearrangement occurs in the vicinity of P7, which would result in its conformational disruption explaining the lack of recognition by mAb 4B1 and the related loss of uphill transport function.
Since previous results had indicated at least a rescue of the conformation of the P7 domain upon refolding of mis-folded LacY after resupplying PE [203, 204, 206], we investigated whether complete rescue of proper topological organization and activity of LacY was possible in vivo. Using strains in which transcription of a single copy of pssA gene is regulated by either ParaB [180] or PLtetO-1 [184], we first grew cells expressing single cysteine replacements in LacY in the absence of inducers for PE synthesis then removed inducer (IPTG) for new LacY synthesis and induced PE synthesis. The result was that mis-oriented LacY assembled in the near absence of PE (2%) reorganized upon synthesis of PE so the TM VII inserted into the membrane and TMs III-VI assumed wild type orientation. However, TM I remained inverted and TM II assumed a mini-loop configuration that did not span the membrane. Introduction of PE post-assembly of LacY also restored uphill transport function. Such large changes in protein organization after membrane insertion and final folding were unprecedented and challenged the dogma of the stable orientation of highly hydrophobic TMs after membrane insertion.
What was the molecular basis for this lipid-dependent topological interconversion of LacY? The availability of strains of E. coli expressing foreign lipids with different physical and chemical properties aided in understanding of the properties of lipids that affected topological organization. Substitution of PE by several foreign lipids that were either uncharged (GlcDAG or GlcGlcDAG) [211, 212] or were zwitterionic but carried no net charge (PC) [185] resulted in assembly of LacY in its wild type orientation. The main property these three lipids have in common is the ability to dilute the high negative charge density of the membrane surface due to PG and CL. Ability to be a hydrogen bond donor (the glycolipids) or non-bilayer prone properties (GlcDAG) versus bilayer prone properties (PC and GlcGlcDAG) was not a factor. On the protein side, increasing the net-positive charge of normally cytoplasmically exposed extra-membrane domains that also contained negative residues prevented mis-orientation of LacY in PE-lacking cells [184]. Increasing the negatively charged residues in these same domains induced inversion of LacY in the presence of PE. Similar results were found with other secondary transporters of E. coli, i.e. the phenylalanine [213], γ-aminobutyrate [214] and sucrose [215] permeases.
These results led to the “Charge Balance Hypothesis” [197]. The vast majority of membrane proteins follow the positive-inside rule where extra-membrane domains with a net positive charge are oriented facing the cytoplasm [216]. In domains with mixtures of positively (cytoplasmic retention signal) and negatively (membrane translocation signal) charged residues, the former are dominant determinants over the latter. However, when the negative charge density of the membrane surface is increase by the lack of PE (or other net-neutral lipids), the negatively charged residues become dominant translocation signals, which result in mis-orientation of some proteins such as the very flexible secondary transporters. When only a part of a protein mis-orients, a flexible transition domain must be formed between the native and mis-oriented TMs. For LacY this is the very hydrophilic TM VII, which comes out of the membrane; increasing the hydrophobicity of TM VII prevents its non-membrane location and inversion of topololgy [184]. Similar transition domains are formed in the other examples of lipid sensitive proteins [213–215]. Therefore, a thermodynamic balance between short- and long-term interactions within the protein and between the protein and the lipid environment determines topological organization, which explains why membrane protein topological prediction methods are only about 85% accurate. A role for lipids in determining membrane protein organization only became apparent after methods were developed to systematically perturb membrane lipid composition.
As noted above, it is important to correlate in vivo experiments with in vitro experiments to establish definitive lipid dependence. There are potentially multiple indirect mechanisms by which lipids could affect LacY topology such as effects on the translocon or protein molecular chaperones or repression of protein quality control systems. To establish a direct effect of lipids in determining topology, LacY was purified from either PE-containing or PE-lacking cells and then reconstituted into proteoliposomes of various lipid compositions. First, LacY properties were completely dependent on the liposome composition and not the source of LacY, i.e. inverted or native in the source cells. As in the in vivo experiments, LacY assumed wild type topological organization in liposomes containing, in addition to PG and CL, net neutral lipids PE or PC [217] and GlcDAG or GlcGlcDAG (Vitrac and Dowhan, unpublished observation). LacY was mis-oriented in liposomes made of PG and CL alone [217]. What about LacY activity? Contrary to earlier in vitro findings [195], we observed robust uphill transport by LacY in E. coli cells containing PC in place of PE [185]. Also mAb 4B1 recognized LacY from these cells by Western blot analysis. Reconstitution of LacY into liposomes made with lipids from the PC-containing E. coli strain also resulted in uphill transport as did commercial PC’s containing at least one saturated fatty (Vitrac and Dowhan, unpublished observation); dioleoyl-PC did not support uphill transport as previously observed [195, 217]. Using the Eastern-Western blotting procedure, E. coli PC and synthetic PC with at least one saturated fatty acid reconstituted recognition by mAb 4B1 [185]. LacY uphill transport function is partially supported in vivo in cells where PE is replaced by GlcDAG but not GlcGlcDAG [211, 212]; a similar result was shown in vitro (Vitrac and Dowhan, unpublished observation). Therefore, the dependence of LacY function on membrane lipid composition is more complex and involves both the correct head group and the correct fatty acid composition. The previous conclusion from in vitro experiments that the free amine of PE of PS is required for uphill transport [195] is not supported by the above more extensive investigation, and the lack of PC support of in vitro uphill transport was due to the fatty acid composition of PC and not the head group chemistry.
The above experiments demonstrate the value of using E. coli as a model system to establish general principles of how lipids influence protein structure and function. At this point in time, such experiments cannot be carried out in even the simplest eukaryotic cells due to the presence of most lipids in all the multiple membranes in such cells. The only possible exception is the mitochondria of Saccharomyces cerevisiae and other simple eukaryotes, which appear to be the only organelles that contain CL and PG. Mitochondrial function is certainly affected in mutants lacking these phospholipids [218–221], but cells are still viable if grown on glucose if necessary so that function can be assessed.
7.3.4. Other phenotypes of cells lacking phosphatidylethanolamine
Dr. Eugenia Mileykovskaya joined my laboratory in the early 1990. She was trained in Russia in the elite laboratories of bioenergetics overseen by Skulachev and has contributed this expertise in our studies of lipid function in both E. coli and yeast. She also has been the driving force in studies involving the role of CL in cell function. She carefully analyzed the bioenergetic properties of inside-out membrane vesicles generated from cells lacking PE [222]. Lack of PE did not affect the magnitude of O2 consumption or proton electrochemical potential generated from added ATP, succinate or lactate, which established the structural integrity of the inner membrane. However, the rate of NADH oxidation by the type II NADH dehydrogenase was reduced by 80%. The level of the dehydrogenase and CoQ8, which couples electron transfer to terminal oxidases, were normal. After eliminating several possible explanations, the conclusion was that lack of PE compromised the interaction of electron carries with the CoQ8 pool.
Not completely unexpected, lack of PE results in activation of stress response systems, one of which is the Cpx system [223], a two-component response system with a membrane sensor component. Making a PE-lacking strain null in the Cpx system corrects many of the phenotypes associated with induction of this system in PE-lacking cells including elevated levels of DegP, an outer membrane associated protease.
A striking phenotype of PE-lacking cells is their failure to synchronously divide with cell growth resulting in filamentous cells ranging in varying lengths. The MinCDE system restricts formation of the Z-ring (formed by FtsZ) to mid-cell in wild type cells followed by sequential organization of additional early cell division proteins on the Z-ring [161]. Expression of fluorescent derivatives of these early cell division proteins in a PE-deficient cell showed proper organization of each at potential division sites along the filamentous cells [224]. However, there was no evidence of constriction at most of these sites, which is the next phase of cell division. The rare occurrence of constriction was observed only with potential division sites at the ends of cells, which might suggest that once a constriction site is passed by during cell growth it is no longer functional when PE is missing. The exact step of cell division compromised in the absence of PE or whether it is lack of PE or increased PG and CL that disrupts constriction is not known. Lack of PE results in increased dwell time of MinD on the membrane surface and disrupts the normal cycling pattern probably due to lack of restriction of anionic lipid enriched domains to the cell poles and division site [162]. Lack of PE also disrupts cytokinesis in eukaryotic cells by loss of its reversible cycling from the inner surface to the outer surface of the plasma membrane, which is required during cell division in Saccharomyces cerevisiae [225] and somatic cells [226]. Therefore, a requirement for PE in cytokinesis is conserved from simple to complex cells.
PE is also required for cell motility and chemotaxis [227]. The defect lies in the inability to induce expression of the master operon regulating the formation of the flagellum. This effect is dose dependent on the amount of PE and not PS, which cannot replace PE.
Although lack of PE does not compromise any absolutely essential E. coli process, it is required for optimal function of many important cell functions. In the case of the mis-organization of secondary transporters (for sugars and amino acids), function is compromised but not completely lost since these transporters still carry out downhill transport. Lack of maintenance of inward gradients essentially turns the membrane into a dialysis bag that specially equilibrates small molecules with the growth medium. PE-lacking cells will grow on lactose as a carbon source at mmolar but not µmolar concentrations [93, 178] and lack of uphill transport by amino acid permeases probably explains the requirement for medium rich in amino acids. There may be other membrane proteins outside of secondary transporters, such as those involved in cell division, whose organization in the membrane is compromised but only results in partial alteration of function. Further analysis of the above partially characterized phenotypes of PE-lacking cells will add additional understanding at the molecular level for the role of specific lipids in cell function.
7.4. What does cardiolipin do in E. coli?
CL is unique among phospholipids in that it contains two phosphatidic acid moieties connected by a glycerol. The central free hydroxyl enters into hydrogen bonds with the two ionizable phosphates, which results in one pKa near 3 and the other greater than 8 [228]. Thus CL has been proposed to act as a proton sink important for energy transducing membranes where CL is almost uniformly found. In yeast, CL is critical to mitochondrial energy transduction in that it is integrated into the structure of all of the components of the electron transport chain [229–231] and acts to organize the individual components into the higher order supercomplex termed the “respirasome” [220, 232–234]. Somatic cells show similar higher order organization, which also involves CL. In yeast and probably somatic cells, mitochondria lacking CL fail to form stable supercomplexes of the respiratory chain and are somewhat compromised in respiration but still grow on non-fermentable carbon sources [220, 221]. Most likely the elevated PG in Δcrd strains (encodes CL synthase) partially substitutes for CL at least for individual components of the respiratory chain. Loss of PG and CL in yeast [218] and somatic cells [235] results in complete loss of respiratory function and massive disruption of mitochondrial organization.
Therefore, it is surprising that in E. coli both ΔclsABC and ΔpgsA mutants (containing appropriate suppressor mutations) do not display such dramatic phenotypes as in eukaryotic cells. However, neither null mutant has been extensively analyzed for its dependence on the source of metabolic energy as was done with the PE-lacking strain [222]. Also no studies under anaerobic conditions have been done. Further analysis of these strains under other stress conditions may uncover additional roles of CL and/or PG.
CL is a non-bilayer anionic prone lipid (NBPAL), which has the potential to organization into domains at the points of membrane curvature associated with cell division and membrane fusion and fission events; for a review of CL involvement in these phenomena see [236]. Mileykovskaya was the first to observe CL-enriched domains at the cell poles and division site in E. coli using NAO as a specific indicator of CL localization [163], which was later verified for the poles by direct measurement of CL [164]. During the cell cycle CL domains undergo dynamic formation at the nascent cell division site followed by development of a mature septal region enriched in CL prior to constriction that forms the new cell poles also enriched in CL [236]. In vitro experiments indicate that the fluorescence emission spectrum of NAO undergoes a green to red shift when it binds to areas concentrated in CL rather than to individual CL molecules [237]. This shift was postulated to be due to π-π interactions of the NAO rings that organize in arrays when bound to a domain formed by CL. NAO binds to other anionic lipids with low affinity and without undergoing a red shift suggesting such interaction is with individual molecules. The NAO domains observed in E. coli show a green to red shift in the emission spectrum [163]. Therefore, CL definitely organizes into lipid-enriched domains in E. coli at sites of increased membrane curvature. Mathematical models of CL domain formation in bacteria suggest that individual CL molecules cannot detect membrane curvature but clusters of CL molecules (distinct from a phase separation that would compromise cell integrity) can organize in areas of membrane curvature [238, 239]. Similar localization of CL has been observed in Gram-positive bacteria [240].
Since cells lacking CL have few dramatic phenotypes, the question remains as to the precise function of such domains and whether they are essential. The fact that ΔpgsA cells still form domains now enriched with enhanced levels of PA and N-acyl PE, both NBPALs, at the cell poles and division site suggests that such domains are important and form even in the absence of CL using lipids with physical and chemical properties similar to CL [167]. N-acyl PE is also elevated in ΔclsABC cells (T. Garrett, E. Mileykovskaya, and W. Dowhan, unpublished observation) where PE levels are normal ruling out an increase in N-acyl PE due to an increase of PE in a ΔpgsA strain. The distribution of NBPAL in the cells lacking CL but still containing PG has not been investigated. Further investigation into the role lipid domains play in E. coli is required before similar understanding reaches the level available in eukaryotic cells, which contain similar lipid-enriched domains termed lipid rafts.
8. Regulation of phospholipid metabolism in E. coli
In spite of the wealth of biochemical and genetic information for E. coli, we know more about what does not rather what does regulate phospholipid metabolism. PssA [87] and PgsA [96] appear to have no preference for CDP-DAG or dCDP-DAG, which are in equal amounts in the cell [82]. Except for the formation of CL from PG, all steps in the pathway are essentially irreversible under physiological conditions. How the ratio of protein to phospholipid in the membrane is regulated must lie at the committed step to phospholipid biosynthesis, i.e. the plsB gene product (sn-glycerol-3-P acyltransferase). Similarly the ratio of PE to PG plus CL must reside at the branch point following the synthesis of CDP-DAG. However, most of the evidence suggests that even though the enzymes of phospholipid metabolism are present on the order of a 1000 molecules per cell, they are already in excess over the amounts necessary to maintain normal flux through the pathways. Overproduction of each of these enzymes from 10 to several 100-fold has only minor effects on the steady state phospholipid composition. However, as expected large changes occur upon reduction in the level of the catalytic capacity of these enzymes [93, 142].
Unfortunately, there is no report of the effects of over production of PlsB on the ratio of protein to phospholipid. Overproduction of PlsB on the order of 50-fold results in formation of intracellular filaments that are largely made up of the enzyme and phospholipid [241], which might reduce the effective catalytic capacity of the overproduced enzyme. However, starvation of the plsB mutant (10-fold higher Km for substrate) for GP ceases phospholipid synthesis immediately, but cells continue to grow for one doubling resulting in a 60% increase in the membrane protein to phospholipid ratio [242]. Addition of GP re-initiated phospholipid synthesis but protein synthesis and cell growth did not resume until the protein to phospholipid ratio returned to normal. Therefore, there is a very loose coupling between membrane protein and phospholipid synthesis. Some evidence exists that PlsB might be regulated by availability of its substrate GP [243]. The initial plsB mutant displayed a 10-fold higher Km for this substrate [24], which could be elevated in cells by supplementation of the growth medium. However, overproduction of the enzyme responsible for GP synthesis did not raise intracellular GP levels or suppress the mutant’s requirement for GP. Therefore, the level of GP is tightly regulated with a secondary regulation of PlsB.
Overproduction of the PssA by 800-fold did not change phospholipid composition [90]. However, curing a ΔpssA strain of a temperature sensitive plasmid carrying the wild type gene resulted in a drop in PE levels beginning at about 25% of wild type enzyme levels indicating an excess of activity over that necessary to maintain PE levels [93]. A similar outcome was observed with overproduction and dilution of PgsA activity during curing of a plasmid copy of pgsA in a ΔpgsA background [142, 143]. Overproduction of PgsA also did not alter phospholipid composition [97]. Although the effects of overproduction of CdsA on lipid composition was not reported, PA levels go from 0.2% to 30% of total phospholipid upon switching a cdsA conditional mutant to its restrictive conditions [109]. The ratio of PE to PG plus CL did change from 3.3 to 4.0 after the switch. The increase in anionic phospholipid content contributed by PA maybe of significance in regulating PssA as discussed below. CL levels increased by overproduction of ClsA but not in the amounts expected based on the level of enzyme overproduction. A 20-fold or 760-fold overproduction of ClsA resulted in only a 2-fold or 4-fold increase in CL mostly at the expense of PG [121], respectively. ClsA levels also increased about 10-fold for cells in stationary phase, which also resulted in a 2-fold increase in CL levels [127].
Some clues to the mode of regulation at the branch point after formation of CDP-DAG was provided by expressing the Bacillus subtilis pssBS gene in an E. coli strain carrying the E. coli ΔpssA allele [244]. As noted earlier, the Bacillus Pss is an integral membrane protein, most likely follows a Bi-Bi reaction mechanism and requires Mg2+ for activity [78]. The E. coli enzyme is an amphitropic protein that reversibly interacts with the membrane by binding to anionic phospholipids [92, 94], follows a Ping-Pong reaction mechanism [86], and does not require Mg2+ for activity. Saha et al. [244] first followed initial rates of incorporation of 32Pi into PE by a short pulse of label and compared wild type cells to several mutants in pgsA with very low PG plus CL levels. PssA content was the same in both cell types, but the rate of incorporation of label into PE was 10-fold lower in the pgsA mutant, which was deficient in anionic phospholipids. Steady state PE content in the pgsA mutant is higher than in wild type cells due to the lack of PG synthesis even though the initial incorporation of label into PE was lower. The pssBS gene under regulation by IPTG was introduced into an E. coli ΔpssA strain. The level of PE at steady state increased with increasing induction of the pssBS gene and with plasmids of increasing copy number in the cell. High overproduction of PssBS was lethal. Therefore, the membrane integrated B. stubtilis Pss in E. coli is not responsive to E. coli regulatory signals, which were suggested to be related to the anionic phospholipid content of the membrane [244]. Anionic phospholipids also activated PssA from E. coli using detergent-lipid mixed micelles [245, 246]. As previously shown, overproduction of PssA has no effect on steady state phospholipid composition or cell viability. These results suggest that PssA reversibly interacts with the membrane through anionic phospholipids and only this membrane bound form is active. Therefore, as anionic phospholipid content increases, it is balanced by an increase in the effective catalytic capacity of the enzyme committed to PE synthesis regardless of its total potential catalytic capacity. Inducing the turnover of phospholipids such as increasing the formation of MDO increases the flux of intermediates through the pathway but not the mix of major phospholipid species [247], which further supports feedback inhibition or activation of the two enzymes at the branch point following CDP-DAG formation.
9. Summary and future perspectives
Establishing the blueprint of phospholipid biosynthesis in all cell types was certainly dominated by Eugene Kennedy and more fully developed in E. coli by his students and postdoctoral fellows. However, John Cronan, Robert Bell and Charles Rock made major contributions to understanding the synthesis of fatty acids and PA in E. coli. As I hopefully emphasized, Isao Shibuya, Akinori Ohta, Kouji Matsumoto, and Hiroshi Hara made important contributions especially in the role of anionic phospholipids in E. coli. These individuals and there descendants continue to make important contributions. I apologize for missing more specific acknowledgement of many others who have made contributions in the area of bacterial phospholipid synthesis and function.
What remains to be done? As note at the end, how phospholipid metabolism is regulated in E. coli remains largely unknown. Why does E. coli have three genes encoding CL synthase activity and what role do CL synthases with different mechanisms play in cell metabolism? The respective gene products are apparently induced or active under different environment conditions and at different points of the cell cycle. What are the functions of each of the pgpABC genes? Why are viable ΔpgsA mutants still temperature sensitive? Little is known about phospholipid metabolism under anaerobic conditions. How do phospholipids presumably made on the cytoplasmic surface of the inner membrane get to the outer surface of the inner membrane and to the outer membrane? Are membrane domains composed of NBPAL essential to cell viability and if so what are their functions? There are a number of phenotypes associated with lack of PE that have not been sufficiently pursued to establish an understanding at the molecular level. Are there other proteins or processes dependent on the proper topological or structural organization of membrane proteins dependent on membrane lipid composition? As shown for the permeases, PE need not be absolutely essential for all functions of a protein, but may only compromise activity to interfere with optimal function. There is only one structure at the atomic level of a bacterial phospholipid biosynthetic enzyme, Pss from Haemophilus influenzae (PDB accession number 3HSI). No detailed structures have been reported for any integral membrane protein responsible for phospholipid biosynthesis from any source. Such information is necessary to fully understand the mechanism of each enzyme in the pathway as a prerequisite to developing new classes of antibiotics. As Raetz pointed out there are hundreds of minor organic solvent soluble phosphate containing compounds in E. coli that have not been identified. These are precursors to and breakdown products of important biological molecules and contain important clues to unknown metabolic processes. Fully answering the above unknowns will provide a complete understanding of phospholipid metabolism and function in bacteria and will provide the molecular basis to investigate similar phenomena in more complex systems. Capitalizing and the unique aspects of bacterial phospholipid metabolism will provide targets for new antibiotics.
Highlights.
Escherichia coli is an excellent system in which to study phospholipid function.
Historical aspects of phospholipid metabolism, enzymology and genetics are presented.
Approaches available to determine specific phospholipid functions in cells are reviewed.
Future studies of metabolic regulation and function of phospholipids are proposed.
Acknowledgements
I wish to dedicate this review to my inspiring and supportive mentor Eugene P. Kennedy and my close friend and colleague Chris Raetz who both passed away in 2011. Special thanks go to Mikhail Bogdanov, Eugenia Mileykovskaya, and Phil Heacock, who have been in my research group for 20 years or more. I wish recognize the NIH, which has supported my research on E. coli phospholipid metabolism and function under grant GM 20478 since 1973 as well as the John Dunn Research Foundation. Jean Chin has been an important proponent at the NIH for membrane protein and lipid research for many years. Her recognition of the importance of this research area is gratefully appreciated by the lipid community.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: ACP, acyl carrier protein; PA, phosphatidic acid; PE, phosphatidylethanolamine; Pi, PO4; GP, glycerophosphate; PG, phosphatidylglycerol; PGP, phosphatidylglycerophosphate: DAG, diacylglycerol; LPS, lipopolysaccharide; MDO, membrane derived oligosaccharide; CL, cardiolipin; PA, phosphatidic acid; PS, phosphatidylserine; PC, phosphatidylcholine, PI, phosphatidylinositol; GlcDAG; monoglucosyl diacylglycerol; GlcGlcDAG, diglucosyl diacylglycerol; PssA, phosphatidylserine synthase; PgsA, phosphatidylglycerophosphate synthase; Pgp, phosphatidylglycerophosphate phosphatase; Cds, CDP-diacylglycerol synthase; Cls, cardiolipin synthase; Ptet, tet operon promoter; IPTG, isopropyl-ß-D-thiogalactoside; PlacOP, lac operon promotor; mAb, monoclonal antibody; TM, transmembrane domain; LacY, lactose permease; mAb, monoclonal antibody; NAO, Nonyl Acridine Orange; NBPAL, non-bilayer prone anionic lipid.
References
- 1.Koga Y, Morii H. Biosynthesis of ether-type polar lipids in Archaea and evolutionary considerations. Microbiol Mol Biol Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chibnall A, Channon H. The ether-soluble substances of cabbage leaf cytoplasm: calcium salts of glyceridephosphoric ccids. Biochem J. 1927;21:233–246. doi: 10.1042/bj0210233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chibnall A, Channon H. The ether-soluble substances of cabbage leaf cytoplasm: preparation and general characters. Biochem J. 1927;21:225–232. doi: 10.1042/bj0210225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levene PA, West CJ. Lecithin. I: "Hydrolecithin" and its bearing on the constitution of cephalin. J Biol Chem. 1918;33:111–117. [Google Scholar]
- 5.Levene PA, West CJ. Cephalin V. Hydrocephalin of the egg yolk. J Biol Chem. 1918;35:285–290. [Google Scholar]
- 6.Chievitz O, Hevesy G. The first radioindicator study in the life sciences with a manmade radionuclide. Nature. 1935;136:1106–1107. [Google Scholar]
- 7.Sinclair RG. The metabolism of the phospholipids: VIII. The passage of elaidic acid into tissue phospholipids. Evidence of the intermediary role of liver phospholipid in fat metabolism. J Biol Chem. 1935;111:515–526. [Google Scholar]
- 8.Artom C, Sarzana G, Perrier C, Santangelo M, Segre E. Rate of organification of phosphorus in animal tissue. Nature. 1937;139:836–837. [Google Scholar]
- 9.Perlman I, Ruben S, Chaikoff IL. Radioactive phosphorus as an indicator of phospholipid metabolism. I. The rate of formation and destruction of phospholipids in the fasting rat. J Biol Chem. 1937–38;122:169–182. [Google Scholar]
- 10.Swanson MA, Artom C. The lipide compostion of the large granules (mitochondria) from rat liver. J Biol Chem. 1950;187:281–287. [PubMed] [Google Scholar]
- 11.Kornberg A, Pricer WE., Jr Enzymatic esterification of α-glycerophosphate by long chain fatty acids. J Biol Chem. 1953;204:345–357. [PubMed] [Google Scholar]
- 12.Kennedy EP. Synthesis of phosphatides in isolated mitochondria. J Biol Chem. 1953;201:399–412. [PubMed] [Google Scholar]
- 13.Kornberg A, Pricer WE., Jr Federation Proc. 1952;11:242. [Google Scholar]
- 14.Kennedy EP. Synthesis of phosphatides in isolated mitochondria. II. Incorporation of choline into lecithin. J Biol Chem. 1954;209:525–535. [PubMed] [Google Scholar]
- 15.Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipids. J Biol Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 16.Kennedy EP. Sailing to Byzantium. Annu Rev Biochem. 1992;61:1–28. doi: 10.1146/annurev.bi.61.070192.000245. [DOI] [PubMed] [Google Scholar]
- 17.Castuma CE, Crooke E, Kornberg A. Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J Biol Chem. 1993;268:24665–24668. [PubMed] [Google Scholar]
- 18.Sekimizu K, Kornberg A. Cardiolipin activation of DnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem. 1988;263:7131–7135. [PubMed] [Google Scholar]
- 19.Rodbell M, Hanahan DJ. Lecithin synthesis in liver. J Biol Chem. 1954;214:607–618. [PubMed] [Google Scholar]
- 20.Agranoff BW, Bradley RM, Brady RO. The enzymatic synthesis of inositol phosphatide. J Biol Chem. 1958;233:1077–1083. [PubMed] [Google Scholar]
- 21.Paulus H, Kennedy EP. The enzymatic synthesis of inositol monophosphatide. J Biol Chem. 1960;235:1303–1311. [PubMed] [Google Scholar]
- 22.Colodzin M, Kennedy EP. Biosynthesis of diphosphoinositide in brain. J Biol Chem. 1965;240:3771–3780. [PubMed] [Google Scholar]
- 23.Voelker DR, Kennedy EP. Cellular and enzymic synthesis of sphingomyelin. Biochemistry. 1982;21:2753–2759. doi: 10.1021/bi00540a027. [DOI] [PubMed] [Google Scholar]
- 24.Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis: macromolecular synthesis in an sn-glycerol 3-phosphate acyltransferase Km mutant. J Bacteriol. 1974;117:1065–1076. doi: 10.1128/jb.117.3.1065-1076.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman J. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J Biol Chem. 1990;265:17215–17221. [PubMed] [Google Scholar]
- 26.Rock CO. Phosphatidic acid biosynthesis in bacteria. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbalip.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumi R, Atomi H, Driessen AJ, van der Oost J. Isoprenoid biosynthesis in Archaea-biochemical and evolutionary implications. Res Microbiol. 2011;162:39–52. doi: 10.1016/j.resmic.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moellering ER, Benning C. Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci. 2011;16:98–107. doi: 10.1016/j.tplants.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. J Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Hermansson M, Hokynar K, Somerharju P. Mechanisms of glycerophospholipid homeostasis in mammalian cells. Prog Lipid Res. 2011;50:240–257. doi: 10.1016/j.plipres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Kanfer J, Kennedy EP. Metabolism and function of bacterial Lipids. I. Metabolism of phospholipids in Escherichia coli B. J Biol Chem. 1963;238:2919–2922. [PubMed] [Google Scholar]
- 33.Jackowski S, Rock CO. Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J Biol Chem. 1986;261:11328–11333. [PubMed] [Google Scholar]
- 34.Rock CO. Turnover of fatty acids in the 1-position of phosphatidylethanolamine in Escherichia coli. J Biol Chem. 1984;259:6188–6194. [PubMed] [Google Scholar]
- 35.Jackowski S, Jackson PD, Rock CO. Sequence and function of the aas gene in Escherichia coli. J Biol Chem. 1994;269:2921–2928. [PubMed] [Google Scholar]
- 36.Kanfer J, Kennedy EP. Metabolism and Function of Bacterial Lipids. Ii. Biosynthesis of Phospholipids in Escherichia Coli. J Biol Chem. 1964;239:1720–1726. [PubMed] [Google Scholar]
- 37.Kanfer JN, Kennedy EP. Synthesis of phosphatidylserine by Escherichia coli. J Biol Chem. 1962;237:PC270–PC271. [PubMed] [Google Scholar]
- 38.Borkenhagen LF, Kennedy EP, Fielding L. Enzymatic formation and decarboxylation of phosphatidylserine. J Biol Chem. 1961;236:PC28–PC30. [Google Scholar]
- 39.Hübscher G, Dils RR, Pover WF. Studies on the biosynthesis of phosphatidyl serine. Biochim Biophys Acta. 1959;36:518–528. doi: 10.1016/0006-3002(59)90194-5. [DOI] [PubMed] [Google Scholar]
- 40.Delhaize E, Hebb DM, Richards KD, Lin JM, Ryan PR, Gardner RC. Cloning and expression of a wheat (Triticum aestivum L.) phosphatidylserine synthase cDNA. Overexpression in plants alters the composition of phospholipids. J Biol Chem. 1999;274:7082–7088. doi: 10.1074/jbc.274.11.7082. [DOI] [PubMed] [Google Scholar]
- 41.Kiyasu JY, Pieringer RA, Paulus H, Kennedy EP. The biosynthesis of phosphatidylglycerol. J Biol Chem. 1963;238:2293–2298. [PubMed] [Google Scholar]
- 42.Chang YY, Kennedy EP. Phosphatidyl glycerophosphate phosphatase. J Lipid Res. 1967;8:456–462. [PubMed] [Google Scholar]
- 43.Lu YH, Guan Z, Zhao J, Raetz CR. Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J Biol Chem. 2011;286:5506–5518. doi: 10.1074/jbc.M110.199265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter JR., Jr Cytidine triphosphate: phosphatidic acid cytidyltransferase in Escherichia coli. J Lipid Res. 1968;9:748–754. [PubMed] [Google Scholar]
- 45.Pangborn MC. Isolation and purification of a serologically active phospholipid from beef heart. J Biol Chem. 1942;142:247–256. [Google Scholar]
- 46.Macfarlane MG. Structure of cardiolipin. Nature. 1958;182:946. doi: 10.1038/182946a0. [DOI] [PubMed] [Google Scholar]
- 47.Stanacev NZ, Chang YY, Kennedy EP. Biosynthesis of cardiolipin in Escherichia coli. J Biol Chem. 1967;242:3018–3019. [PubMed] [Google Scholar]
- 48.Hostetler KY, van den Bosch H, van Deenen LL. The mechanism of cardiolipin biosynthesis in liver mitochondria. Biochim Biophys Acta. 1972;260:507–513. doi: 10.1016/0005-2760(72)90065-3. [DOI] [PubMed] [Google Scholar]
- 49.Hirschberg CB, Kennedy EP. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69:648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CRH, Guan Z. Discovery of a novel cardiolipin synthase in Escherichia coli utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1212797109. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okonogi K, Shibuya I, Maruo B. Isolation of Escherichia coli mutants with temperature sensitive synthesis of phosphatidylethanolamine. (in Japanese) Seikagaku. 1971;43:665. [Google Scholar]
- 52.Ohta A, Okonogi K, Shibuya I, Maruo B. Isolation of Escherichia coli mutants with temperature-sensitive formation of phosphatidylethanolamine. J Gen Appl Microbiol. 1974;20:21–32. [Google Scholar]
- 53.Ohta A, Shibuya I. Membrane phospholipid synthesis and phenotypic correlation of an Escherichia coli pss mutant. J Bacteriol. 1977;132:434–443. doi: 10.1128/jb.132.2.434-443.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohta A, Shibuya I, Maruo B. Escherichia coli mutants with temperature-sensitive phosphatidylserine synthetase: genetic analysis. Agric Biol Chem. 1975;39:2443–2445. [Google Scholar]
- 55.Ohta A, Shibuya I, Maruo B, Ishinaga M, Kito M. An extremely labile phosphatidylserine synthetase of an Escherichia coli mutant with the temperature-sensitive formation of phosphatidylethanolamine. Biochim Biophys Acta. 1974;348:449–454. [PubMed] [Google Scholar]
- 56.Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km defective sn-glycerol-3-phosphate acyltransferase activities. J Biol Chem. 1975;250:7147–7152. [PubMed] [Google Scholar]
- 57.Raetz CR. Isolation of Escherichia coli mutants defective in enzymes of membrane lipid synthesis. Proc Natl Acad Sci U S A. 1975;72:2274–2278. doi: 10.1073/pnas.72.6.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raetz CR. Phosphatidylserine synthetase mutants of Escherichia coli. Genetic mapping and membrane phospholipid composition. J Biol Chem. 1976;251:3242–3249. [PubMed] [Google Scholar]
- 59.Ganong BR, Leonard JM, Raetz CR. Phosphatidic acid accumulation in the membranes of Escherichia coli mutants defective in CDP-diglyceride synthetase. J Biol Chem. 1980;255:1623–1629. [PubMed] [Google Scholar]
- 60.Icho T, Raetz CR. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol. 1983;153:722–730. doi: 10.1128/jb.153.2.722-730.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raetz CR, Wermuth MM, McIntyre TM, Esko JD, Wing DC. Somatic cell cloning in polyester stacks. Proc Natl Acad Sci U S A. 1982;79:3223–3227. doi: 10.1073/pnas.79.10.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Homann MJ, Carman GM. Detection of phospholipid biosynthetic enzyme activities in Saccharomyces cerevisiae by colony autoradiography. Anal Biochem. 1983;135:447–452. doi: 10.1016/0003-2697(83)90710-8. [DOI] [PubMed] [Google Scholar]
- 63.Hawrot E, Kennedy EP. Biogenesis of membrane lipids: mutants of Escherichia coli with temperature-sensitive phosphatidylserine decarboxylase. Proc Natl Acad Sci U S A. 1975;72:1112–1116. doi: 10.1073/pnas.72.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawrot E, Kennedy EP. Conditional lethal phosphatidylserine decarboxylase mutants of Escherichia coli. Mapping of the structural gene for phosphatidylserine decarboxylase. Mol Gen Genet. 1976;148:271–279. doi: 10.1007/BF00332901. [DOI] [PubMed] [Google Scholar]
- 65.Hawrot E, Kennedy EP. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J Biol Chem. 1978;253:8213–8220. [PubMed] [Google Scholar]
- 66.Pluschke G, Hirota Y, Overath P. Function of phospholipids in Escherichia coli. Characterization of a mutant deficient in cardiolipin synthesis. J Biol Chem. 1978;253:5048–5055. [PubMed] [Google Scholar]
- 67.Dowhan W, Wickner WT, Kennedy EP. Purification and properties of phosphatidylserine decarboxylase from Escherichia coli. J Biol Chem. 1974;249:3079–3084. [PubMed] [Google Scholar]
- 68.Warner TG, Dennis EA. Action of the highly purified, membrane-bound enzyme phosphatidylserine decarboxylase Escherichia coli toward phosphatidylserine in mixed micelles and erythrocyte ghosts in the presence of surfactant. J Biol Chem. 1975;250:8004–8009. [PubMed] [Google Scholar]
- 69.Carman GM, Deems RA, Dennis EA. Lipid signaling enzymes and surface dilution kinetics. J Biol Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 70.Satre M, Kennedy EP. Identification of bound pyruvate essential for the activity of phosphatidylserine decarboxylase of Escherichia coli. J Biol Chem. 1978;253:479–483. [PubMed] [Google Scholar]
- 71.Li QX, Dowhan W. Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J Biol Chem. 1988;263:11516–11522. [PubMed] [Google Scholar]
- 72.Riley WD, Snell EE. Histidine decarboxylase of Lactobacillus 30a. V. Origin of enzyme-bound pyruvate and separation of nonidentical subunits. Biochemistry. 1970;9:1485–1491. doi: 10.1021/bi00809a002. [DOI] [PubMed] [Google Scholar]
- 73.Tyhach RJ, Hawrot E, Satre M, Kennedy EP. Increased synthesis of phosphatidylserine decarboxylase in a strain of Escherichia coli bearing a hybrid plasmid. Altered association of enzyme with the membrane. J Biol Chem. 1979;254:627–633. [PubMed] [Google Scholar]
- 74.Li QX, Dowhan W. Studies on the mechanism of formation of the pyruvate prosthetic group of phosphatidylserine decarboxylase from Escherichia coli. J Biol Chem. 1990;265:4111–4115. [PubMed] [Google Scholar]
- 75.Clancey CJ, Chang SC, Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem. 1993;268:24580–24590. [PubMed] [Google Scholar]
- 76.Trotter PJ, Pedretti J, Voelker DR. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem. 1993;268:21416–21424. [PubMed] [Google Scholar]
- 77.Voelker DR. Phosphatidylserine decarboxylase. Biochim Biophys Acta. 1997;1348:236–244. doi: 10.1016/s0005-2760(97)00101-x. [DOI] [PubMed] [Google Scholar]
- 78.Matsumoto K. Phosphatidylserine synthase from bacteria. Biochim Biophys Acta. 1997;1348:214–227. doi: 10.1016/s0005-2760(97)00110-0. [DOI] [PubMed] [Google Scholar]
- 79.Bae-Lee MS, Carman GM. Phosphatidylserine synthesis in Saccharomyces cerevisiae. Purification and characterization of membrane-associated phosphatidylserine synthase. J Biol Chem. 1984;259:10857–10862. [PubMed] [Google Scholar]
- 80.Raetz CR, Kennedy EP. The association of phosphatidylserine synthetase with ribosomes in extracts of Escherichia coli. J Biol Chem. 1972;247:2008–2014. [PubMed] [Google Scholar]
- 81.Raetz CR, Kennedy EP. Partial purification and properties of phosphatidylserine synthetase from Escherichia coli. J Biol Chem. 1974;249:5083–5045. [PubMed] [Google Scholar]
- 82.Raetz CR, Kennedy EP. Function of cytidine diphosphate-diglyceride and deoxycytidine diphosphate-diglyceride in the biogenesis of membrane lipids in Escherichia coli. J Biol Chem. 1973;248:1098–1105. [PubMed] [Google Scholar]
- 83.Raetz CR, Hirschberg CB, Dowhan W, Wickner WT, Kennedy EP. A membrane-bound pyrophosphatase in Escherichia coli catalyzing the hydrolysis of cytidine diphosphate-diglyceride. J Biol Chem. 1972;247:2245–2247. [PubMed] [Google Scholar]
- 84.Raetz CR, Dowhan W, Kennedy EP. Partial purification and characterization of cytidine 5'-diphosphate-diglyceride hydrolase from membranes of Escherichia coli. J Bacteriol. 1976;125:855–863. doi: 10.1128/jb.125.3.855-863.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bulawa CE, Hermes JD, Raetz CR. Chloroform-soluble nucleotides in Escherichia coli. Role of CDP-diglyceride in the enzymatic cytidylylation of phosphomonoester acceptors. J Biol Chem. 1983;258:14974–14980. [PubMed] [Google Scholar]
- 86.Raetz CR, Carman GM, Dowhan W, Jiang RT, Waszkuc W, Loffredo W, Tsai MD. Phospholipids chiral at phosphorus. Steric course of the reactions catalyzed by phosphatidylserine synthase from Escherichia coli and yeast. Biochemistry. 1987;26:4022–4027. doi: 10.1021/bi00387a042. [DOI] [PubMed] [Google Scholar]
- 87.Larson TJ, Dowhan W. Ribosomal-associated phosphatidylserine synthetase from Escherichia coli: purification by substrate-specific elution from phosphocellulose using cytidine 5'-diphospho-1,2-diacyl-sn-glycerol. Biochemistry. 1976;15:5212–5218. doi: 10.1021/bi00669a003. [DOI] [PubMed] [Google Scholar]
- 88.Clarke L, Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 89.Raetz CR, Larson TJ, Dowhan W. Gene cloning for the isolation of enzymes of membrane lipid synthesis: phosphatidylserine synthase overproduction in Escherichia coli. Proc Natl Acad Sci U S A. 1977;74:1412–1416. doi: 10.1073/pnas.74.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohta A, Waggoner K, Louie K, Dowhan W. Cloning of genes involved in membrane lipid synthesis. Effects of amplification of phosphatidylserine synthase in Escherichia coli. J Biol Chem. 1981;256:2219–2225. [PubMed] [Google Scholar]
- 91.Louie K, Dowhan W. Investigations on the association of phosphatidylserine synthase with the ribosomal component from Escherichia coli. J Biol Chem. 1980;255:1124–1127. [PubMed] [Google Scholar]
- 92.Louie K, Chen YC, Dowhan W. Substrate-induced membrane association of phosphatidylserine synthase from Escherichia coli. J Bacteriol. 1986;165:805–812. doi: 10.1128/jb.165.3.805-812.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeChavigny A, Heacock PN, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 94.Carman GM, Dowhan W. Phosphatidylserine synthase from Escherichia coli. The role of Triton X-100 in catalysis. J Biol Chem. 1979;254:8391–8397. [PubMed] [Google Scholar]
- 95.Larson TJ, Hirabayshi T, Dowhan W. Phosphatidylglycerol biosynthesis in Bacillus licheniformis. Resolution of membrane-bound enzymes by affinity chromatography on cytidinediphospho-sn-1,2-diacylglycerol Sepharose. Biochemistry. 1976;15:974–979. doi: 10.1021/bi00650a005. [DOI] [PubMed] [Google Scholar]
- 96.Hirabayashi T, Larson TJ, Dowhan W. Membrane-associated phosphatidylglycerophosphate synthetase from Escherichia coli: purification by substrate affinity chromatography on cytidine 5'-diphospho-1,2-diacyl-sn-glycerol sepharose. Biochemistry. 1976;15:5205–5211. doi: 10.1021/bi00669a002. [DOI] [PubMed] [Google Scholar]
- 97.Ohta A, Waggoner K, Radominska-Pyrek A, Dowhan W. Cloning of genes involved in membrane lipid synthesis: effects of amplification of phosphatidylglycerophosphate synthase in Escherichia coli. J Bacteriol. 1981;147:552–562. doi: 10.1128/jb.147.2.552-562.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nishijima M, Raetz CR. Membrane lipid biogenesis in Escherichia coli: identification of genetic loci for phosphatidylglycerophosphate synthetase and construction of mutants lacking phosphatidylglycerol. J Biol Chem. 1979;254:7837–7844. [PubMed] [Google Scholar]
- 99.Gopalakrishnan AS, Chen YC, Temkin M, Dowhan W. Structure and expression of the gene locus encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J Biol Chem. 1986;261:1329–1338. [PubMed] [Google Scholar]
- 100.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 101.Usui M, Sembongi H, Matsuzaki H, Matsumoto K, Shibuya I. Primary structures of the wild-type and mutant alleles encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J Bacteriol. 1994;176:3389–3392. doi: 10.1128/jb.176.11.3389-3392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lightner VA, Bell RM, Modrich P. The DNA sequences encoding plsB and dgk loci of Escherichia coli. J Biol Chem. 1983;258:10856–10861. [PubMed] [Google Scholar]
- 103.Icho T, Bulawa CE, Raetz CR. Molecular cloning and sequencing of the gene for CDP-diglyceride hydrolase of Escherichia coli. J Biol Chem. 1985;260:12092–12098. [PubMed] [Google Scholar]
- 104.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nishijima M, Raetz CR. Characterization of two membrane-associated glycolipids from an Escherichia coli mutant deficient in phosphatidylglycerol. J Biol Chem. 1981;256:10690–10696. [PubMed] [Google Scholar]
- 106.Raetz CR. Molecular genetics of membrane phospholipid synthesis. Annu Rev Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- 107.Dowhan W. The Raetz pathway for lipid A biosynthesis: Christian Rudolf Hubert Raetz, MD PhD, 1946–2011. J Lipid Res. 2011;52:1857–1860. doi: 10.1194/jlr.E020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wickner WT, Stubbe J, Hirschberg CB, Garrett T, Dowhan W. Chris Raetz, scientist and enduring friend. Proc Natl Acad Sci U S A. 2011;108:17255–17256. doi: 10.1073/pnas.1114405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ganong BR, Raetz CR. Massive accumulation of phosphatidic acid in conditionally lethal CDP-diglyceride synthetase mutants and cytidine auxotrophs of Escherichia coli. J Biol Chem. 1982;257:389–394. [PubMed] [Google Scholar]
- 110.Sparrow CP, Raetz CR. Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. J Biol Chem. 1985;260:12084–12091. [PubMed] [Google Scholar]
- 111.Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisia e is essential for cell growth. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- 112.Wu L, Niemeyer B, Colley N, Socolich M, Zuker CS. Regulation of PLCmediated signalling in vivo by CDP-diacylglycerol synthase. Nature. 1995;373:216–222. doi: 10.1038/373216a0. [DOI] [PubMed] [Google Scholar]
- 113.Weeks R, Dowhan W, Shen H, Balantac N, Meengs B, Nudelman E, Leung DW. Isolation and expression of an isoform of human CDP-diacylglycerol synthase cDNA. DNA Cell Biol. 1997;16:281–289. doi: 10.1089/dna.1997.16.281. [DOI] [PubMed] [Google Scholar]
- 114.Icho T. Membrane-bound phosphatases in Escherichia coli: sequence of the pgpA gene. J Bacteriol. 1988;170:5110–5116. doi: 10.1128/jb.170.11.5110-5116.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Icho T. Membrane-bound phosphatases in Escherichia coli: sequence of the pgpB gene and dual subcellular localization of the pgpB product. J Bacteriol. 1988;170:5117–5124. doi: 10.1128/jb.170.11.5117-5124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Touze T, Blanot D, Mengin-Lecreulx D. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J Biol Chem. 2008;283:16573–16583. doi: 10.1074/jbc.M800394200. [DOI] [PubMed] [Google Scholar]
- 117.Funk CR, Zimniak L, Dowhan W. The pgpA and pgpB genes of Escherichia coli are not essential: evidence for a third phosphatidylglycerophosphate phosphatase. J Bacteriol. 1992;174:205–213. doi: 10.1128/jb.174.1.205-213.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Osman C, Haag M, Wieland FT, Brügger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 2010;29:1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CR, Dowhan W, Dixon JE. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ohta A, Obara T, Asami Y, Shibuya I. Molecular cloning of the cls gene responsible for cardiolipin synthesis in Escherichia coli and phenotypic consequences of its amplification. J Bacteriol. 1985;163:506–514. doi: 10.1128/jb.163.2.506-514.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hiraoka S, Nukui K, Uetake N, Ohta A, Shibuya I. Amplification and substantial purification of cardiolipin synthase of Escherichia coli. J Biochem. 1991;110:443–449. doi: 10.1093/oxfordjournals.jbchem.a123600. [DOI] [PubMed] [Google Scholar]
- 122.Quigley BR, Tropp BE. E. coli cardiolipin synthase: function of N-terminal conserved residues. Biochim Biophys Acta. 2009;1788:2107–2113. doi: 10.1016/j.bbamem.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 123.Guo D, Tropp BE. A second Escherichia coli protein with CL synthase activity. Biochim Biophys Acta. 2000;1483:263–274. doi: 10.1016/s1388-1981(99)00193-6. [DOI] [PubMed] [Google Scholar]
- 124.Shibuya I, Yamagoe S, Miyazaki C, Matsuzaki H, Ohta A. Biosynthesis of novel acidic phospholipid analogs in Escherichia coli. J Bacteriol. 1985;161:473–477. doi: 10.1128/jb.161.2.473-477.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nishijima S, Asami Y, Uetake N, Yamagoe S, Ohta A, Shibuya I. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J Bacteriol. 1988;170:775–780. doi: 10.1128/jb.170.2.775-780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Michaelis AM, Gitai Z. Dynamic polar sequestration of excess MurG may regulate enzymatic function. J Bacteriol. 2010;192:4597–4605. doi: 10.1128/JB.00676-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hiraoka S, Matsuzaki H, Shibuya I. Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 1993;336:221–224. doi: 10.1016/0014-5793(93)80807-7. [DOI] [PubMed] [Google Scholar]
- 128.Serricchio M, Bütikofer P. An essential bacterial-type cardiolipin synthase mediates cardiolipin formation in a eukaryote. Proc Natl Acad Sci U S A. 2012;109:E954–E961. doi: 10.1073/pnas.1121528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shibuya I, Miyazaki C, Ohta A. Alteration of phospholipid composition by combined defects in phosphatidylserine and cardiolipin synthases and physiological consequences in Escherichia coli. J Bacteriol. 1985;161:1086–1092. doi: 10.1128/jb.161.3.1086-1092.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van Golde LMG, Schulman H, Kennedy EP. Metabolism of membrane phospholipids and its relation to a novel class of oligosaccharides in Escherichia coli. Proc Natl Acad Sci U S A. 1973;70:1368–1372. doi: 10.1073/pnas.70.5.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kennedy EP, Rumley MK, Schulman H, Van Golde LM. Identification of sn glycero-1-phosphate and phosphoethanolamine residues linked to the membrane-derived Oligosaccharides of Escherichia coli. J Biol Chem. 1976;251:4208–4213. [PubMed] [Google Scholar]
- 132.Schneider JE, Reinhold V, Rumley MK, Kennedy EP. Structural studies of the membrane-derived oligosaccharides of Escherichia coli. J Biol Chem. 1979;254:10135–10138. [PubMed] [Google Scholar]
- 133.Schulman H, Kennedy EP. Identification of UDP-glucose as an intermediate in the biosynthesis of the membrane-derived oligosaccharides of Escherichia coli. J Biol Chem. 1977;252:6299–6303. [PubMed] [Google Scholar]
- 134.Jackson BJ, Kennedy EP. The biosynthesis of membrane-derived oligosaccharides. A membrane-bound phosphoglycerol transferase. J Biol Chem. 1983;258:2394–2398. [PubMed] [Google Scholar]
- 135.Raetz CR, Newman KF. Diglyceride kinase mutants of Escherichia coli: inner membrane association of 1,2-diglyceride and its relation to synthesis of membrane-derived oligosaccharides. J Bacteriol. 1979;137:860–868. doi: 10.1128/jb.137.2.860-868.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miller KJ, Kennedy EP, Reinhold VN. Osmotic adaptation by gram-negative bacteria: possible role for periplasmic oligosaccharides. Science. 1986;231:48–51. doi: 10.1126/science.3941890. [DOI] [PubMed] [Google Scholar]
- 137.Weissborn AC, Rumley MK, Kennedy EP. Isolation and characterization of Escherichia coli mutants blocked in production of membrane-derived oligosaccharides. J Bacteriol. 1992;174:4856–4859. doi: 10.1128/jb.174.14.4856-4859.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gupta SD, Dowhan W, Wu HC. Phosphatidylethanolamine is not essential for the N-acylation of apolipoprotein in Escherichia coli. J Biol Chem. 1991;266:9983–9986. [PubMed] [Google Scholar]
- 139.Reynolds CM, Kalb SR, Cotter RJ, Raetz CR. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem. 2005;280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 140.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, Datsenko KA, Nakayashiki T, Tomita M, Wanner BL, Mori H. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Heacock PN, Dowhan W. Construction of a lethal mutation in the synthesis of the major acidic phospholipids of Escherichia coli. J Biol Chem. 1987;262:13044–13049. [PubMed] [Google Scholar]
- 143.Heacock PN, Dowhan W. Alteration of the phospholipid composition of Escherichia coli through genetic manipulation. J Biol Chem. 1989;264:14972–14977. [PubMed] [Google Scholar]
- 144.Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–446. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Miyazaki C, Kuroda M, Ohta A, Shibuya I. Genetic manipulation of membrane phospholipid composition in Escherichia coli: pgsA mutants defective in phosphatidylglycerol synthesis. Proc Natl Acad Sci U S A. 1985;82:7530–7534. doi: 10.1073/pnas.82.22.7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Asai Y, Katayose Y, Hikita C, Ohta A, Shibuya I. Suppression of the lethal effect of acidic-phospholipid deficiency by defective formation of the major outer membrane lipoprotein in Escherichia coli. J Bacteriol. 1989;171:6867–6869. doi: 10.1128/jb.171.12.6867-6869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu HC. Biosynthesis of lipoproteins. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1005–1014. [Google Scholar]
- 148.Suzuki M, Hara H, Matsumoto K. Envelope disorder of Escherichia coli cells lacking phosphatidylglycerol. J Bacteriol. 2002;184:5418–5425. doi: 10.1128/JB.184.19.5418-5425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kikuchi S, Shibuya I, Matsumoto K. Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J Bacteriol. 2000;182:371–376. doi: 10.1128/jb.182.2.371-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nagahama H, Sakamoto Y, Matsumoto K, Hara H. RcsA-dependent and - independent growth defects caused by the activated Rcs phosphorelay system in the Escherichia coli pgsA null mutant. J Gen Appl Microbiol. 2006;52:91–98. doi: 10.2323/jgam.52.91. [DOI] [PubMed] [Google Scholar]
- 151.Shiba Y, Yokoyama Y, Aono Y, Kiuchi T, Kusaka J, Matsumoto K, Hara H. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J Bacteriol. 2004;186:6526–6535. doi: 10.1128/JB.186.19.6526-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shiba Y, Miyagawa H, Nagahama H, Matsumoto K, Kondo D, Matsuoka S, Hara H. Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiology. 2012;158:1238–1248. doi: 10.1099/mic.0.056945-0. [DOI] [PubMed] [Google Scholar]
- 153.de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- 154.Kusters R, Dowhan W, de Kruijff B. Negatively charged phospholipids restore prePhoE translocation across phosphatidylglycerol-depleted Escherichia coli inner membranes. J Biol Chem. 1991;266:8659–8662. [PubMed] [Google Scholar]
- 155.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 156.Yung BY, Kornberg A. Membrane attachment activates DnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xia W, Dowhan W. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Garner J, Durrer P, Kitchen J, Brunner J, Crooke E. Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J Biol Chem. 1998;273:5167–5173. doi: 10.1074/jbc.273.9.5167. [DOI] [PubMed] [Google Scholar]
- 159.Kitchen JL, Li Z, Crooke E. Electrostatic interactions during acidic phospholipid reactivation of DnaA protein, the Escherichia coli initiator of chromosomal replication. Biochemistry. 1999;38:6213–6221. doi: 10.1021/bi982733q. [DOI] [PubMed] [Google Scholar]
- 160.Li Z, Kitchen JL, Boeneman K, Anand P, Crooke E. Restoration of growth to acidic phospholipid-deficient cells by DnaA(L366K) is independent of its capacity for nucleotide binding and exchange and requires DnaA. J Biol Chem. 2005;280:9796–9801. doi: 10.1074/jbc.M413923200. [DOI] [PubMed] [Google Scholar]
- 161.Mileykovskaya E, Dowhan W. Role of membrane lipids in bacterial division-site selection. Curr Opin Microbiol. 2005;8:135–142. doi: 10.1016/j.mib.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 162.Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W. Effects of phospholipid composition on MinD-membrane interactions i n vitro and in vivo. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- 163.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koppelman CM, Den Blaauwen T, Duursma MC, Heeren RM, Nanninga N. Escherichia coli minicell membranes are enriched in cardiolipin. J Bacteriol. 2001;183:6144–6147. doi: 10.1128/JB.183.20.6144-6147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mazor S, Regev T, Mileykovskaya E, Margolin W, Dowhan W, Fishov I. Mutual effects of MinD-membrane interaction: I. Changes in the membrane properties induced by MinD binding. Biochim Biophys Acta. 2008;1778:2496–2504. doi: 10.1016/j.bbamem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mazor S, Regev T, Mileykovskaya E, Margolin W, Dowhan W, Fishov I. Mutual effects of MinD-membrane interaction: II. Domain structure of the membrane enhances MinD binding. Biochim Biophys Acta. 2008;1778:2505–2511. doi: 10.1016/j.bbamem.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA. Phosphatidic acid and N-acylphosphatidylethanolamine form membrane domains in Escherichia coli mutant lacking cardiolipin and phosphatidylglycerol. J Biol Chem. 2009;284:2990–3000. doi: 10.1074/jbc.M805189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Raetz CR, Kantor GD, Nishijima M, Newman KF. Cardiolipin accumulation in the inner and outer membranes of Escherichia coli mutants defective in phosphatidylserine synthetase. J Bacteriol. 1979;139:544–551. doi: 10.1128/jb.139.2.544-551.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li Q-X. Department of Biochemistry and Molecular Biology, vol. Ph.D. Houston, TX: University of Texas Health Science Center at Houston. Graduate School of Biomedical Sciences; 1989. Structure and biosynthesis of a pyruvate-dependent enzyme: Phosphatidylserine decarboxylase from Escherichia coli; p. 110. [Google Scholar]
- 170.Rietveld AG, Killian JA, Dowhan W, de Kruijff B. Polymorphic regulation of membrane phospholipid composition in Escherichia coli. J Biol Chem. 1993;268:12427–12433. [PubMed] [Google Scholar]
- 171.Cullis PR, de Kruijff B. The polymorphic phase behaviour of phosphatidylethanolamines of natural and synthetic origin. A 31P NMR study. Biochim Biophys Acta. 1978;513:31–42. doi: 10.1016/0005-2736(78)90109-8. [DOI] [PubMed] [Google Scholar]
- 172.Cullis PR, Verkleij AJ, Ververgaert PH. Polymorphic phase behaviour of cardiolipin as detected by 31P NMR and freeze-fracture techniques. Effects of calcium, dibucaine and chlorpromazine. Biochim Biophys Acta. 1978;513:11–20. doi: 10.1016/0005-2736(78)90107-4. [DOI] [PubMed] [Google Scholar]
- 173.Vasilenko I, De Kruijff B, Verkleij AJ. Polymorphic phase behaviour of cardiolipin from bovine heart and from Bacillus subtilis as detected by 31P-NMR and freeze-fracture techniques. Effects of Ca2+, Mg2+, Ba2+ and temperature. Biochim Biophys Acta. 1982;684:282–286. doi: 10.1016/0005-2736(82)90018-9. [DOI] [PubMed] [Google Scholar]
- 174.Rietveld AG, Chupin VV, Koorengevel MC, Wienk HL, Dowhan W, de Kruijff B. Regulation of lipid polymorphism is essential for the viability of phosphatidylethanolamine-deficient Escherichia coli cells. J Biol Chem. 1994;269:28670–28675. [PubMed] [Google Scholar]
- 175.Chang CF, Shuman H, Somlyo AP. Electron probe analysis, X-ray mapping, and electron energy-loss spectroscopy of calcium, magnesium, and monovalent ions in log-phase and in dividing Escherichia coli B cells. J Bacteriol. 1986;167:935–939. doi: 10.1128/jb.167.3.935-939.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262:12570–12574. [PubMed] [Google Scholar]
- 177.Raetz CR, Foulds J. Envelope composition and antibiotic hypersensitivity of Escherichia coli mutants defective in phosphatidylserine synthetase. J Biol Chem. 1977;252:5911–5915. [PubMed] [Google Scholar]
- 178.Wikström M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander Ä, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 179.Kanipes MI, Lin S, Cotter RJ, Raetz CR. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-D-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J Biol Chem. 2001;276:1156–1163. doi: 10.1074/jbc.M009019200. [DOI] [PubMed] [Google Scholar]
- 180.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bertram R, Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol. 2008;1:2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bogdanov M, Dowhan W. Lipid-dependent generation of a dual topology for a membrane protein. J Biol Chem. 2012 doi: 10.1074/jbc.M112.404103. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xia W, Dowhan W. Phosphatidylinositol cannot substitute for phosphatidylglycerol in supporting cell growth of Escherichia coli. J Bacteriol. 1995;177:2926–2928. doi: 10.1128/jb.177.10.2926-2928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology. 2004;150:45–51. doi: 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 188.Bogdanov M, Xie J, Dowhan W. Lipid-protein interactions drive membrane protein topogenesis in accordance with the positive inside rule. J Biol Chem. 2009;284:9637–9641. doi: 10.1074/jbc.R800081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Carter JR, Fox CF, Kennedy EP. Interaction of sugars with the membrane protein component of the lactose transport system of Escherichia coli. Proc Natl Acad Sci U S A. 1968;60:725–732. doi: 10.1073/pnas.60.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fox CF, Carter JR, Kennedy EP. Genetic control of the membrane protein component of the lactose transport system of Escherichia coli. Proc Natl Acad Sci U S A. 1967;57:698–705. doi: 10.1073/pnas.57.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kennedy EP, Fox CF, Carter JR. Membrane structure and function. J Gen Physiol. 1966;49:347–354. doi: 10.1085/jgp.49.6.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fox CF, Kennedy EP. Specific labeling and partial purification of the M protein, a component of the beta-galactoside transport system of Escherichia coli. Proc Natl Acad Sci U S A. 1965;54:891–899. doi: 10.1073/pnas.54.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification and reconstitution of functional lactose carrier from Escherichia coli. J Biol Chem. 1981;256:11804–11808. [PubMed] [Google Scholar]
- 194.Newman MJ, Wilson TH. Solubilization and reconstitution of the lactose transport system from Escherichia coli. J Biol Chem. 1980;255:10583–10586. [PubMed] [Google Scholar]
- 195.Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J Biol Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- 196.Seto-Young D, Chen CC, Wilson TH. Effect of different phospholipids on the reconstitution of two functions of the lactose carrier of Escherichia coli. J Membr Biol. 1985;84:259–267. doi: 10.1007/BF01871389. [DOI] [PubMed] [Google Scholar]
- 197.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bogdanov M, Dowhan W. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J Biol Chem. 1995;270:732–739. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- 199.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 200.Sun J, Wu J, Carrasco N, Kaback HR. Identification of the epitope for monoclonal antibody 4B1 which uncouples lactose and proton translocation in the lactose permease of Escherichia coli. Biochemistry. 1996;35:990–998. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 201.Frillingos S, Kaback HR. Monoclonal antibody 4B1 alters the pKa of a carboxylic acid at position 325 (helix X) of the lactose permease of Escherichia coli. Biochemistry. 1996;35:10166–10171. doi: 10.1021/bi960995r. [DOI] [PubMed] [Google Scholar]
- 202.Frillingos S, Wu J, Venkatesan P, Kaback HR. Binding of ligand or monoclonal antibody 4B1 induces discrete structural changes in the lactose permease of Escherichia coli. Biochemistry. 1997;36:6408–6414. doi: 10.1021/bi970233b. [DOI] [PubMed] [Google Scholar]
- 203.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 204.Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. Embo J. 1998;17:5255–5264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J Biol Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 206.Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 207.Kaback HR, Sahin-Toth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 208.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bogdanov M, Heacock PN, Dowhan W. Study of polytopic membrane protein topological organization as a function of membrane lipid composition. Methods Mol Biol. 2010;619:79–101. doi: 10.1007/978-1-60327-412-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM™): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wikström M, Kelly AA, Georgiev A, Eriksson HM, Klement MR, Bogdanov M, Dowhan W, Wieslander Ä. Lipid-engineered Escherichia coli membranes reveal critical lipid headgroup size for protein function. J Biol Chem. 2009;284:954–965. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem. 2006;281:19172–19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 214.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 215.Vitrac H, Bogdanov M, Heacock P, Dowhan W. Lipids and topological rules of membrane protein assembly: balance between long and short range lipid-protein interactions. J Biol Chem. 2011;286:15182–15194. doi: 10.1074/jbc.M110.214387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nilsson I, von Heijne G. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell. 1990;62:1135–1141. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- 217.Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. Embo J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 219.Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 220.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 221.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mileykovskaya EI, Dowhan W. Alterations in the electron transfer chain in mutant strains of Escherichia coli lacking phosphatidylethanolamine. J Biol Chem. 1993;268:24824–24831. [PubMed] [Google Scholar]
- 223.Mileykovskaya E, Dowhan W. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol. 1997;179:1029–1034. doi: 10.1128/jb.179.4.1029-1034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mileykovskaya E, Sun Q, Margolin W, Dowhan W. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J Bacteriol. 1998;180:4252–4257. doi: 10.1128/jb.180.16.4252-4257.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Iwamoto K, Kobayashi S, Fukuda R, Umeda M, Kobayashi T, Ohta A. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells. 2004;9:891–903. doi: 10.1111/j.1365-2443.2004.00782.x. [DOI] [PubMed] [Google Scholar]
- 226.Emoto K, Umeda M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J Cell Biol. 2000;149:1215–1224. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shi W, Bogdanov M, Dowhan W, Zusman DR. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J Bacteriol. 1993;175:7711–7714. doi: 10.1128/jb.175.23.7711-7714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kates M, Syz JY, Gosser D, Haines TH. pH-dissociation characteristics of cardiolipin and its 2'-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 229.Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy (EM) J Biol Chem. 2012 doi: 10.1074/jbc.M112.367888. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 231.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. Embo J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. Embo J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Stuart RA. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Biol Chem. 2008;40:411–417. doi: 10.1007/s10863-008-9168-4. [DOI] [PubMed] [Google Scholar]
- 234.Wittig I, Schägger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim Biophys Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 235.Kawasaki K, Kuge O, Chang SC, Heacock PN, Rho M, Suzuki K, Nishijima M, Dowhan W. Isolation of a chinese hamster ovary (CHO) cDNA encoding phosphatidylglycerophosphate (PGP) synthase, expression of which corrects the mitochondrial abnormalities of a PGP synthase-defective mutant of CHO-K1 cells. J Biol Chem. 1999;274:1828–1834. doi: 10.1074/jbc.274.3.1828. [DOI] [PubMed] [Google Scholar]
- 236.Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 2001;507:187–190. doi: 10.1016/s0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- 238.Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 241.Wilkison WO, Walsh JP, Corless JM, Bell RM. Crystalline arrays of the Escherichia coli sn-glycerol-3-phosphate acyltransferase, an integral membrane protein. J Biol Chem. 1986;261:9951–9958. [PubMed] [Google Scholar]
- 242.McIntyre TM, Chamberlain BK, Webster RE, Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis. Effects of cessation and reinitiation of phospholipid synthesis on macromolecular synthesis and phospholipid turnover. J Biol Chem. 1977;252:4487–4493. [PubMed] [Google Scholar]
- 243.Clark D, Lightner V, Edgar R, Modrich P, Cronan JE, Jr, Bell RM. Regulation of phospholipid biosynthesis in Escherichia coli. Cloning of the structural gene for the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J Biol Chem. 1980;255:714–717. [PubMed] [Google Scholar]
- 244.Saha SK, Nishijima S, Matsuzaki H, Shibuya I, Matsumoto K. A regulatory mechanism for the balanced synthesis of membrane phospholipid species in Escherichia coli. Biosci Biotechnol Biochem. 1996;60:111–116. doi: 10.1271/bbb.60.111. [DOI] [PubMed] [Google Scholar]
- 245.Linde K, Gröbner G, Rilfors L. Lipid dependence and activity control of phosphatidylserine synthase from Escherichia coli. FEBS Lett. 2004;575:77–80. doi: 10.1016/j.febslet.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 246.Rilfors L, Niemi A, Haraldsson S, Edwards K, Andersson AS, Dowhan W. Reconstituted phosphatidylserine synthase from Escherichia coli is activated by anionic phospholipids and micelle-forming amphiphiles. Biochim Biophys Acta. 1999;1438:281–294. doi: 10.1016/s1388-1981(99)00060-8. [DOI] [PubMed] [Google Scholar]
- 247.Jackson BJ, Gennity JM, Kennedy EP. Regulation of the balanced synthesis of membrane phospholipids. Experimental test of models for regulation in Escherichia coli. J Biol Chem. 1986;261:13464–13468. [PubMed] [Google Scholar]
- 248.Conover GM, Martinez-Morales F, Heidtman MI, Luo ZQ, Tang M, Chen C, Geiger O, Isberg RR. Phosphatidylcholine synthesis is required for optimal function of Legionella pneumophila virulence determinants. Cell Microbiol. 2008;10:514–528. doi: 10.1111/j.1462-5822.2007.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]