Abstract
Purpose
Pancreatic cancer (PC) is the fourth cause of death from cancer in the western world. Majority of patients present with advanced unresectable disease responding poorly to most chemotherapeutic agents. Chemotherapy for PC might be improved by adjusting it to individual genetic profiles. We attempt to identify genetic predictors of chemosensitivity to broad classes of anticancer drugs.
Experimental Design
Using a panel of genetically defined human PC cell lines, we tested gemcitabine (anti-metabolite), docetaxel (anti-microtubule), mitomycin C (alkylating), irinotecan (topoisomerase I inhibitor), cisplatin (crosslinking), KU0058948 (Parp1 inhibitor), triptolide (terpenoid drug) and artemisinin (control).
Results
All PC cell lines were sensitive to triptolide and docetaxel. Most PC cells were also sensitive to gemcitabine and MMC. The vast majority of PC cell lines were insensitive to cisplatin, irinotecan, and a Parp1 inhibitor. However, individual cell lines were often sensitive to these compounds in unique ways. We found that DPC4/SMAD4 inactivation sensitized PC cells to cisplatin and irinotecan by 2–4 fold, but they were modestly less sensitive to gemcitabine. PC cells were all sensitive to triptolide and 18% were sensitive to the Parp1 inhibitor. P16/CDKN2A inactivated PC cells were 3–4 fold less sensitive to gemcitabine and MMC.
Conclusions
Chemosensitivity of PC cells correlated with some specific genetic profiles. These results support the hypothesis that genetic subsets of pancreatic cancer exist, and these genetic backgrounds may permit one to personalize the chemotherapy of PC in the future. Further work will need to confirm these responses and determine their magnitude in vivo.
Keywords: exome wide analysis, cytotoxicity screening, chemotherapeutic drugs, pancreatic cancer, personalizing chemotherapy
Introduction
Pancreatic cancer (PC) is the fourth leading cause of cancer death in the USA and leads to an estimated 227,000 deaths per year worldwide (1). 5-year survival rates for PC are estimated to be less than 5% because of its advanced stage at diagnosis, aggressive growth and resistance to most anticancer drugs (2). Despite this, systemic chemotherapy is generally used to treat patients with PC. Gemcitabine is a nucleoside analog and historically the standard first-line choice for the treatment of advanced PC. The median overall survival of single agent gemcitabine is approximately 6 months with a response rate of 10% in patients with metastatic disease (3). Recently, the combination of biweekly administered fluorouracil, leucovorin, irinotecan and oxaliplatin (FOLFIRINOX) was studied in a phase III trial yielding a response rate of 31.6% and a median overall survival of 11.1 months (4). De Jesus-Acosta, et al also demonstrated a substantial survival benefit with gemcitabine, docetaxel and capecitabine (GTX) chemotherapy in patients with metastatic and locally advanced PC, and achieved a median survival of 11.3 and 25.0 months, respectively (5).
Recent studies in human PC have revealed complex genetic alterations in its pathogenesis and progression. Jones et al (6) sequenced 20,661 protein-coding genes in 24 PCs and found alterations in KRAS (>95%), p16/CDKN2A (95%), TP53 (50–75%) and DPC4/SMAD4 (55%), the four high-frequency PC driver genes (6–8). Recent studies reported that inactivation of DPC4/SMAD4 was associated with poorer prognosis in patients with surgically resected PC and a widely metastatic phenotype in unresected patients (9, 10). Chemoresistance of tumor cells can be enhanced by mutations in oncogenes, loss of tumor suppressors or dysregulation of genes involved in DNA repair, cell cycle, cell proliferation, signal transduction, angiogenesis or apoptosis (11, 12).
Using genetic alterations to predict chemotherapeutic response may prove to be one of the most important concepts in the therapy of human cancer. The field of targeted therapy is rapidly expanding with more than 800 new agents developed, 140 of which are under clinical evaluation. For example, Her2/neu is closely associated with aggressive clinical behavior and poor outcome (13), and trastuzumab was the first Her2-targeted therapy approved by the FDA in 1998 for the treatment of metastatic Her2-mutated patients (14). Parp1 is a critical enzyme involved in cell proliferation and repair of single stranded DNA breaks and multiple PARP1 inhibitors have been tested clinically with encouraging results, particularly in triple negative BRCA1/2-mutated breast cancer (15). Extrapolation of this finding to other BRCA1/2 defective cancers may not necessarily and directly follow (16). The cytogenetic lesion of chronic myelogenous leukemia (CML) is the Philadelphia chromosome, which arises from a balanced reciprocal translocation between the long arms of chromosomes 9 and 22 (17). This translocation produces a constitutively active tyrosine kinase (BCR-ABL) (18) for which imatinib mesylate, a first-generation tyrosine kinase inhibitor (TKI), is a potent inhibitor (19). However, although numerous studies have addressed chemosensitivity testing in a variety of malignancies, yet there are few studies in PC (20–22).
We wished to test the hypothesis that the genetic mutations in PC might predict chemotherapeutic response. For these initial studies, we selected a panel of broadly acting anticancer agents including, an anti-metabolite drug (gemcitabine), an anti-microtubule drug (docetaxel), an alkylator (mitomycin C), a topoisomerase I inhibitor (irinotecan), a crosslinking agent (cisplatin), a Parp inhibitor (KU0058948), a terpenoid (triptolide) and a negative control drug (artemisinin). We tested this panel of drugs for cytotoxicity using a panel of genetically defined human PC cells. We also verified some initial findings using DPC4/SMAD4 and TP53 isogenic pairs of cancer cells. The preliminary data demonstrate that genetic subsets of PC correlate with in vitro chemosensitivity, and these genetic backgrounds may be useful for personalizing chemotherapy of PC patients in the future.
Materials and Methods
Human PC cell lines, human pancreatic ductal epithelial cell line, human DPC4/SMAD4 isogenic PC cell lines, human TP53 isogenic colon cancer cell lines and cell culture
Thirty-four human pancreatic cancer cell lines used in this study were isolated from the tumor samples of patients in Departments of Pathology and Oncology, Johns Hopkins School of Medicine (Supplementary Table S1). Among them, nineteen cell lines were used for the Discovery Screen and ten cell lines were used for the Prevalence Screen of the original PC sequencing study (6). The sequences of 23,219 transcripts representing 20,661 protein-coding genes were determined, and somatic mutations identified in the discovery and prevalence screen, homozygous deletions, and SAGE gene expression data have been published in detail elsewhere (6). Twelve of the patients contributing samples to this study had a family history of the disease, defined as having at least two first degree relatives affected with pancreatic cancer. The use of human tissue was approved by the local IRB (John Hopkins University). All human pancreatic cancer cell lines were cultured in MEM medium (Gibco, Grand Island, NY) supplemented with 10% Fetal Bovine Serum (FBS) (Gibco), 100 U/ml Penicillin and 100 ug /ml Streptomycin.
An HPV transfected normal human pancreatic ductal epithelial cell line (HPDE), generously provided by Dr. Ming-Sound Tsao, was also analyzed. Apart from slightly aberrant expression of p53, molecular profiling of this cell line has shown that expression of other proto-oncogenes and tumor suppressor genes are normal (23). The HPDE cell line was grown in keratinocyte serum free-media (Invitrogen, Grand Island, NY) supplemented with bovine pituitary extract and recombinant epidermal growth factor.
Pa01C and Pa02C parental cell lines were both generated from liver metastases of PC patients, which both had homozygous deletions of DPC4. Pa01C and Pa02C cells were stably transfected with either a cDNA expression plasmid to overexpress DPC4 (pcDNA3.1-DPC4) or an empty plasmid (pcDNA3.1Mock) using the Attractene transfection agent (Qiagen, Valencia, CA). The Pa01C and Pa02C derivative isogenic cells were both analyzed as single clones. All of the genetically complemented clones express DPC4, while the control cells do not (data not shown). TGFβ pathway activities were tested for these isogenic pairs using a previously reported luciferase assay (24).These cell lines were cultured in DMEM medium (Gibco) supplemented with 10% FBS (Gibco), 100 U/ml Penicillin, 100 ug /ml Streptomycin, and 0.35 mg/mL G418 sulfate (Mediatech, Manassas, VA). Supplementary Table S2 summarizes the DPC4/SMAD4 isogenic cell lines.
The parental colorectal cancer cell lines HCT116, SW48, and RKO were from ATCC. All the isogenic cell pairs for TP53 were created with genetic knockout / knockin method, as was previously reported (25). These cell lines were cultured in McCoy’s 5A medium (Gibco) supplemented with 10% FBS (Gibco), 100 U/ml Penicillin and 100 ug /ml Streptomycin. Supplementary Table S3 shows the characteristics of TP53 isogenic cell lines.
All cells were maintained at 37 ° in a humidified atmosphere with 5% CO2.
Preparation of anticancer drugs
Triptolide, docetaxel, MMC, cisplatin, irinotecan and artemisinin were purchased from Sigma (St Louis, MO). Gemcitabine was obtained from Net Qem (Research Triangle Park, NC). Parp1 inhibitor (KU0058948) was synthesized in Dr. Vogelstein’s lab. Artemisinin (an anti-malarial drug) was selected as a negative control drug since it possessed no known anticancer activity. The eight drugs were dissolved in DMSO (triptolide, docetaxel, cisplatin, irinotecan and artemisinin) or PBS (MMC and gemcitabine), stock solutions of 10mM were made and they were stored at −80 °.
In vitro cell growth assay for drug screening
Cells were plated in 96-well plates at a density of 3,000 cells per well using standard culture media (described above). Edge wells of the 96-well plate were filled with 1 X PBS. After overnight growth, media were removed and 200ul of medium with drug was added into each well. Drugs were serially diluted to desired final concentrations with medium containing 10% serum and each dose had 6 replicate wells. Cells incubated in the medium with the vehicle DMSO or PBS served as controls. Plates were incubated with drugs for 72 hours. Media were aspirated and each well was washed with PBS 3 times to remove cellular debris. 100ul of ddH20 was added to each well and plates were incubated at room temperature for 1 hour to lyse the cells. 100ul of deionized H2O containing 0.15% of SYBR green I solution (Molecular Probes, cat # S7567) were added to wells and mixed 10 times. Fluorescence was measured by using BMG FluoStar Galaxy (BMG Labtechnologies, USA)(excitation at 480nm and measurement at 520nm) (25).
Human PC cell lines and HPDE were tested initially at the following doses: 0nM, 1nM, 10nM, 100nM, 1uM, 10uM and 100uM. Positive drug effects and isogenic cells drugs were tested using final concentrations: 0nM, 1nM, 3nM, 10nM, 30nM, 100nM, 300nM, 1uM, 3uM and 10uM in three independent experiments (18 replicate wells totally per dose).
Gene set enrichment analysis for gene expression data (Supplementary Materials and Methods)
Statistical analysis
IC50 values were calculated by fitting sigmoid dose-response curves with GraphPad Prism 5.0 (GraphPad Software, Inc.). IC50 distributions were compared by genetic status (mutations, deletions or both) and family history status with Wilcoxon rank sum tests. A drug response was defined as an IC50 value below the median or below the highest IC50 that did not exceed 10000 (this happened for drugs where more than 50% of cell lines had IC50 values greater than or equal to 10000). The relationship between response and genetic status was summarized with an odds ratio from a logistic regression model and Fisher’s exact test. When comparing the IC50 values between different drugs or to fold change in pathway activity, Spearman’s rank based correlation was used. IC50 values between isogenic pairs were compared with a paired Student’s t test. Chemosensitivity correlation analysis of clustered cell lines was examined with heatmaps and a network map, which is a visual depiction of how cell lines cluster based on whether the Spearman correlation of the within-cell line standardized IC50 values between a pair of cell lines was 0.65 or higher. Statistical analyses were performed using GraphPad Prism 5.0 and R (version 2.13.1). Due to the exploratory nature of the study, p-values are not corrected for multiple comparisons and are included for descriptive purposes only.
Results
Substantial variation of PC cell lines to broad classes of chemotherapeutic agents
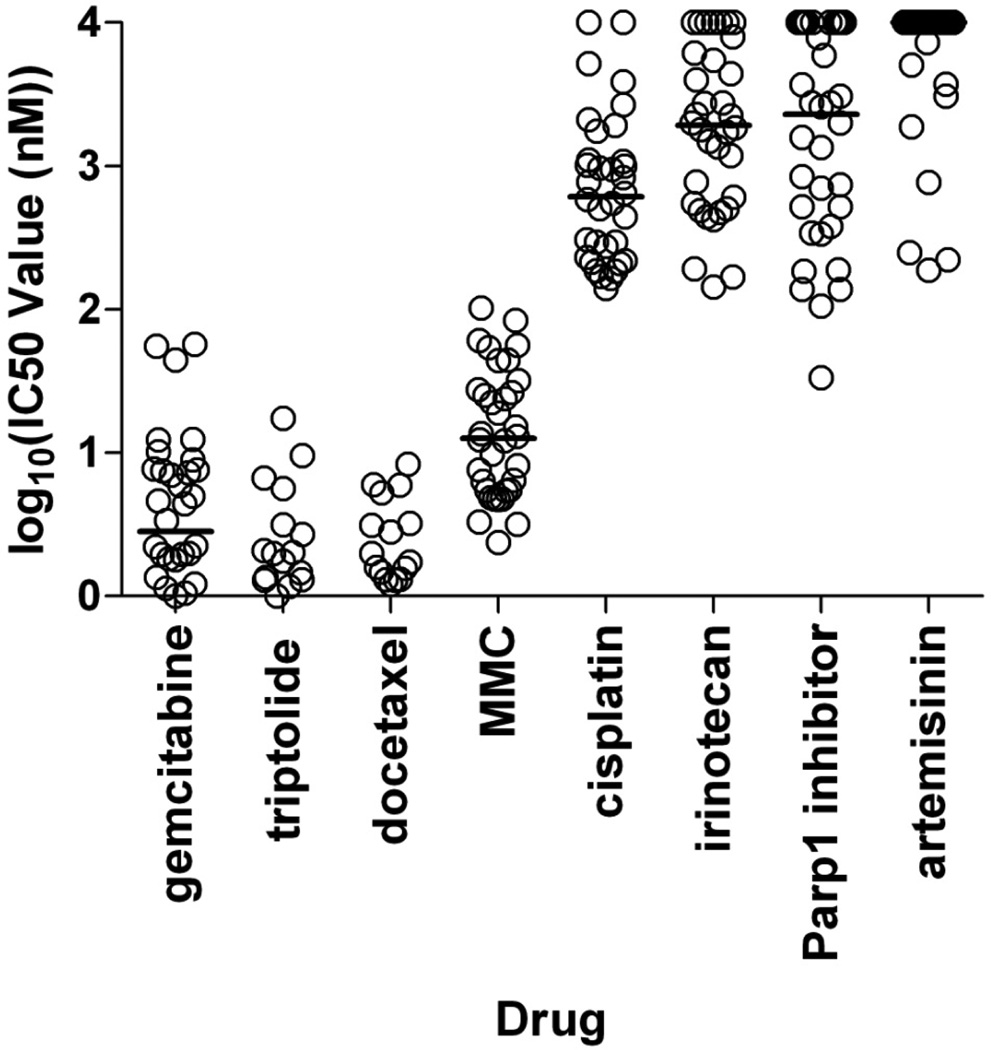
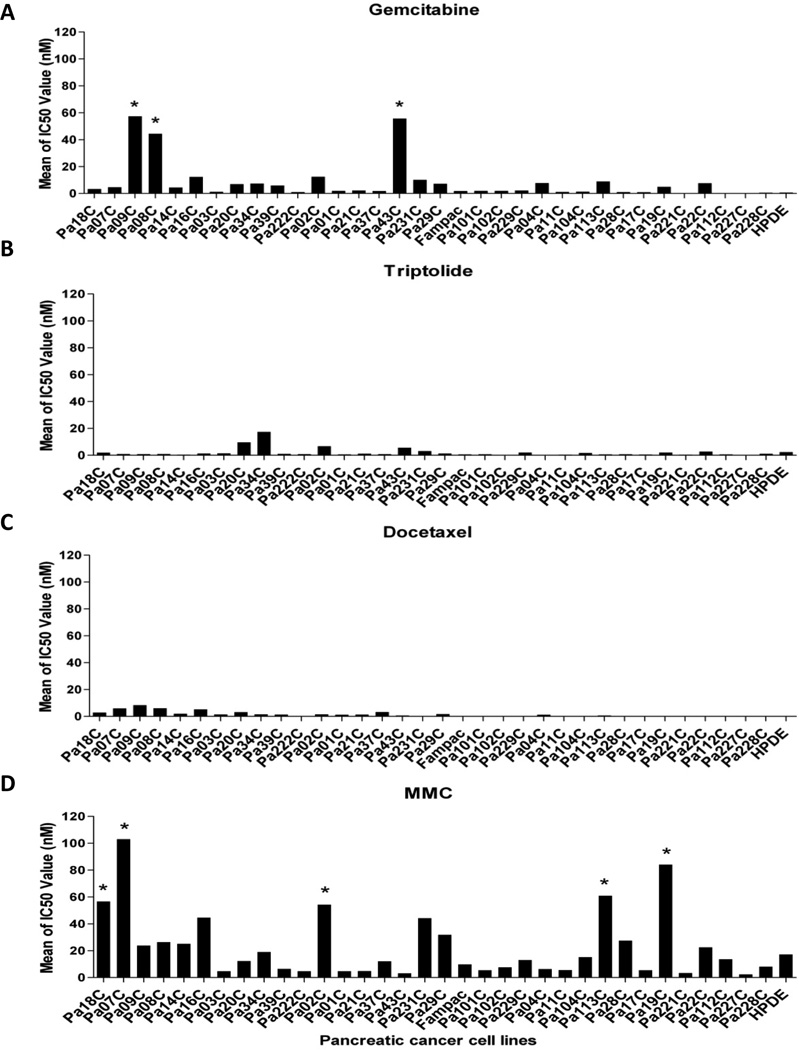
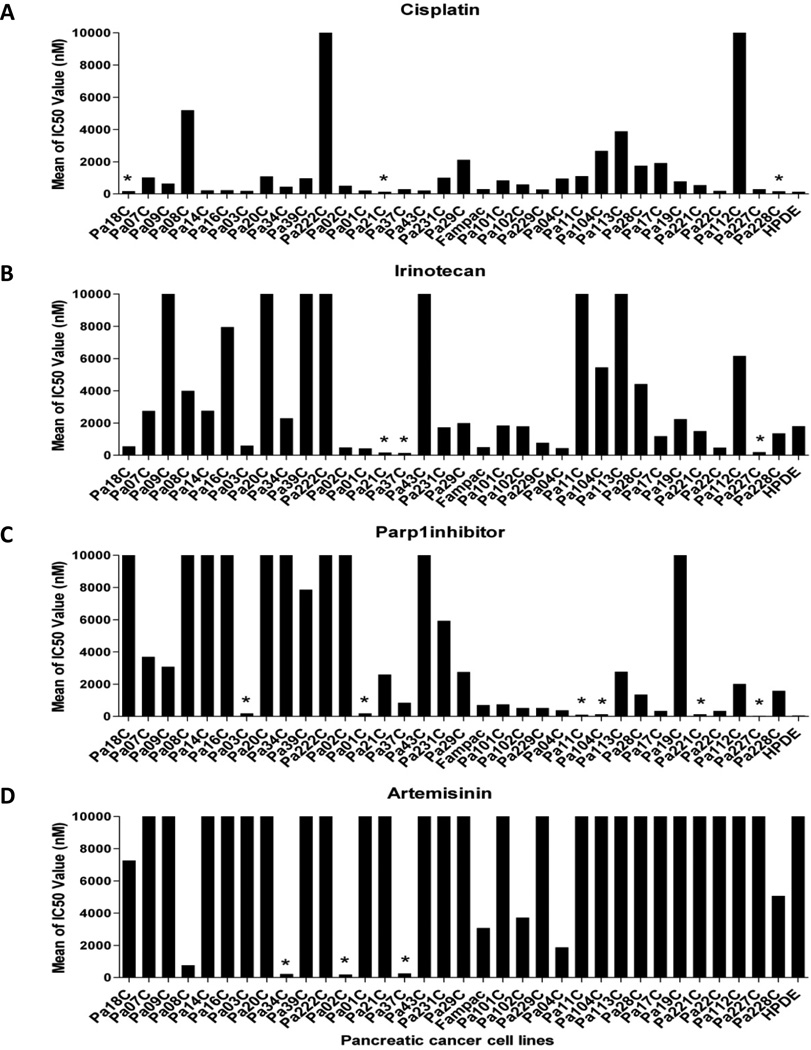
We screened 29 genetically defined PC cell lines with the seven broad classes of anticancer drugs listed above. Pancreatic cancer cells were exposed to increasing concentrations of the drugs in 6 replicate wells and IC50 values calculated (Figure 1 and Supplementary Table S4, S5). Most pancreatic cancer cell lines were sensitive to gemcitabine, triptolide, docetaxel and MMC. The majority of pancreatic cancer cell lines were insensitive to cisplatin, irinotecan, Parp1 inhibitor and the negative control drug. Among the generally sensitive drugs, some cell lines were notably less sensitive (Figure 2A, 2D). For example, with gemcitabine, the vast majority of cell lines were sensitive with IC50s in the single digit nanomolar range, whereas three cell lines (Pa09C, Pa08C and Pa43C) were significantly less sensitive (IC50s: 40 to 60 nM). Similarly, for MMC, most of cell lines were sensitive with IC50s under 50 nM, whereas five cell lines (Pa18C, Pa07C, Pa02C, Pa113C and Pa19C) were substantially resistant. Surprisingly, among the generally ineffective drugs (Figure 1), individual cell lines were uniquely sensitive (Figure 3). For example, with cisplatin, most cell lines were quite insensitive, whereas a few (Pa18C, Pa21C and Pa228C) had relatively lower IC50 values (100 to 200 nM). Similarly, for irinotecan, three cell lines (Pa21C, Pa37C and Pa227C) demonstrated IC50 values less than 200nM, and for the Parp1 inhibitor, six cell lines (Pa03C, Pa01C, Pa11C, Pa104C, Pa221C and Pa227C) had IC50s less than 200nM. In summary, among the sensitive drugs, outlier PC cell lines were more resistant, while among the resistant drugs, unique cell lines were more sensitive.
Figure 1. Chemosensitivity of PC cell lines to broad classes of chemotherapeutic agents.
Drug responses (Log10 of the IC50 values) of 34 PC cell lines tested with 7 chemotherapeutic drugs representing different mechanisms of action, and the control drug (artemisinin). Each cell line result is represented by a single circle. Horizontal lines indicate the median IC50 value for each drug for all cell lines. The four drugs on the left are generally sensitive drugs, and the four drugs on the right are generally inactive.
Figure 2. Reponses of individual PC cell lines to the 4 sensitive drugs (note Y axis scale, 0 to 120nM).
Cytotoxic effects of gemcitabine (A), triptolide (B), docetaxel (C) and MMC (D) on PC cells. Gemcitabine (A) demonstrated generally good responses, although unique cell lines showed more than 100 fold less sensitivities (* indicates outlier cell lines). Triptolide (B) and docetaxel (C) demonstrated nearly universal toxicities at doses in the single digit nanomolar concentration. MMC (D) showed more variation in response, and unique cell lines (*) showed more than 40 fold less sensitivity.
Figure 3. Reponses of individual PC cell lines to the 4 insensitive drugs (note Y axis scale, 0 to 10,000nM).
Cytotoxic effects of cisplatin (A), irinotecan (B) and Parp1 inhibitor(C) on pancreatic cancer cell viability demonstrated variable drug responses, and unique cell lines showed more than 200 fold variation in their sensitivities to these drugs (*indicates uniquely sensitive cell line). Artemisinin (D) showed the nearly invariant resistance at doses in greater than 10,000 nanomolar, although unique cell lines (*) were sensitive.
DPC4/SMAD4 inactivation sensitized PC cells to cisplatin and irinotecan but reduced their sensitivity to gemcitabine
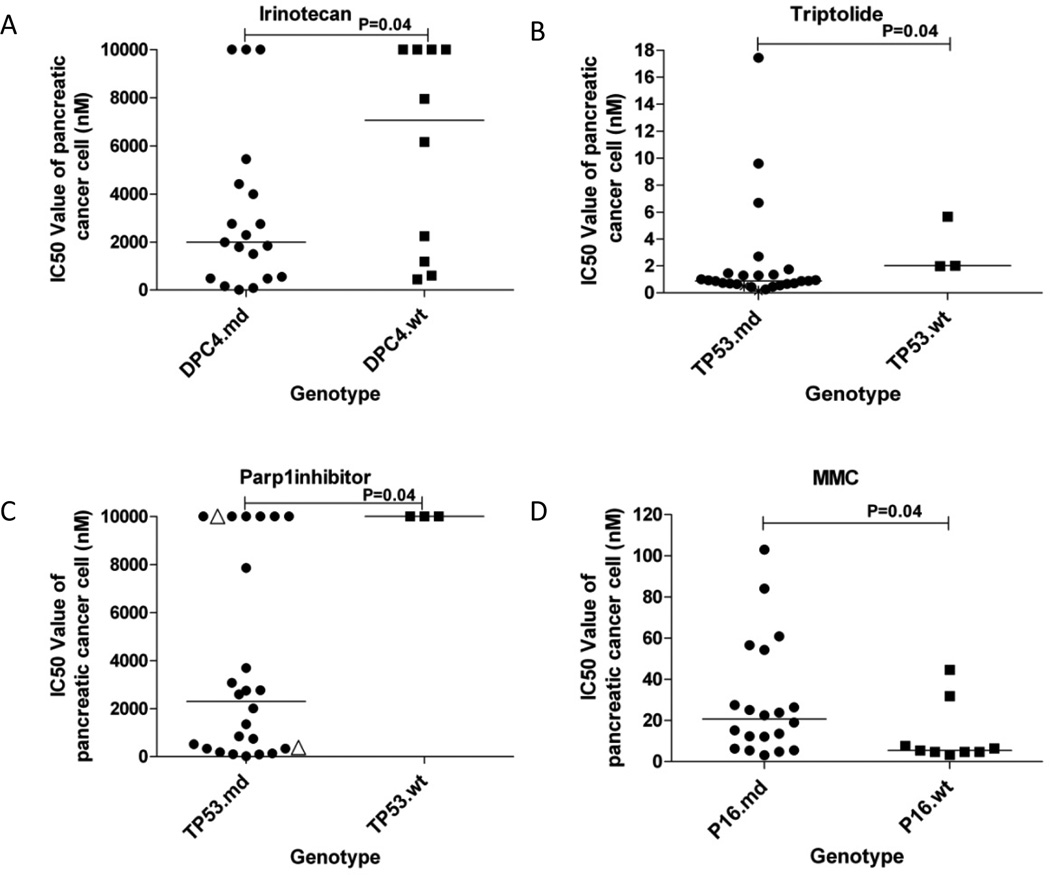
We attempted to explain the variation in drug response noted above by correlating it with the genetic status of these lines. We checked all the somatic mutations and homozygous deletions which existed in the 19 PC cell lines included in the discovery screen and the 10 PC cell lines included in the prevalence screen (Supplementary Table S1) (6). We attempted to correlate drug responses to genotype (Figure 4 and Supplementary Figure S1, S2), considering each gene as either mutated only (mut), deleted only (del), mutated or deleted combined (md), or wildtype (wt). We initially focused on DPC4/SMAD4 (Supplementary Table S6, S7).
Figure 4. Correlations of drug response to genotype.
Cytotoxic effect of irinotecan (A) was related to DPC4/SMAD4 inactivation, those of triptolide (B) and Parp1 inhibitor (C) were related to TP53 inactivation, and that of MMC (D) was related to P16 inactivation. Wilcoxon rank sum tests were used to compare differences in the median IC50 by mutation status. “md” indicates either mutation or deletion and the “wt” indicates wildtype. For TP53.md group in panel C, solid circles indicate TP53 mutated PC cell lines, and triangles indicate TP53 deleted PC cell lines.
PC cells with DPC4 homozygous deletions were modestly (2–fold) more sensitive to cisplatin compared to those with wild type genotype (P=0.04) (Supplementary Figure S1A and Supplementary Table S6). Cells with the deletion of any of four SMAD Pathway genes (SMAD3, SMAD4, TGFβR2 or TGFβR3) were 2.2-fold more sensitive to cisplatin (P=0.02) (Supplementary Table S6). In logistic regression analysis, comparisons of DPC4 deleted or SMADPath gene deleted or SMADPath gene defective (mutated or deleted) PC cells to wildtype cells treated with cisplatin confirmed this finding (odds ratios: 11.93, 15.07 and 7.56, P values: 0.01, 0.001 and 0.05, respectively) (Supplementary Table S7).
One concern about this study is that of multiple comparisons. Accordingly, to confirm these findings, we constructed isogenic cell lines from Pa01C and Pa02C with or without homozygous DPC4 deletions. With cisplatin, we found that IC50 values of DPC4 deficient cell lines for cisplatin were 1.8-fold and 1.2-fold lower than for DPC4-containing cell lines (Figure 5A and Supplementary Table S8). This was less significant in another isogenic cell line pair derived from Pa02C, but this cell line showed lower TGFβ signaling even when DPC4 was present (Supplementary Figure S3A). Because all the pairs of isogenic cells had different levels of TGFβ pathway signaling (Supplementary Figure S3A), we attempted to correlate the cisplatin IC50 values with TGFβ pathway activities in these cells. Our results confirmed that IC50 values of cisplatin were positively correlated with TGFβ activity (r=0.86, 95%CI: 0.39 to 0.97, P=0.007), although this relationship is largely driven by the two cell lines with highest TGFβR activity (Supplementary Figure S3B and Supplementary Table S9). The analysis of expression data showed that cytotoxicity of cisplatin was closely positively associated with MAP3K12 (an activator of the SAPK/JNK pathway), MAPK11 (most closely related to p38 MAP kinase), and MAP4K2 (involved in activation of MAP3K1/MEKK1) (Supplementary Table S10).
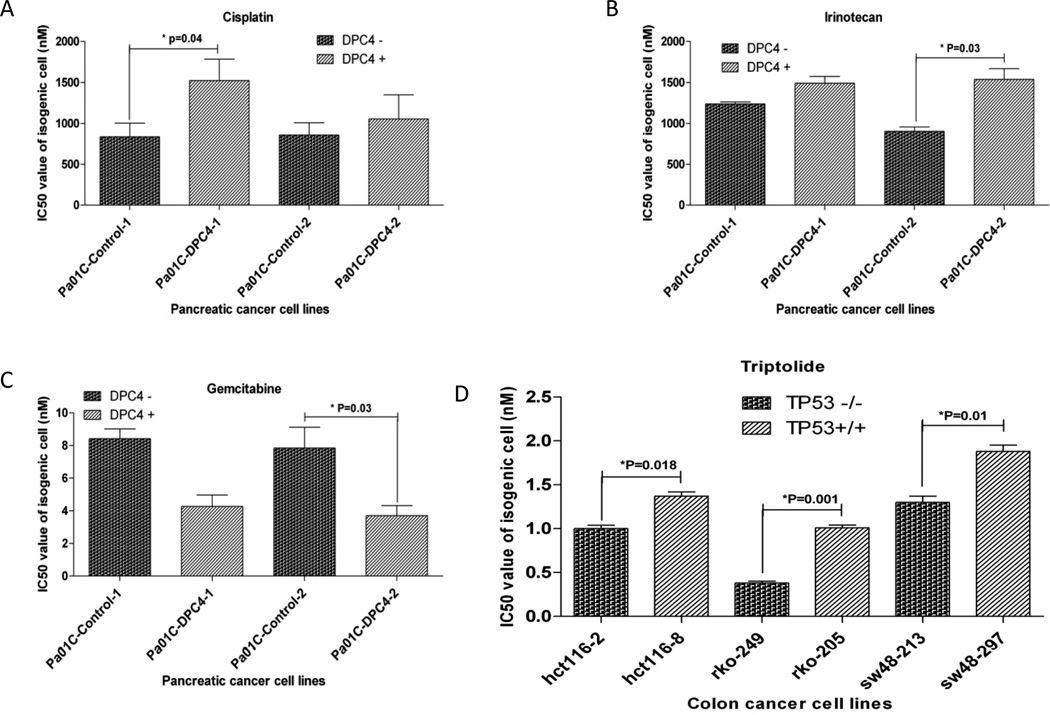
Figure 5. Reponses of isogenic cancer cell lines to anticancer drugs.
Two pairs of isogenic pancreatic cancer cell lines for DPC4/SMAD4 were tested with cisplatin (A), irinotecan (B) and gemcitabine (C). Results for Pa01C isogenic cells are shown, as they demonstrate higher TGFβ functional complementation. Three pairs of isogenic colon cancer cell lines for TP53 demonstrated differences in response for triptolide (D). Differences of IC50 value for isogenic pairs were compared with paired t-test analysis and significant P values indicated. IC50s were calculated for each experiment using 6 replicates per dose, and three separate experiments were used to calculate the mean IC50 for each drug and cell line combination (18 replicate wells totally per dose).
PC cells with DPC4 defects (mutations or deletion combined) were 4.5-fold more sensitive to irinotecan than those with wildtype DPC4 (P=0.04) (Figure 4A and Supplementary Table S6). In logistic regression analysis, comparison of DPC4 defective cells to wildtype cells treated with irinotecan produced an odds ratio of 8.64 and a P value of 0.02, and when only DPC4 deleted cells were compared to wildtype cells, a P value of 0.02 was obtained (Supplementary Table S7). In order to verify these findings, we tested the pairs of DPC4 isogenic cell lines with irinotecan. We found IC50 values of DPC4 deficient cell lines were 1.2-fold and 1.7-fold lower than those of DPC4 complemented cell lines (Figure 5B and Supplementary Table S8). Correlation analysis for IC50 values and TGFβ pathway activities was not significant (P=0.067) (Supplementary Table S9)
We tested two pairs of DPC4 isogenic cells for possible synergy between cisplatin and irinotecan at a constant ratio of 1:1 with simultaneous treatment. The combination indices (CI) were calculated using CompuSyn (26). Simultaneous treatment with the two drugs resulted in antagonistic interactions with DPC4 defective cells and synergistic interactions in DPC4 complemented clones at ID50, but these cell lines all showed antagonistic or additive interactions at ID75 (Supplementary Table S11).
Despite the lack of correlation between DPC4 status and gemcitabine response in the panel of PC cell lines (Supplementary Figure S1C, S1E), we tested the DPC4 isogenic cell lines with gemcitabine, and found that DPC4 defective cells were about 2-fold less sensitive than wildtype cells (P=0.058 and 0.033) (Figure 5C and Supplementary Table S8). We made the correlation analysis for IC50 values and TGFβ pathway activities in these isogenic cells. Our results confirmed that gemcitabine IC50s were negatively correlated with TGFβ pathway activities (P=0.015) (Supplementary Figure S3C and Supplementary Table S11).
Chemotherapeutic effects of triptolide and a Parp1 inhibitor were related to TP53 inactivation
PC cells with TP53 defects were 2.3-fold more sensitive to triptolide than wildtype TP53 cells, P=0.04 (Figure 4B and Supplementary Table S6). We confirmed these results in logistic regression analysis, P=0.02 (Supplementary Table S7), and confirmed a modest effect (1.5–2.7 fold) using three pairs of TP53 isogenic colon cancer cells (1.4–2.7 fold) (p<0.05) (Figure 5D and Supplementary Table S12).
PC cells with defective TP53 were 4.3-fold more sensitive to Parp1 inhibitor than those with wildtype TP53 alleles, P=0.04 (Figure 4C and Supplementary Table S6). In this analysis, only three cell lines were wildtype, and the response of TP53 defective cell lines was clearly bimodal with one highly resistant population (IC50s: > 10,000 nM), and a second variably sensitive population (IC50s: 20 to 4,000 nM). Logistic regression analysis confirmed that a subset of TP53 defective cancer cells were more sensitive to the Parp1 inhibitor (an odds ratio of 11.93 for TP53 mutation versus wild type, P=0.02, and a p value of 0.01 for TP53 mutation or deletion versus wild type) (Supplementary Table S7). We investigated possible explanations for this effect (Supplementary Table S13, S14), and identified a possible association between sensitivity and wild type P16 status, although this was not statistically significant. We attempted to confirm these findings using three pairs of TP53 isogenic colon cancer cell lines (Supplementary Figure S3D and Supplementary Table S12), however the opposite effect was seen, possibly because the different genetic and cellular backgrounds of the PC cells and the colon cancer cell lines.
P16/CDKN2A inactivation made human PC cells less sensitive to gemcitabine and MMC
PC cells with P16 homozygous deletions demonstrated approximately 3.1-fold higher IC50 values for gemcitabine than those with wildtype P16 (P=0.04) (Supplementary Figure S2A and Supplementary Table S6), although this was less significant when mutations and deletions were considered together (P=0.11) (Supplementary Figure S2C). Since wildtype alleles for DPC4 and P16 were each associated with gemcitabine sensitivity, we also compared PC cells having both wildtype P16 and DPC4 with those having both P16 and DPC4 defects, and found cells with wildtype DPC4 and P16 were 4-fold more sensitive to gemcitabine than DPC4 and P16 defects, although this difference did not achieve statistical significance (P=0.15) (Supplementary Figure S2D). PC cells with wildtype P16 alleles were 3.8-fold more sensitive to MMC than those with either P16 mutation or deletion (P=0.04) (Figure 4D and Supplementary Table S5). We confirmed this in logistic regression analysis (an odds ratio of 0.19 for P16 defects versus wildtype, P=0.05) (Supplementary Table S7). We attempted to explain the variation in drug response of the P16 defective cancer cells, but were unable to identify cooperating mutations in other genes (Supplementary Table S15).
Other findings in the anticancer drug screening
We selected the anti-malarial drug, artemisinin, as a negative control drug, and the majority (88.2%) of cancer cells demonstrated no response, as expected. To our surprise, three PC cell lines showed sensitive responses with IC50s less than 300 nM (Figure 3D).We examined the SAGE expression data in attempt to explain these results. We noted significant the lack of expression of P16, p15 and a series of mitogen-activated protein kinase genes in the sensitive lines (Supplementary Table S16).
Other drug response-genotype associations, drug response-gene expression profile correlations and DPC4/SMAD4, TP53 or P16/CDKN2A status-gene expression profile correlations are presented in Supplementary Results (Supplementary Table S6, S7, S10, S16, S17, S18, S19, S20, S21 and Supplementary Figure S4, S5). With regard to gemcitabine sensitivity, we were unable to correlate it with quasimesenchymal subtype (27) or ZEB1/2 expression (data not shown).
Cluster analysis for anticancer drug sensitivities
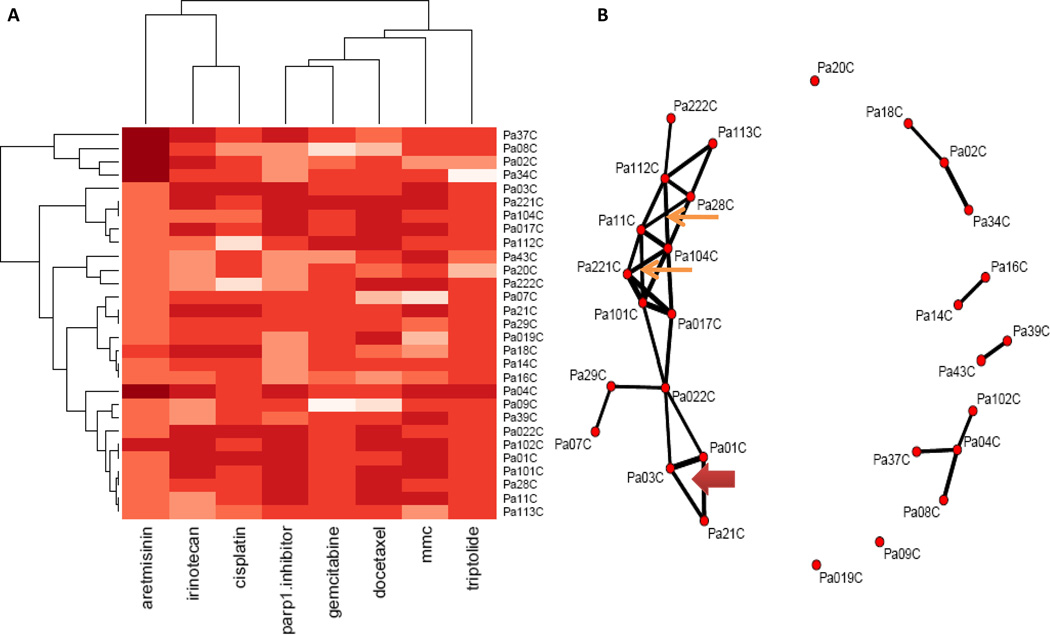
We also analyzed the chemosensitivity without consideration of the genetics using cluster analysis. Using an evolutionary cluster analysis, it appears that the drug pattern is dominant over the cell line clustering. As expected, sensitive drugs clustered on the right branch, and insensitive drugs clustered on the left branch (Figure 6A). We found that cisplatin with irinotecan, and gemcitabine with Parp1 inhibitors had similar drug response profiles. Some PC cell lines clustered together forming several groups with similar drug response profiles for 8 drugs (Figure 6B). For example, Pa01C, Pa03C and Pa21C in a triangle network showed similar drug responses, and similar different gene expression profiles that distinguished them from other PC cell lines (Supplementary Table S22). Other attempts to identify genes responsible for the clustering were not successful.
Figure 6. Chemosensitivity correlation analysis of PC cell lines using evolutionary trees and cluster analysis.
Cluster analysis was employed to analyze chemosensitivity correlation of PC cell lines, and a threshold of 0.65 (spearman correlation) indicates a positive correlation. Panel A is a clustered heatmap and panel B is a network map for PC cell lines. Different drugs clustered because of similar drug response profiles for PC cell lines (A). In panel B, the dots represent individual cell lines. Cell lines are connected by a line to every other sample with which it was correlated by at least 0.65. Related cancer cells are demonstrated by “boxes”(thin arrows) with “Xs” in the center for 4 cell lines and triangles between 3 cell lines (thick arrows). The line thickness indicates the strength of the correlation. Individual points mean that sample does not correlate with anyone else.
Discussion
Personalized chemotherapy based on the pattern of genetic alterations and gene expression in a tumor can, in theory, identify drugs to which the tumor will be sensitive as well as those unlikely to provide a therapeutic response. Comprehensive germline and somatic changes in patients and their cancers can now be determined at a reasonable cost in 2012. However, the bioinformatics to translate these data to chemoprediction is years behind. Our study attempts to fill this gap. In our study, we screened a set of genetically defined PCs to determine whether there were correlations between genetic and transcriptomic profiles and drug sensitivities. We found that somatic mutations or homozygous deletions of DPC4/SMAD4, TP53 and P16/CDKN2A genes correlated with in vitro drug responses, though the differential effects were not pronounced. Additional studies will be necessary to determine whether these correlations can be reproduced in pre-clinical animal models and in patients.
We identified an association between DPC4/SMAD4 defects and sensitivity to cisplatin and irinotecan in the initial panel of PC cell lines, and confirmed these effects using isogenic cell lines. The observation that cisplatin cytotoxicity was correlated with alteration of MAP kinases indicates that cytotoxic mechanism of cisplatin in PC cell may be partly linked to noncanonical TGFβ pathways. The activation of mitogen-activated protein kinase (MAPK) pathways by TGFβ depends upon the cell type, often in a DPC4-independent manner (28). For irinotecan, we observed that DPC4 inactivation sensitized PC cells to this drug. DNA topoisomerase I (Top1) is essential in metazoans for the relaxation of DNA supercoiling by forming Top1 cleavage complexes (Top1cc) (29), and Top1 inhibitors can stabilize Top1cc and Top1-associated DNA damage (30). TGFβ signaling influences cellular proliferation through inhibition of G1/S transition by inducing expression of cyclin kinase inhibitors such as P15, P21 and P27 while TGFβ signaling repressing c-Myc expression (31). Consistent with these prior studies, we observed downregulation of P15/CDKN1B and upregulation of CDCA4 and CDC2L6 with DPC4 inactivation in our panel of pancreatic cancer cells (Supplementary Table S19 and Supplementary Figure S5A, S35B). In a recent study, Bornstein et al (32) reported increased genomic instability in both normal head and neck tissues and HNSCC from HN-mice with selective inactivation of DPC4/SMAD4 in head and neck epithelium, which correlated with downregulated expression and function of genes encoding proteins in the Fanconi anemia/Brca DNA repair pathway. Under inactivation of DPC4, cell cycle checkpoint arrest is reduced, thereby increasing genomic instability and resulting in apoptosis, which may contribute to sensitivity of DNA damaging drugs in PC cells. In our study, we also found that wildtype DPC4 was a sensitive factor for gemcitabine. PCs are represented by distinct genetic subtypes with significantly different patterns of failure (9). So it may be valuable to determine DPC4 status at initial diagnosis and stratify patients into personalized chemotherapy.
We found substantial variation in Parp1 inhibitor sensitivity among PC cell lines, where sensitivity correlated with inactivated TP53. All TP53 wild type PC cells were highly resistant to the Parp1 inhibitor while there was considerable variation among TP53 inactivated cells. This variation was not correlated with the type of p53 mutation or mutation status at a second gene (Supplementary Table S13). Nor could we explain these results through differences in expression (Supplementary Table S14). TP53 downregulates homologous recombination (HR) either directly (33) or by interacting and/or regulating HR-related proteins, such as Rad51 (34). In this sense, TP53 inactivation results in increased exacerbated spontaneous or damage induced HR (35, 36). TP53-deficient mice exhibit high HR frequency at different developmental stages (37) and TP53 mutations correlate with elevated recombination rates (38, 39). Moreover, TP53 preferentially suppresses both spontaneous and DSB-induced nonconservative intrachromosomal HR between imperfect homologous sequences (ICHR), susceptible of generating gross rearrangements such as deletions, duplications or inversions (40). In our study, it was observed that RAD51 and other HR genes were upregulated in TP53 inactivated PC cells, which indicated a high HR frequency in TP53 deficient PC (Supplementary Table S20 and Supplementary Figure S5C). The correct balance between desirable and undesirable repair is attained by the finely tuned HR regulation of TP53, as evidenced by the fact that both excessive and defective HR are associated with cancer predisposition and resistance to therapeutic drugs (41).Therefore, TP53 inactivation may result in an imbalance between desirable and undesirable repair because of the lack of proper regulation by TP53 in HR. Some studies report that cells lacking PARP1 also show a hyper-recombinogenic phenotype (an increased frequency of HR) (42–44), which has also been observed in mice using the pun assay (45). We propose that TP53 inactivation may have a synthetic lethality with Parp1 inhibitor in PC cells because TP53 controls the fidelity of Rad51-dependent HR and represses aberrant processing of replication forks after stalling at unrepaired DNA lesions. In our study, we also found that the sensitivity of Parp1 inhibitor was related to expression profiles of some DNA damage and repair pathway genes (ERCC3, RAD17, SUMO1, MUTYH, CRY1, HSP90B1, CDC37, RXRA, USP5) in (Supplementary Tables S10, S14, S16).
Triptolide covalently binds to human XPB (also known as ERCC3), a subunit of the transcription factor TFIIH, and inhibits its DNA-dependent ATPase activity, which leads to the inhibition of RNA polymerase II–mediated transcription and likely nucleotide excision repair (46). In our study, we found that the expression level of ERCC3 was upregulated in triptolide sensitive PC cells and triptolide was more sensitive to PC cells with TP53 inactivation (Supplementary Table S16 and Supplementary Figure S4A). This indicates that ERCC3 may exert a synthetic lethality with TP53. Recent reports also indicate that triptolide causes PC cell death in vitro and in vivo by inducing apoptosis via inhibition of HSP70 which can inhibit apoptosis in cancer cells by attenuating cytosolic calcium and stabilizing lysosomes (47, 48). In our study, it was found that elevated expressions of HSF1 and HSPA9 were related to the triptolide sensitivities in PC cell lines (Supplementary Table S16).
Additionally, we showed that PC cells with P16 inactivation were less sensitive to gemcitabine and MMC. These findings are consistent with those of Halloran, who reported that gemcitabine combined with adenoviral-mediated reintroduction of P16 greatly enhanced cytotoxicity in Panc-1 pancreatic adenocarcinoma cells (49). P16/CDKN2A is a tumor suppressor and plays a pivotal role in regulating the G1/S cell cycle checkpoint (50). Gemcitabine and MMC can both cause DNA damage by incorporation of nucleoside analogs or production of crosslinks and wild type P16 may facilitate the process for cycle checkpoint arrest and apoptosis in PC cells. Cell cycle checkpoints are important in cellular response to DNA damage by arresting the cell cycle to provide time for repair and by inducing transcription of genes that facilitate repair (51). This is consistent with a clinical trial which suggested that the ability of cells to progress into the S phase after gemcitabine and radiation treatment was the key factor in radiosensitization (52). A recent study indicated that the polymorphic genotypes of p16 were associated with significantly shorter time to tumor progression and poorer response to therapy (53). Our analysis of expression data also showed that cytotoxicity of gemcitabine was closely associated with the expression of genes regulating the cell cycle, apoptosis, autophagy, DNA repair, tumor suppressor gene and anticancer drug metabolism pathway (Supplementary Table S16). So we hypothesize that mutations and deletions in cell cycle genes such as P16 may modulate the response to treatment with gemcitabine or MMC-based chemotherapy.
We also analyzed the chemosensitivity data by ignoring the genetics. For active drugs, we found uniquely insensitive lines, and among inactive drugs, including even the control drug, we found unique sensitive lines. By using cluster analysis, we found that some anticancer drugs have similar drug response profiles for PC cell lines, and some PC cell lines clustered together forming several groups with similar drug response profiles. Consistent with the findings of Collisson, et al (27), this provides evidence that functional subsets of PC exist, however, despite our best effort, we were unable to correlate clusters in Figure 6B with specific genetic defects. Drugs that closely cluster may indicate that they target a common pathway or downstream effector.
In vitro studies, such as ours, have limitations. Several hypotheses have been suggested to explain why purified malignant cells may exhibit sensitivities in vitro that do not extrapolate to in vivo tumors. Malignant cells may possess biologic properties that are lost in tissue culture. Alternatively, the resistance to systemic therapies in PC patients might also be partly explained by inefficient drug delivery to malignant cells, because stromal microenvironment may create a hypovascular penetration barrier that impairs effective chemotherapeutic delivery (54). Further, the degree of genetic heterogeneity of a tumor is also likely to be an important determinant of therapeutic outcome (55), the reconstruction of tumor clonal architectures and the identification of early driver mutations may provide more robust biomarkers and therapeutic approaches (56).
In a conclusion, we screened broad classes of anticancer drugs for their antiproliferative properties in a panel of human PC cell lines. These results suggest that genetic subsets of pancreatic cancer exist, and that many are significantly correlated with chemosensitivity to specific agents. These genetic subsets may be valuable for personalizing PC chemotherapy in the future.
Supplementary Material
Statement of Translational Relevance.
The costs of next generation sequencing are rapidly decreasing, and the $1,000 genome is near. This “new tool” may permit more rational and effective use of chemotherapeutic drugs. We studied the chemosensitivity of a broad panel of genetically defined pancreatic cancers to seven chemotherapeutic drugs with distinct mechanisms of cytotoxicity. We found seven different gene-drug correlations in vitro. Additional work will be needed to confirm and determine the magnitude of these effects in vivo. These findings provide an initial framework for personalized chemotherapy of this nearly lethal malignancy.
Acknowledgments
We thank Scott E. Ken, M.D., Christopher L. Wolfgang, M.D., Ph.D., F.A.C.S. and Leslie Cope, Ph.D. for helpful discussion and Ming-Sound Tsao, Ph.D. for generously providing the HPDE cell line in this project.
Grant Support:
These studies were supported by PanCan (JRE), CA130938 (JRE), CA140599 (CID), Lustgarten (BV), the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation for Pancreatic Cancer Research (AM, CID, JRE).
Footnotes
Disclosure of Potential Conflicts of Interest:
The authors disclose a patent licensed to Myriad Genetics (Drs. Hruban and Eshleman); Advisory Board membership in Inostics, Morphotek, Exact Sciences and PGDx (Dr. Vogelstein); Ownership Interest in Personal Genome Diagnostics and Inostics (Dr. Diaz); and Advisory Board membership in Roche Molecular Diagnostics (Dr. Eshleman). No potential conflicts of interest were declared by other authors.
References
- 1.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009;6:699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 2.Shaib YH, Davila JA, El-Serag HB. The epidemiology of pancreatic cancer in the United States: changes below the surface. Aliment Pharmacol Ther. 2006;24:87–94. doi: 10.1111/j.1365-2036.2006.02961.x. [DOI] [PubMed] [Google Scholar]
- 3.Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 4.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.De Jesus-Acosta A, Oliver GR, Blackford A, Kinsman K, Flores EI, Wilfong LS, et al. A multicenter analysis of GTX chemotherapy in patients with locally advanced and metastatic pancreatic adenocarcinoma. Cancer Chemother Pharmacol. 69:415–424. doi: 10.1007/s00280-011-1704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 8.Iacobuzio-Donahue CA, Song J, Parmiagiani G, Yeo CJ, Hruban RH, Kern SE. Missense mutations of MADH4: characterization of the mutational hot spot and functional consequences in human tumors. Clin Cancer Res. 2004;10:1597–1604. doi: 10.1158/1078-0432.ccr-1121-3. [DOI] [PubMed] [Google Scholar]
- 9.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackford A, Serrano OK, Wolfgang CL, Parmigiani G, Jones S, Zhang X, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15:4674–4679. doi: 10.1158/1078-0432.CCR-09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W, Lockhart AC, Kim RB, Rothenberg ML. Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. Oncologist. 2005;10:104–111. doi: 10.1634/theoncologist.10-2-104. [DOI] [PubMed] [Google Scholar]
- 12.Kleeff J, Michalski C, Friess H, Buchler MW. Pancreatic cancer: from bench to 5-year survival. Pancreas. 2006;33:111–118. doi: 10.1097/01.mpa.0000229010.62538.f2. [DOI] [PubMed] [Google Scholar]
- 13.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 14.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 15.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 16.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 17.Heisterkamp N, Stephenson JR, Groffen J, Hansen PF, de Klein A, Bartram CR, et al. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 18.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 19.Buchdunger E, Zimmermann J, Mett H, Meyer T, Muller M, Druker BJ, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 20.Dan S, Tsunoda T, Kitahara O, Yanagawa R, Zembutsu H, Katagiri T, et al. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002;62:1139–1147. [PubMed] [Google Scholar]
- 21.Zembutsu H, Ohnishi Y, Tsunoda T, Furukawa Y, Katagiri T, Ueyama Y, et al. Genome-wide cDNA microarray screening to correlate gene expression profiles with sensitivity of 85 human cancer xenografts to anticancer drugs. Cancer Res. 2002;62:518–527. [PubMed] [Google Scholar]
- 22.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 24.Sohn TA, Su GH, Ryu B, Yeo CJ, Kern SE. High-throughput drug screening of the DPC4 tumor-suppressor pathway in human pancreatic cancer cells. Ann Surg. 2001;233:696–703. doi: 10.1097/00000658-200105000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Jr, Kinzler KW, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 27.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 29.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009;8:1008–1014. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng B, Fleming JB, Breslin T, Grau AM, Fojioka S, Abbruzzese JL, et al. Suppression of tumorigenesis and induction of p15(ink4b) by Smad4/DPC4 in human pancreatic cancer cells. Clin Cancer Res. 2002;8:3628–3638. [PubMed] [Google Scholar]
- 32.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janz C, Susse S, Wiesmuller L. p53 and recombination intermediates: role of tetramerization at DNA junctions in complex formation and exonucleolytic degradation. Oncogene. 2002;21:2130–2140. doi: 10.1038/sj.onc.1205292. [DOI] [PubMed] [Google Scholar]
- 34.Linke SP, Sengupta S, Khabie N, Jeffries BA, Buchhop S, Miska S, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63:2596–2605. [PubMed] [Google Scholar]
- 35.Mekeel KL, Tang W, Kachnic LA, Luo CM, DeFrank JS, Powell SN. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 36.Saintigny Y, Rouillard D, Chaput B, Soussi T, Lopez BS. Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene. 1999;18:3553–3563. doi: 10.1038/sj.onc.1202941. [DOI] [PubMed] [Google Scholar]
- 37.Bishop AJ, Hollander MC, Kosaras B, Sidman RL, Fornace AJ, Jr, Schiestl RH. Atm-, p53-, and Gadd45a-deficient mice show an increased frequency of homologous recombination at different stages during development. Cancer Res. 2003;63:5335–5343. [PubMed] [Google Scholar]
- 38.Bertrand P, Rouillard D, Boulet A, Levalois C, Soussi T, Lopez BS. Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene. 1997;14:1117–1122. doi: 10.1038/sj.onc.1200931. [DOI] [PubMed] [Google Scholar]
- 39.Akyuz N, Boehden GS, Susse S, Rimek A, Preuss U, Scheidtmann KH, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudenhoffer C, Rohaly G, Will K, Deppert W, Wiesmuller L. Specific mismatch recognition in heteroduplex intermediates by p53 suggests a role in fidelity control of homologous recombination. Mol Cell Biol. 1998;18:5332–5342. doi: 10.1128/mcb.18.9.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 42.Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, et al. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31:4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldman AS, Waldman BC. Stimulation of intrachromosomal homologous recombination in mammalian cells by an inhibitor of poly(ADP-ribosylation) Nucleic Acids Res. 1991;19:5943–5947. doi: 10.1093/nar/19.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Claybon A, Karia B, Bruce C, Bishop AJ. PARP1 suppresses homologous recombination events in mice in vivo. Nucleic Acids Res. 38:7538–7545. doi: 10.1093/nar/gkq624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, et al. XPB a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol. 7:182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudeja V, Mujumdar N, Phillips P, Chugh R, Borja-Cacho D, Dawra RK, et al. Heat shock protein 70 inhibits apoptosis in cancer cells through simultaneous and independent mechanisms. Gastroenterology. 2009;136:1772–1782. doi: 10.1053/j.gastro.2009.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips PA, Dudeja V, McCarroll JA, Borja-Cacho D, Dawra RK, Grizzle WE, et al. Triptolide induces pancreatic cancer cell death via inhibition of heat shock protein 70. Cancer Res. 2007;67:9407–9416. doi: 10.1158/0008-5472.CAN-07-1077. [DOI] [PubMed] [Google Scholar]
- 49.Halloran CM, Ghaneh P, Shore S, Greenhalf W, Zumstein L, Wilson D, et al. 5-Fluorouracil or gemcitabine combined with adenoviral-mediated reintroduction of p16INK4A greatly enhanced cytotoxicity in Panc-1 pancreatic adenocarcinoma cells. J Gene Med. 2004;6:514–525. doi: 10.1002/jgm.540. [DOI] [PubMed] [Google Scholar]
- 50.Macaluso M, Montanari M, Cinti C, Giordano A. Modulation of cell cycle components by epigenetic and genetic events. Semin Oncol. 2005;32:452–457. doi: 10.1053/j.seminoncol.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 51.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 52.Ostruszka LJ, Shewach DS. The role of cell cycle progression in radiosensitization by 2',2'- difluoro-2'-deoxycytidine. Cancer Res. 2000;60:6080–6088. [PubMed] [Google Scholar]
- 53.Chen J, Li D, Killary AM, Sen S, Amos CI, Evans DB, et al. Polymorphisms of p16, p27, p73, and MDM2 modulate response and survival of pancreatic cancer patients treated with preoperative chemoradiation. Ann Surg Oncol. 2009;16:431–439. doi: 10.1245/s10434-008-0220-8. [DOI] [PubMed] [Google Scholar]
- 54.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 55.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012:323–324. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 56.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.