SUMMARY
Expansions of simple DNA repeats cause numerous hereditary diseases in humans. We analyzed the role of DNA polymerases in the instability of Friedreich’s ataxia (GAA)n repeats in a yeast experimental system. The elementary step of expansion corresponded to ~160 bp in the wild type strain, matching the size of Okazaki fragments in yeast. This step increased when DNA polymerase α was mutated suggesting a link between the scale of expansions and Okazaki fragment size. Expandable repeats strongly elevated the rate of mutations at substantial distances around them, a phenomenon we call repeat-induced mutagenesis (RIM). Notably, defects in the replicative DNA polymerases δ and ε strongly increased rates for both repeat expansions and RIM. The increases in repeat-mediated instability observed in DNA polymerase δ mutants depended on translesion DNA polymerases. We conclude that repeat expansions and RIM are two sides of the same replicative mechanism.
INTRODUCTION
Expansions of DNA microsatellites are responsible for several dozens of hereditary neurological and developmental disorders in humans, including fragile X syndrome, Friedreich’s ataxia, Huntington’s disease, myotonic dystrophy and others (reviewed in (Lopez Castel et al., 2010; Mirkin, 2007; Orr and Zoghbi, 2007). These repeats are stably inherited when short, but become highly unstable when their length exceeds a threshold of ~150 bps. Hundreds, and in some cases thousands of copies of the repeat could be added in a few generations (Ashley and Warren, 1995). Notably, the longer the repeat, the higher is its propensity to expand.
Molecular mechanisms of repeat expansions were studied in many model experimental systems including bacteria, yeast, Drosophila, mice and cultured human cells (reviewed in (Kovtun and McMurray, 2008). These studies implicated DNA replication (reviewed in (Cleary and Pearson, 2005; Mirkin, 2006), DNA repair (reviewed in (McMurray, 2008, 2010), DNA recombination (reviewed in (Wells et al., 2005) and transcription (reviewed in (Dion and Wilson, 2009) in the expansion process. As a rule, expansions observed in these systems were relatively short-scale (addition of 10-to-20 repeats), differently from what was observed in human pedigrees. To bridge the gap between the model system and human data, we developed a system to analyze large-scale repeat expansions in budding yeast, S. cerevisiae (Shishkin et al., 2009). To this end, Friedreich’s ataxia (GAA)n repeats were cloned into an intron of the artificially split URA3 gene. Expansions of the repeat that increased the intron’s length beyond ~1.1 kb blocked URA3 splicing, thus making cells 5-FOA resistant. Using this approach, we indeed observed large-scale repeat expansions, up to hundreds of triplets. Consistent with data from human pedigrees, longer repeats in our system expanded with a higher propensity than shorter ones. Preliminary genetic analysis led us to propose that these expansions occur during replication through the repeat and possibly involve template switching at the replication fork (Shishkin et al., 2009).
Prima facie, a replication model for repeat instability implies that replicative DNA polymerases at the fork should be key players in the expansion process. Surprisingly however, there is a lack of data on this topic with the exception of just one study that investigated the effects of DNA polymerase mutations on the instability of (CAG)n repeats (Schweitzer and Livingston, 1999). We, therefore, decided to analyze the rates and elementary steps of large-scale expansions of (GAA)n repeats in yeast strains carrying defects in each of the three replicative DNA polymerases: alpha (α), delta (δ) and epsilon (ε), as well as in mutants with compromised translesion DNA synthesis (TLS). We found that the rate of repeat expansions increased in all replicative but not TLS polymerase mutants, and most significantly in those affecting Polδ and Polε. A mutation in the PCNA precluding the K164 ubiqiutylation, a signal critical for TLS, moderated these increases. An elementary expansion step for different (GAA)n repeats in the wild-type strain fell between 44 and 63 repeats (122–189 bps), but it became much bigger, 80-to-85 triplets, in Polα mutants. These results show a link between the size of an Okazaki fragment and the scale of expansions.
We have previously noticed that (GAA)n repeats also elevate the rate of mutagenesis in cis at a significant distance from their location (Shishkin et al., 2009). Here we confirmed the phenomenon of repeat-induced mutagenesis (RIM) and analyzed the role of the DNA polymerase mutations in this process. Tellingly, the effects of various replication mutants on RIM were qualitatively similar to their effects on repeat expansions. Altogether, these data demonstrate the pivotal role of DNA replication in various forms of genome instability attributed to DNA repeats, including their expansions and mutagenesis.
RESULTS
Characteristics of primary expansions of (GAA)n repeats in the wild-type yeast
Our approach to detect large-scale repeat expansions in yeast was based on the fact that introns longer than ~1.1 kb cannot be efficiently spliced in S. cerevisiae (Yu and Gabriel, 1999). We reasoned therefore that large-scale expansions of a repeat within the intron of an artificially split URA3 gene would cause its inactivation, allowing yeast to grow on 5-FOA-containing media. This approach has indeed allowed us to detect large-scale expansions of the FRDA (GAA)n repeats in our system: up to 300 triplets could be added to the starting (GAA)100 repeat (Shishkin et al., 2009). Our original experimental system, however, appeared to have a caveat: introns carrying different (GAA)n repeats were much shorter (700-to-850 bp) than the splicing threshold and also were not of a uniform length between different repeats. This caveat led to a situation, in which only longer-size expansions pushed the intron length beyond the splicing threshold (~1.1 kb) and were recovered on selective media. These longer-sized expansions could be secondary expansions that followed a primary expansion event. Consequently, we could not easily distinguish between the primary and secondary events and were unable to estimate an elementary expansion step for different repeats.
To address these problems, we adjusted the lengths of each intron carrying different (GAA)n repeats by adding non-repetitive sequences and making them sufficiently long (900 to 1,000 bps). As a result, the addition of a relatively few (25-to-30 copies) triplets to an initial (GAA)n repeat during the primary expansion event increased the overall length of the intron beyond the splicing threshold (Fig. S1). This design effectively eliminated the selective pressure in favor of longer, secondary expansions, making it possible to establish the rates and scales of the primary expansion events. URA3 cassettes with 52, 78, 100, 123, or 150 (GAA)n repeats with balanced introns were integrated into the chromosome III, ~1 kb downstream of ARS306 (Fig. 1A). In all cases, the (GAA)n runs were located in the sense strand for transcription, as in the human FXN gene (Campuzano et al., 1996), and in the lagging strand template, i.e. in the orientation causing replication fork stalling (Krasilnikova and Mirkin, 2004).
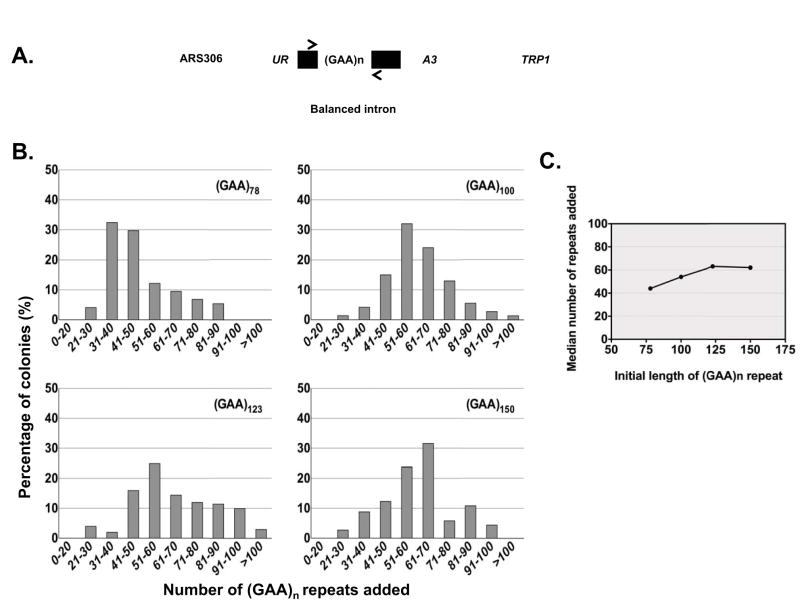
Fig. 1.
Yeast model system to monitor repeat expansions. A. Selectable cassette for the detection of large-scale expansions of the (GAA)n repeats. Arrows show positions of primers used for the PCR analysis of repeat lengths. B. Distributions of repeats added during the expansion of various (GAA)n runs. C. Primary steps of repeat expansion for various (GAA)n runs. Each point represents a median of the corresponding distribution shown above.
Fig. 1B shows the length distributions for expanded repeats originating from different-sized (GAA)n runs. These distributions resulted from the PCR analysis of roughly 100 expansions for each initial repeat length. One can see that each distribution is relatively narrow, continuous and unimodal. A combination of primary and secondary expansion events would result in a much broader, possibly multimodal distributions, which we observed for our original, unbalanced cassettes. Hence we conclude that our data reflect primary expansion events.
Using the Kernell density plot method, the median values of added repeats for various (GAA)n repeats were determined (Fig. 1C). The median expansion step for the original 78, 100, 123 and 150 (GAA)n repeats corresponded to 44, 54, 63 and 62 triplets respectively. Notably, the primary expansion step reached a plateau of ~60 triplets for longer repeats (Fig. 1C). Most importantly, the entire range of the primary expansion steps for different lengths repeats appeared to be quite narrow - 44 to 63 triplets. Okazaki fragment size in S. cerevisae was recently estimated biochemically (Smith and Whitehouse, 2012) and genetically (Waisertreiger et al., 2012) to be ~165 nts. Our expansion steps are in near-perfect agreement with this estimate.
The rates of the primary expansion events for various (GAA)n repeats determined using the method of mutant accumulation (Drake, 1991) are shown in Fig. S1. Note that we were unable to detect expansions for the (GAA)52 repeat, indicating that its expansion rate is less than 10−8 per replication. Thus, large-scale expansions do not occur until the repeat’s length exceeds Okazaki fragment size. For longer repeats, the rates of expansions increased exponentially with their lengths varying from 4.5×10−6 per replication for the (GAA)78 run to 1.4×10−4 per replication for the (GAA)150 run. In other words, doubling the repeat’s size increased its expansion rate by 30-fold. Overall, these data are in accord with what is known for families with Friedreich’s ataxia (Montermini et al., 1997), where the expansions threshold falls between 40 and 60 repeats, while expansion rates drastically increase with the repeat’s length.
Effect of mutations in DNA polymerase genes on repeat expansions
To analyze the roles of different DNA polymerases in repeat expansions, we chose a particular repetitive run, (GAA)100. Expansions were analyzed in yeast strains carrying mutations in the genes encoding for each of the three replicative DNA polymerases: α, δ and ε. These were missense mutations resulting in amino acid changes within the active sites of the corresponding DNA polymerases, directly affecting their efficiency and/or fidelity.
POL1 encodes the catalytic subunit of DNA polymerase in the Polα-primase complex involved in the initiation of the DNA synthesis at origins and at the beginning of Okazaki fragments. Mutations pol1-L868F and pol1-Y869A are a leucine-to-phenylalanine and a tyrosine-to-alanine substitution, respectively, in the enzyme’s catalytic domain. Mutation pol1-L868F leads to a slow growth combined with the strong mutator phenotype (Niimi et al., 2004), constitutively mono-ubiquitinated PCNA (Suzuki et al., 2009), and accumulation of long stretches of single-stranded DNA in the Okazaki initiation zone of the fork (Suzuki et al., 2009). Mutation pol1-Y869A also leads to a slow growth, sensitivity to hydroxyurea (HU) and increases the mutation rate, particularly in the MMR deficient strains (Pavlov et al., 2001).
POL3 encodes the catalytic subunit of Polδ, which is the main replicative and repair DNA polymerase, and the one well-suited for replication of the lagging strand (Kunkel and Burgers, 2008). We studied two mutations, pol3-Y708A and pol3-t, both of which are thought to increase polymerase slippage and/or dissociation from the template. Mutation pol3-Y708A is an alanine substitution for a tyrosine within the binding pocket for the nascent base pair in the polymerase active site (Pavlov et al., 2001; Swan et al., 2009). It slows down yeast growth, increases its HU and methylmethane sulfonate (MMS) sensitivity, and confers a mutator phenotype dependent on the TLS polymerase Polζ (Northam et al., 2006; Pavlov et al., 2001). A temperature-sensitive mutation pol3-t is an asparagine substitution for aspartate 643 in vicinity of polymerase active site (Tran et al., 1999). At semi-permissive temperatures, it impairs yeast growth and grossly elevates deletions between direct repeats (Gordenin et al., 1992; Kokoska et al., 1998).
POL2 encodes the catalytic subunit of Polε, which participates in the leading strand synthesis (Kunkel and Burgers, 2008; Pavlov and Shcherbakova, 2010). Mutation pol2–9 is a methionine-to-isoleucine substitution in the polymerase active site. At restrictive temperatures, this temperature-sensitive mutation impairs DNA synthesis likely due to the mutant polymerase dissociating from its template (Araki et al., 1992).
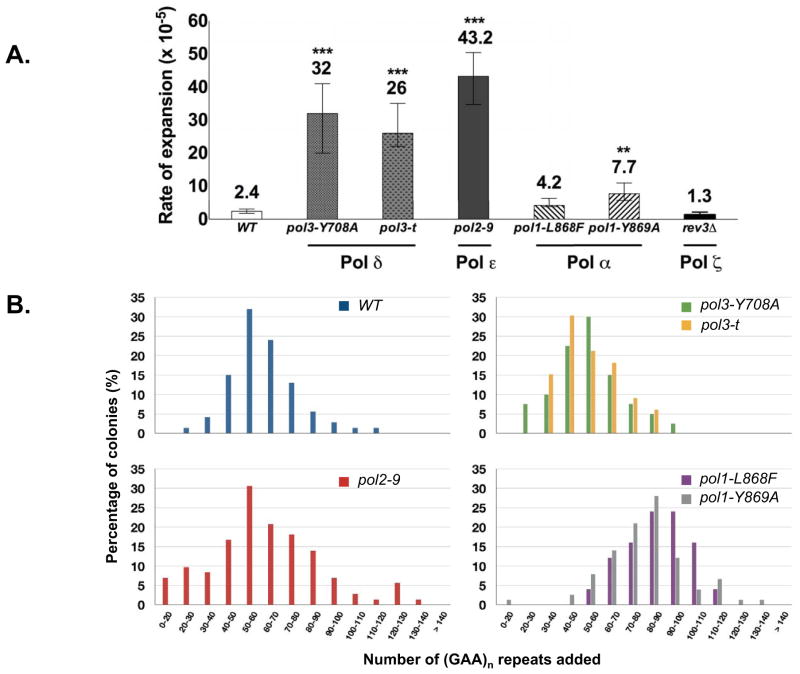
The rates of repeat expansions in these replication mutants are presented in Fig. 2A. One can see that Polε mutation causes the strongest (18-fold) increase in the expansion rate of the (GAA)100 repeat. Both Polδ mutations also strongly (~12-fold) increase repeat expansion rates. At the same time, the Polα mutations resulted in a much more modest (2-to-3-fold) increase.
Fig. 2.
Rates (A) and scales (B) of (GAA)100 repeat expansions in various replication mutants.
We then analyzed the scale of repeat expansions in various DNA polymerase mutant strains. To this end, expansions of the original (GAA)100 run in the wild-type and mutant strains was compared via PCR analysis of the expanded repeats. Fig. 2B shows the length distributions of expanded repeats in various mutants, generated upon the analysis of roughly 75 expansions in each case. The length distributions are indistinguishable in the wild-type, Polδ and Polε mutant strains. The median expansion step in all these cases is about 55 triplets and the interquartile range is 44-to-65 triplets. In contrast, the distributions of expanded repeats in both Polα mutant strains are dramatically different: the median expansion step grew to 80 or 85 triplets (depending on the mutant), while its interquartile range varies from 65-to-90 triplets. Statistical analysis of these distributions using two-sample Kolmogorov–Smirnov test confirms that the differences of the expansion scales between the wild-type and Polα mutants are highly significant (Fig. S2). Summarizing, defects in the major leading and lagging strand DNA polymerases increase the rate, but not the scale of repeat expansions, while mutations in Polα, responsible for the proper initiation of Okazaki fragments, increase the scale, but no so much the rate of expansions for the (GAA)n repeats.
To analyze the pattern of the replication fork progression through our repeat-containing URA3 cassette in these polymerase mutants, we carried out 2-dimensional analysis of yeast replication intermediates (Fig. S3). In the wild-type strain, the replication fork progression is smooth, as is evident from the shape of the Y-arc, with the exception of a stall site coinciding with the location of the (GAA)100 repeat. In the pol2–9 mutant, fork stalling at the repeat is grossly exacerbated; in fact, the stalling is so profound that the Y-arc past the repeat looks severely constricted. In the pol3-Y708A mutant, fork progression is also compromised as is evident from the dramatic decrease in the amount of intermediates in which fork passed the repeat. The latter effect seems also be true for pol1 mutants, albeit it is less pronounced. Note that in all DNA polymerase mutants, the Y-arcs look fuzzier in the first dimension of the electrophoresis, reflecting the accumulation of replication intermediates of similar shapes but different molecular weights caused by replication fork indiscretions.
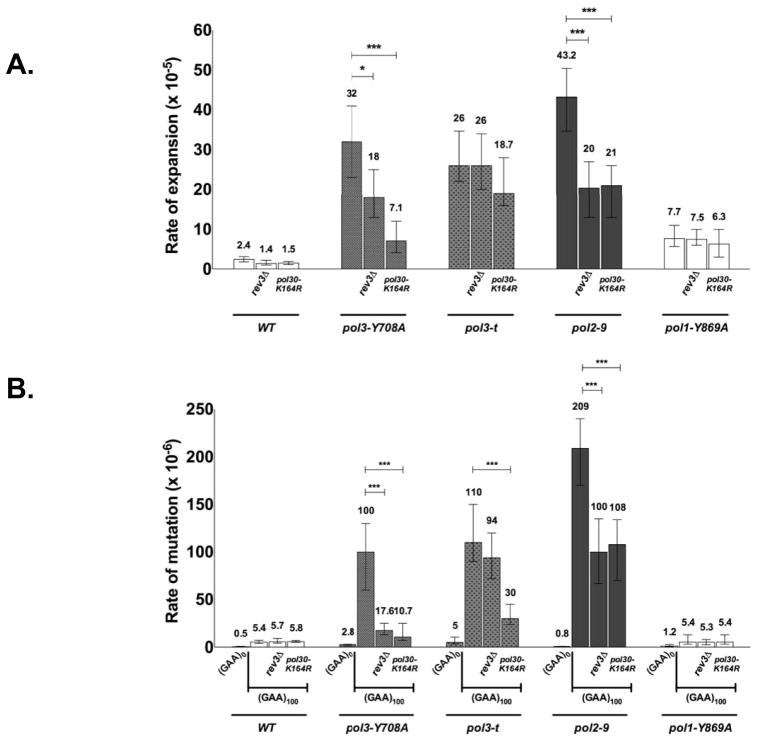
Next we analyzed the effects of two mutations, which affect the DNA damage tolerance, on the repeat expansion rates. REV3 encodes the catalytic subunit of Polζ that mediates TLS allowing the replisome to bypass DNA lesions (Prakash et al., 2005). Inactivation of this polymerase ablates nearly all DNA damage-induced mutagenesis. Recruitment of TLS polymerases to the fork is regulated by monoubiquitylation of PCNA at its lysine 164 residue. Mutation pol30-K164R is a lysine-to-arginine substitution, which abolishes ubiquitin modification thereby preventing a switch from the replicative to TLS polymerase and damage-induced mutagenesis (Ulrich, 2011). One can see that neither Polζ knockout (Fig. 2A), nor the PCNA mutation alone (Fig. 3A) has an effect on repeat expansions.
Fig. 3.
Effect of TLS deficiency on repeat expansions (A) and repeat-induced mutagenesis (B).
As discussed above, the phenotypes of several DNA polymerase mutants studied by us are known to depend on TLS polymerases. We were wondering, therefore, whether elevated rates of repeat expansions in our polymerase mutants could be attributed to the TLS. To address this question, we looked at the expansion rates in double mutants, in which a replicative polymerase defect was combined with either the pol30-K164R mutation or REV3 deletion. The surge in expansions caused by the pol3-Y708A mutation drastically decreased when combined with the pol30-K164R mutation, showing that TLS was indeed involved (Fig. 3A). More specifically, Polζ was at least partially responsible for this expansion surge. In the case of pol2–9, addition of either pol30-K164R or rev3Δ mutations decreased expansion rate by 2-fold, indicating that TLS carried out by Polζ accounts for the half of the expansion spurt. At the same time, TLS does not seem be a factor in pol3-t as well as in the pol1-Y869A mutant, since expansion rates are only mildly and statistically insignificantly decreased in the double mutants, compared to the polymerase mutants alone.
We believe, therefore, that the erratic progression of the replication fork through repeats accentuated in replicative polymerase mutants is a prime factor leading to their expansions. TLS may or may not be involved in the process depending on the properties of the particular DNA polymerase mutation.
Effect of mutations in DNA polymerase genes on repeat-induced mutagenesis in cis
We have previously found that besides expanding, (GAA)n repeats enthused mutations in cis in the body of the URA3 cassette. This repeat-induced mutagenesis (RIM) was detected among 5-FOA-resistant colonies, in which the repeat lengths were unchanged relative to those in the starting URA+ clones (Shishkin et al., 2009). Furthermore, this mutagenesis depended on the repeat’s length: a 2-fold increase the repeat’s length (from 78 to 150 triplets) resulted in a 10-fold increase in the mutation rate (Shishkin et al., 2009). It remained unclear, however, whether expansions and mutations occur in the course of the same process, and what is the nature of the repeat-induced mutations.
We thus studied the effects of the same polymerase mutations (used to study expansions) on RIM. We chose a cassette with the (GAA)100 repeat and calculated the rates of 5-FOA-resistant events, in which the repeat’s length was not substantially changed. We also sequenced the whole URA3 cassette from a significant fraction of such clones to assure that they do have mutations, which turned out to be the case. Our data in Fig. 3B show that in the wild-type strain, the (GAA)100 repeat increases mutagenesis in cis by an order of magnitude compared to the repeat-less situation. The (GAA)100 repeat stimulates mutagenesis 230-fold in the Polε mutant, which is a 23x surge relatively to the wild-type. In Polδ mutants, mutators in their own right, the repeat elevates mutation rates 22-to-36-folds, which is modestly (2-to-4x) higher than in the wild type. Finally, the rate of RIM is unaffected in the Polα mutant. These effects of the polymerase mutations on RIM are in a qualitative agreement with our data on expansions, where Polε mutants had the strongest, Polδ-intermediate and Polα - the weakest effects.
We investigated the role of TLS in RIM (Fig. 3B). In the wild-type genetic background, TLS is not responsible for this mutagenesis, as it is not affected by either a pol30-K164R mutation, or a REV3 deletion. In contrast, RIM is primarily Polζ-dependent in the pol3-Y708A mutant. The situation in the pol2–9 mutant is intermediate between these two extremes. In the pol3-t mutant, RIM is Polζ-independent, but requires ubiquitylation of PCNA, as it is strongly reduced by the pol30-K164R mutation. Again, these effects are qualitatively similar to what we described above for repeat expansions. The pol1-L868F mutation elevated the rate of mutagenesis more than 100-fold even without the repeat, making it unfeasible for studying RIM.
Our preliminary analysis of mutations accumulating during RIM in the wild-type and mutant strains are presented in Fig. S4. Importantly, the mutations accumulate both upstream and downstream of the repeat relative to the direction of replication. They are detected at substantial distances from the repeat: up to 500 bp upstream and 1 kb downstream of the repeat. Finally, while the spectrum of mutations in the wild-type strain includes various base pair changes, the spectrum in the pol3-Y708A mutant shows a strong bias for C-to-G transversions, which is a clear-cut mutational signature of Polζ (Northam et al., 2010; Zhong et al., 2006).
DISCUSSION
We analyzed the roles of replicative and translesion DNA synthesis in repeat-mediated genome instability, including large-scale expansions and RIM. Defects in replicative polymerases strongly affected both processes. The role of TLS was prominent only when a replicative polymerase was compromised. The pol3-Y708A mutant best illustrates the latter situation as repeat expansions and RIM almost completely depend on the TLS machinery.
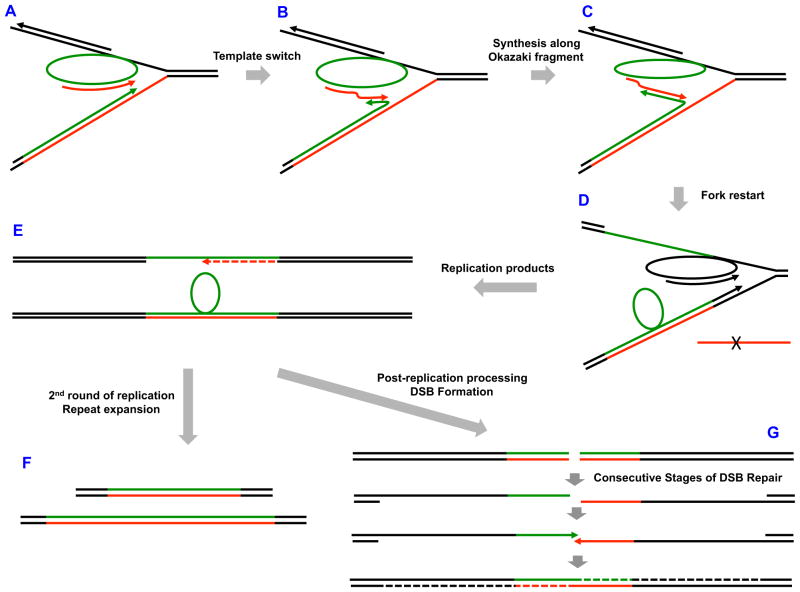
Previous analysis of large-scale repeat expansions in yeast led us to propose the model for the repeat expansions based on template switching (Cherng et al., 2011; Shishkin et al., 2009). It stipulated that while replication fork progresses through a long repetitive run (Fig. 4A), either leading or lagging strand DNA polymerase can inadvertently switch from its template onto the corresponding nascent strand (Fig. 4B). This is followed by DNA polymerization along a nascent strand, which is presented as the leading strand extension in Fig. 4C. When the extended strand returns to its original template, the fork can restart (Fig. 4D). This scenario would result in the accumulation of extra repeats as a loop out in the nascent leading strand (Fig. 4E), which can convert into expansions during the next replication round (Fig. 4F).
Fig. 4.
Model depicting different outcomes of template-switching during replication of an expandable repeat (see text for details). A. Progression of the replication fork through a long repetitive run. B. Switching from template to nascent strand during replication of the repeat. C. Extension of the leading strand along the repetitive Okazaki fragment. D. Leading strand switch to its original template and fork restart. E. Replication products containing repetitive loop-outs and gaps. F. Repeat expansions after the second round of replication. G. Post replicative formation of a double-strand break within the repeat and its repair leading to RIM. Complementary strands of the repeat are shown in green and red; 3′-ends of growing DNA chains are shown by arrows.
An important prediction of this model is that mutations in the replicative DNA polymerases that destabilize the fork, should favor template switching, increasing the rate of expansions. Our data are in excellent agreement with this prediction: both expansion rates and fork stalling at the repeat increased the most by the pol2–9 mutation, followed by the two Polδ mutations, and then Polα mutations.
Another prediction of our model is that template switching becomes much more feasible when the length of a repetitive run exceeds the size of an Okazaki fragment, i.e. when both leading and lagging strand DNA polymerases traverse through the repeat. The median size of an Okazaki fragment in yeast corresponds to ~165 nts (Smith and Whitehouse, 2012). Indeed, we could never detect large-scale expansions for the (GAA)52 run, which is shorter than an average Okazaki fragment.
Our model postulates the existence of a link between the expansion step and the size of an Okazaki fragment (Fig. 4C). Thus, mutations affecting the size of an Okazaki fragment in yeast should affect the expansion step accordingly. In the wild-type yeast, the primary expansion step for various (GAA)n repeats appeared to be 44-to-63 triplets, which is close to an average Okazaki fragment size. Remarkably, the primary expansion step got much bigger, 80-to-85 triplets in Polα mutants. Both the activity and fidelity of Polα are compromised in these mutants, the net result of which is delays in accumulation of RNA/DNA hybrid primers and concomitant increase of the single-stranded DNA region at the fork (Okazaki initiation zone) together with the size of an Okazaki fragment. For pol1-L868F mutant an increase in size of an Okazaki initiation zone over the wild-type has indeed been demonstrated (Suzuki et al., 2009). We propose, therefore, that a larger expansion step detected in Polα mutants is likely due to the increased average size of an Okazaki fragment in those mutants in agreement with our predictions.
Overall, our data are in excellent agreement with the template-switch model for repeat expansions. Note, however, that the finer mechanisms of template switching remain to be elucidated. For example, pol30-K164R mutation in the PCNA appears to have no effect on repeat expansions in the wild-type yeast. This is intriguing, since polyubiquitination of K164 in the PCNA is important at least for one form of template switching driven by the Rad5 protein (Ulrich, 2011). We found that TLS leads to an increased rate of expansions in some strains with defective replicative DNA polymerases. The fork progression impairment in mutants of replicative DNA polymerases is known to recruit Pol ζ (Northam et al., 2006; Northam et al., 2010; Suzuki et al., 2009). We speculate, therefore, that the template switching could be easier, when the primer ends are handled by Pol ζ. That said, expansions in the pol3-t strain do not involve translesion DNA synthesis (Fig. 3). One explanation could be that the altered catalytic subunit of DNA polymerase δ acquires some properties of polymerase ζ that promote template switching.
Besides expanding, (GAA)n repeats induced mutagenesis at a distance in cis (RIM), a phenomenon which also depended on the repeat’s length (Shishkin et al., 2009). Here we show that RIM is detected up to 1.0 kb away from the repeat, and this is probably an underestimation attributable to the size of our selectable URA3 cassette. Studying the role of polymerase mutations in RIM shed light on its mechanisms. The effects of the polymerase mutations on mutagenesis mirrored their effects on expansions: the rate of RIM was elevated the most in the Polε mutant, followed by Polδ mutants, and then Polα mutants. Furthermore, RIM did not depend on translesion DNA synthesis in the wild-type genetic setting, but became Polζ dependent in pol3-Y708A mutant, again similar to the expansion data. We conclude, therefore, that RIM is results from the same replication problem that leads to repeat expansions. That said, for the very long (GAA)n repeat (n ≥ 230), RIM becomes independent of DNA replication (Tom Petes, personal communication).
As shown in Fig. 4E, two replication products result from template switching within the repeat: one contains a large (Okazaki fragment-size) repetitive loop-out in the nascent leading strand, while another, unless filled, has a repetitive single-stranded gap within the repeat. These single-stranded segments may also fold onto the surrounding repetitive duplex forming triplex DNA structures (Krasilnikova and Mirkin, 2004). We hypothesize that post-replicative processing of these complex structures can produce double-strand breaks. We have, in fact, previously demonstrated the formation of double-stranded breaks at (GAA)n repeats in the course of DNA replication (Kim et al., 2008). DSB repair, which involves extensive end-resection, strand reannealing and repair DNA synthesis, is known to be error-prone (Hicks et al., 2010). We believe that this scenario could be responsible for RIM (Fig. 4G). It nicely explains two groups of our data: (i) the remarkable similarity in the effects of polymerase mutations on expansions and mutagenesis and (ii) the fact that we observe RIM both upstream and downstream of the repeat with regard to the replication direction. While this idea seems plausible, other mechanisms accounting for RIM cannot be ruled out and warrant further investigation.
MATERIALS AND METHODS
Strains and Plasmids
S. cerevisiae CH1585 strain (MATa leu2-Δ1, trp1-Δ63, ura3–52, his3–200) was used to create all mutants employed in this study. Mutations in essential replication genes were introduced by the POP-IN/POP-OUT approach as described in supplemental methods.
Fluctuation assay and rates
Fluctuation assays were carried out as previously described (Shishkin et al., 2009).
2-dimensional electrophoretic analysis of replication intermediates
Replication analysis was carried out as previously described in (Shishkin et al., 2009).
Supplementary Material
HIGHLIGHTS.
Elementary step of repeat expansion corresponds to size of an Okazaki fragment
Mutated DNA polymerase á leads to an increase in expansion step
Mutated DNA polymerases ä and å lead to elevated expansion rates
Repeats induce mutagenesis, which is further elevated in polymerase mutants
Acknowledgments
We are grateful to Tom Petes and members of Tufts RRR seminar for many useful comments, suggestions, and discussions. Statistical consulting was provided by Durwood Marshall at Tufts University Information Techology, Research and Geospatial Technology Services. This study was supported by the NIH grants GM60987 to S.M.M, CA129925 to Y.I.P and ES015869 to P.V.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araki H, Ropp PA, Johnson AL, Johnston LH, Morrison A, Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992;11:733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley C, Jr, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan L, Matera R, Sarkar PS, Ashizawa T, Freudenreich CH, Mirkin SM. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci USA. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Pearson CE. Replication fork dynamics and dynamic mutations: The fork-shift model of repeat instability. Trends Genet. 2005;21:272–280. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin DA, Malkova AL, Peterzen A, Kulikov VN, Pavlov YI, Perkins E, Resnick MA. Transposon Tn5 excision in yeast: influence of DNA polymerases alpha, delta, and epsilon and repair genes. Proc Natl Acad Sci USA. 1992;89:3785–3789. doi: 10.1073/pnas.89.9.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Narayanan V, Mieczkowski PA, Petes TD, Krasilnikova MM, Mirkin SM, Lobachev KS. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska RJ, Stefanovic L, Tran HT, Resnick MA, Gordenin DA, Petes TD. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad 27) and DNA polymerase delta (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184:27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork-20 years later. Mutat Res. 2010;685:45–53. doi: 10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Schweitzer JK, Livingston DM. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics. 1999;152:953–963. doi: 10.1093/genetics/152.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, Krasilnikova MM, Narayanan V, Lobachev KS, Mirkin SM. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Niimi A, Limsirichaikul S, Tomida S, Miao Huang Q, Izuta S, Usukura J, Itoh Y, Hishida T, Akashi T, et al. PCNA mono-ubiquitination and activation of translesion DNA polymerases by DNA polymerase alpha. J Biochem. 2009;146:13–21. doi: 10.1093/jb/mvp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Degtyareva NP, Gordenin DA, Resnick MA. Genetic factors affecting the impact of DNA polymerase delta proofreading activity on mutation avoidance in yeast. Genetics. 1999;152:47–59. doi: 10.1093/genetics/152.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585:2861–2867. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Waissertreiger ISR, Liston VG, Menezes MR, Kim HM, Lobachev KS, Stepchenkova EI, Tahirov TH, Rogozin IB, Pavlov YI. Modulation of mutagenesis in eukaryotes by replication fork dynamics and quality of nucleotide pools. Environ Mol Mutagen. 2012 doi: 10.1002/em.21735. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.