Abstract
In a cross-sectional study, we assessed effects of calcineurin inhibitor (CNI) or rapamycin on T-regulatory (Treg) cells from children with stable liver (n=53) or kidney (n=9) allografts several years post-transplant. We analyzed Treg number, phenotype, suppressive function, and methylation at the Treg-specific demethylation region (TSDR) using Tregs and peripheral blood mononuclear cells. 48 patients received CNI (39 as monotherapy) and 12 patients received rapamycin (9 as monotherapy). Treg numbers diminished over time on either regimen, but reached significance only with CNI (r=−0.424, p=0.017). CNI levels inversely correlated with Treg number (r=−0.371, p=0.026), and positively correlated with CD127+ expression by Tregs (r=0.437, p=0.023). Patients with CNI levels >3.6 ng/ml had weaker Treg function than those with levels <3.6 ng/ml, whereas rapamycin therapy positively correlated with Treg numbers (r=0.628, p=0.029) and their expression of CTLA4 (r=0.726, p=0.041). Overall, CTLA4 expression, TSDR demethylation and an absence of CD127 were important for Treg suppressive function. We conclude that rapamycin has beneficial effects on Treg biology, whereas long-term and high dose CNI use may impair Treg number, function and phenotype, potentially acting as a barrier to attaining host hyporesponsiveness to an allograft.
Keywords: Immunosuppression, immunoregulation, clinical transplantation
Introduction
Though essential for allograft survival in most recipients, immunosuppression can cause opportunistic infections, malignancies, cardiovascular diseases and nephrotoxicity (1). Despite excellent one-year graft survival rates, chronic allograft dysfunction persists, perhaps reflecting, at least in part, impairment of FOXP3+ Tregs (2). CNI use, central to most immunosuppressive regimens (3), blocks IL-2 production and may adversely affect Tregs (4–6). Experimentally, rapamycin enhances de novo conversion, proliferation and suppressive function of Tregs (5, 6), and may increase Treg numbers clinically (7, 8), though ongoing deterioration in renal function occurred in some (9) but not other series (10). Given this uncertainty, we developed a composite approach to simultaneously analyze 4 key Treg parameters: numbers, phenotype, suppressive function under highly standardized conditions, and FOXP3 epigenetic status. We show the clinical utility of our approach by determining the effects of CNI or rapamycin therapy in pediatric transplant (Tx) recipients.
Materials and Methods
Healthy normal donors
Healthy volunteer donors (n=13, 7 males) provided peripheral blood mononuclear cells (PBMC) via the University of Pennsylvania Human Immunology Core.
Patients
We studied 62 patients with stable liver (n=53) or kidney (n=9) allografts (Tables 1–2). CNI (tacrolimus) blood levels were 0.7–7.4 ng/ml (mean=3.61±0.21 ng/ml), and rapamycin levels were 1–9.1 ng/ml (mean=4.1±0.69 ng/ml). Patients were similarly distributed for gender, race, time post-Tx, donor type (living or deceased), proportions receiving liver or kidney allografts, primary diseases leading to Tx, and concomitant medications, but varied in ages (CNI=13.1±0.5 and Rapa=10.7±0.9 years, p=0.03). As ages were not correlated with data except time post-Tx (r=0.561, p<0.001), rapamycin and CNI groups were suitable for comparisons.
Table 1.
Clinical and demographic data of enrolled patients
| Sitea | Study ID | Sex | Age yrs | Yrs post-Tx | Graft | Donor type | No. acute rejections | Immune suppression (IS)b | Primary disease |
|---|---|---|---|---|---|---|---|---|---|
| CHOP | 01-01L | M | 6.2 | 5.3 | liver | Deceased | 1 | Rapa | Biliary atresia |
| CHOP | 01-02L | M | 13.3 | 1.8 | liver | Deceased | 0 | CNI | Biliary atresia |
| CHOP | 01-03L | F | 7.5 | 6.8 | liver | Living | 0 | No IS | Biliary atresia |
| CHOP | 01-04L | F | 15.4 | 14.3 | liver | Deceased | 0 | CNI | Biliary atresia |
| CHOP | 01-05L | F | 17.6 | 6.3 | liver | Deceased | 0 | CNI | Biliary atresia |
| CHOP | 01-06L | F | 11.5 | 10.9 | liver | Deceased | 3 | CNI | Biliary atresia |
| CHOP | 01-07L | M | 9.8 | 9.5 | liver | Deceased | 1 | CNI | Biliary atresia |
| CHOP | 01-08L | M | 6.5 | 5.8 | liver | Deceased | 0 | CNI | Biliary atresia |
| CHOP | 01-09L | F | 10.7 | 9.8 | liver | Living | 0 | Rapa | Biliary atresia |
| CHOP | 01-10L | F | 9.2 | 7.0 | liver | Deceased | 0 | Rapa | Biliary atresia |
| CHOP | 01-12L | M | 16.9 | 12.9 | liver | Deceased | 1 | CNI | Indeterminate |
| HSC | 02-01L | F | 15.6 | 13.6 | liver | Deceased | 6 | CNI | Indeterminate |
| HSC | 02-02L | M | 15.8 | 10.2 | liver | Deceased | 4 | CNI | Tyrosinemia |
| HSC | 02-03L | M | 16.3 | 8.2 | liver | Deceased | 1 | CNI | Urea cycle disorder |
| HSC | 02-04L | F | 12.4 | 5.1 | liver | Deceased | 1 | CNI | Alagille syndrome |
| HSC | 02-05L | F | 6.2 | 6.1 | liver | Living | 0 | Rapa | Hepatoblastoma |
| HSC | 02-06L | M | 7.3 | 5.8 | liver | Living | 2 | CNI | Biliary atresia |
| HSC | 02-07L | F | 13.0 | 12.8 | liver | Deceased | 0 | CNI | Biliary atresia |
| HSC | 02-08L | F | 17.0 | 15.9 | liver | Deceased | 0 | CNI | Biliary atresia |
| HSC | 02-09L | F | 13.4 | 11.1 | liver | Deceased | 0 | CNI | Biliary atresia |
| HSC | 02-10L | M | 8.8 | 5.9 | liver | Deceased | 0 | CNI | Hepatoblastoma |
| HSC | 02-11L | M | 10.3 | 10.0 | liver | Living | 0 | Rapa | Bile acid synthesis defect |
| HSC | 02-12L | M | 7.1 | 4.3 | liver | Deceased | 3 | Rapa | Biliary atresia |
| HSC | 02-13L | M | 14.5 | 8.8 | liver | Deceased | 0 | CNI | Biliary atresia |
| HSC | 02-14L | F | 14.2 | 13.0 | liver | Living | 2 | CNI | Indeterminate |
| HSC | 02-15L | F | 16.4 | 14.8 | liver | Living | 0 | CNI | Biliary atresia |
| HSC | 02-16L | M | 10.8 | 6.1 | liver | Living | 0 | CNI | Urea cycle disorder |
| HSC | 02-17L | F | 10.0 | 9.2 | liver | Living | 0 | CNI | Biliary atresia |
| HSC | 02-18L | M | 16.5 | 16.2 | liver | Deceased | 0 | CNI(Cs)* | Biliary atresia |
| HSC | 02-19L | F | 12.3 | 10.6 | liver | Living | 0 | CNI | Biliary atresia |
| CCHMC | 03-01L | M | 12.3 | 12.0 | liver | Deceased | 1 | Rapa | Biliary atresia |
| CCHMC | 03-02L | F | 10.2 | 10.1 | liver | Deceased | 0 | CNI | Hemangioendothelioma |
| CCHMC | 03-03L | F | 10.5 | 9.8 | liver | Living | 0 | CNI | Biliary atresia |
| CCHMC | 03-04L | M | 13.4 | 13.2 | liver | Deceased | 1 | CNI | Cryptogenic cirrhosis |
| CCHMC | 03-05L | F | 16.2 | 14.6 | liver | Living | 2 | CNI | Biliary atresia |
| CCHMC | 03-06L | F | 15.9 | 15.1 | liver | Deceased | 0 | CNI(Cs)* | Biliary atresia |
| CCHMC | 03-07L | F | 8.7 | 6.3 | liver | Living | 1 | CNI | Biliary atresia |
| CCHMC | 03-08L | F | 10.6 | 4 | liver | Deceased | 0 | CNI | Alpha-1-antitrypsin |
| CCHMC | 03-09L | M | 12.7 | 12.1 | liver | Deceased | 0 | CNI | Biliary atresia, Hepatoblastoma |
| CCHMC | 03-10L | M | 9.6 | 7.1 | liver | Deceased | 0 | CNI | Biliary atresia |
| CCHMC | 03-11L | F | 13.9 | 5 | liver | Deceased | 1 | CNI+MMF/MPA | Biliary atresia |
| CCHMC | 03-12L | M | 15.4 | 5.8 | liver | Deceased | 1 | CNI+MMF/MPA | Other tumor |
| CCHMC | 03-13L | F | 9.3 | 5.9 | liver | Deceased | 1 | CNI | Hepatoblastoma |
| CCHMC | 03-14L | F | 11.3 | 10.6 | liver | Deceased | 0 | CNI | Biliary atresia |
| CCHMC | 03-15L | M | 10.8 | 7 | liver | Deceased | 0 | CNI | Glycogen storage disease |
| CCHMC | 03-16L | M | 17.2 | 13 | liver | Deceased | 4 | CNI+MMF/MPA | Biliary atresia |
| CCHMC | 03-17L | M | 9.8 | 8 | liver | Deceased | 0 | Rapa | Tyrosinemia |
| CCHMC | 03-18L | M | 10.7 | 10 | liver | Deceased | 2 | CNI | Neonatal hepatitis, MDR 3 (PFIC) disease |
| CCHMC | 03-19L | F | 10.0 | 9.1 | liver | Deceased | 0 | CNI | Biliary atresia |
| CCHMC | 03-20L | F | 17.8 | 16.1 | liver | Deceased | 0 | Rapa+P | Autoimmune hepatitis |
| CCHMC | 03-21L | M | 10.9 | 7 | liver | Living | 0 | Rapa | Other tumor |
| CCHMC | 03-22L | M | 17.0 | 15.2 | liver | Deceased | 1 | No IS | Alagille syndrome |
| CCHMC | 03-25L | M | 16.5 | 13.3 | liver | Deceased | 1 | CNI | Hepatoblastoma |
| CHOP | 01-01K | M | 13.4 | 8.2 | kidney | Living | 0 | Rapa+P | Cortical necrosis |
| CHOP | 01-02K | F | 6.1 | 4.2 | kidney | Living | 0 | CNI | Congenital nephrotic syndrome |
| CHOP | 01-03K | M | 15.9 | 6.0 | kidney | Deceased | 0 | CNI,MMF/MPA, P | Posterior urethral valves |
| CHOP | 01-04K | F | 17.9 | 10.3 | kidney | Living | 0 | CNI+Aza, P | Interstitial nephritis |
| CHOP | 01-05K | M | 17.3 | 6.6 | kidney | Living | 0 | CNI+Aza, P | Interstitial nephritis |
| CHOP | 01-06K | M | 13.0 | 9.8 | kidney | Living | 0 | CNI,MMF/MPA, P | Interstitial nephritis |
| CHOP | 01-07K | M | 14.8 | 5.5 | kidney | Living | 0 | CNI+Aza, P | FSGS |
| CCHMC | 03-01K | F | 10.5 | 8.8 | kidney | Living | 0 | Rapa+MMF/MPA | Congenital nephrotic syndrome |
| CCHMC | 03-02K | M | 14.2 | 3.8 | kidney | Living | 2 | Rapa,MMF/MPA, P | Medullary cystic kidney disease |
| Total: | |||||||||
| N = 62: 18 CHOP 19 HSC 25 CCHMC |
32 M 30 F |
Mean age 12.5±0.4 |
Yrs post-Tx 9.3±0.5 |
Liver 53 Kidney 9 |
Deceased 40 Living 22 |
39 of 62 with no AR |
39 CNI monotherapy (2 cyclosporin, 37 tacrolimus) 9 Rapa monotherapy 8 combined therapy with CNI 4 combined therapy with Rapa 2 no immunosuppression |
||
Sites: CCHMC, Cincinnati Children's Hospital Medical Center; CHOP, Children's Hospital of Philadelphia; HSC, Hospital for Sick Children.
Immunosuppression: Aza, azathioprine; CNI, calcineurin inhibitors when CNI(Cs) is cyclosporin, all other CNI patients received tacrolimus; P, prednisone; Rapa, rapamycin.
Table 2.
The enrolment criteria of current study
| Inclusion criteria | Exclusion criteria |
|---|---|
|
|
No patient received intravenous immunoglobulin, rituximab or cytogam, or had undergone plasmapheresis.
Cellular studies
PBMC were isolated from 32 ml of blood and shipped overnight at 4 °C to Children's Hospital of Philadelphia (CHOP). For standardization, CHOP samples were also kept at 4°C overnight. Cell numbers and viability were determined, and Tregs isolated using magnetic beads (CD4+CD25+Treg isolation kit, Miltenyi-Biotec). Aliquots of Tregs were immediately tested in suppression assays (11), and frozen for flow cytometry and DNA isolation. To ensure suppression assays reflected effects of immunosuppression on Tregs and not conventional T effector (Teff) cells, the same healthy donor CFSE-labeled T-cells were used in each assay. In preliminary studies, these cells were tested with different healthy donor Tregs to ensure absence of variability due to HLA differences (data not shown). In parallel, we assessed proliferation of patient CFSE-labeled CD3-stimulated CD4+CD25− Teff cells. Aliquots of isolated Tregs were stained for CD4, CD25, CD127, CTLA4, FOXP3 (PCH101, eBioscience) and LIVE/DEAD Kit (Invitrogen). PBMC stimulated overnight with CD3/CD28 mAb-coated beads 1:1 (Invitrogen) and 100 U/ml IL-2 (Roche) were stained with CD4, CD25, CD31, CD39, Helios and FOXP3 mAbs. Staining for FOXP3, CTLA4 and Helios were performed using FOXP3 Fix/Perm kit (eBioscience). FOXP3 TSDR-methylation assays used DNA from 60–100×103 Tregs or 100–400×103 PBMC (12). TSDR-methylation was evaluated separately by gender, but also percentages of demethylated Tregs in girls were calculated as %Tregs = (100-HM)*2, where HM is %TSDR-hypermethylation.
Statistical analysis
Data are shown as mean ± SEM. For continuous variables with normal distribution, statistical significance was assessed by Student's t-test to compare two groups, and Pearson correlation assay used to test correlations, unless specified. In some cases, we performed partial correlation assays with FOXP3 as a controlling variable. For testing association in two-way tables, we used chi-square or Fisher exact test. To test correlations between variables not normally distributed, we used nonparametric Spearman test. To standardize results of each suppression assay, we calculated area-under-curve as described (11) and shown (Figure S1). A two-tailed p value of <0.05 was considered statistically significant. For all calculations, all available data without any type of variable reductions (outliers etc.) or adjustments were used.
Results
Transplant recipients often display altered Treg phenotypes
Patient Tregs isolated using CD4+CD25+ beads had consistently high purity (85.8±5%), but unlike in healthy donors, had variable FOXP3+ expression (Figure S2A). To assess contamination by CD4+CD25+CD127+ activated Teffs, we analyzed FOXP3+ and FOXP3− cells within CD4+CD25+ isolates. About 50% of patients had FOXP3+ cells mostly within the CD25highCD127− subset, and FOXP3− cells occurred mostly within the CD25−/lowCD127+ subset (Figure S2B, patients a, d), like in normal donors. However, additional unusual distributions of FOXP3+ and FOXP3− cells were noted, including increases in CD25+CD127+FOXP3+ cells (Figure S2B, patients b & c) or CD25+/highCD127−/lowFOXP3− cells (Figure S2B, patients e & f). In healthy donor Tregs, CD25high or CD25highCD127− gating strategies showed 90–96% FOXP3+ cells, but in Tx recipients with unusual FOXP3 distributions this led to only 75–80% FOXP3+ cells and extremely low yields (20%–50% of yields with magnetic beads, data not shown). Given limited blood volumes available from pediatric recipients, sorting of CD4+CD25+CD127− cells would not generate enough Tregs for functional and methylation studies, and would provide the same variable FOXP3 purity as achieved by bead isolation. We therefore stained aliquots of each isolated Treg sample for CD4, CD25, FOXP3, CD127 and CTLA4, and adjust our data according to FOXP3 expression, as shown in figure legends and text. Thus, we calculated numbers of CD4+CD25+ isolated Tregs and CD4+CD25+FOXP3+ Tregs and described both statistics. When two groups of Tregs were compared (e.g. CNI & rapamycin), we tested differences in FOXP3 expression. We also evaluated correlations of various markers in isolated Tregs with suppressive function, FOXP3 epigenetic status and clinical variables, and used CD4+CD25+FOXP3+ phenotype to exclude effects of varying FOXP3 expression. During Treg isolation, calculation of viable CD4+ cells and CD4+CD25+ Tregs collected from 1×106 PBMC allowed us to evaluate Treg numbers with regard to number of isolated CD4+CD25+ Tregs, number of CD4+CD25+FOXP3+ Tregs, and number of TSDR-demethylated Tregs. TSDR-demethylation data was adjusted by gender as described in Methods, but also analyzed separately for boys and girls.
Treg numbers decreased over time on CNI immunosuppression
The number of CD4+CD25+ Tregs and CD4+CD25+FOXP3+ Tregs inversely correlated with years post-Tx (r=–0.357, p=0.02 for CD4+CD25+ Tregs in liver recipients). If rapamycin-treated patients were excluded, this correlation increased (r=−0.424, p=0.017). There was no significant correlation for rapamycin (r=−0.30, p=0.43), and no correlation between patient age and Treg number, indicating an effect specific for chronic CNI use. These data suggest continuous CNI therapy is linked with progressive decline in Treg numbers.
CNI but not rapamycin use led to impaired Treg numbers and function
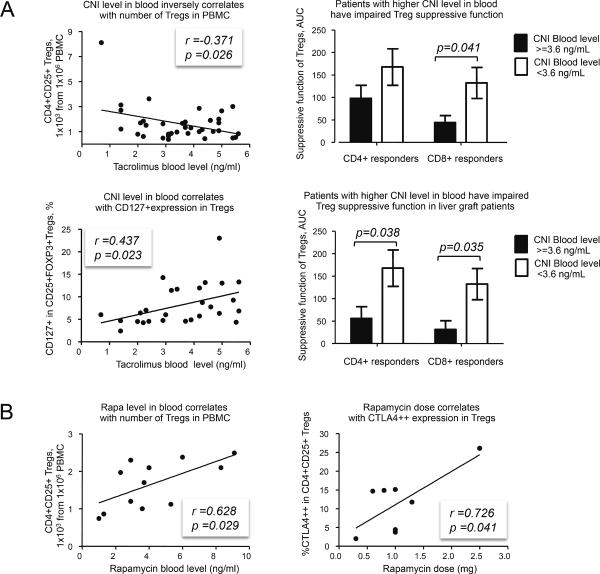
CNI blood levels correlated inversely with the number of Tregs (and FOXP3+ Tregs) in PBMC, numbers of Tregs in the CD4+ subset, and numbers of TSDR-demethylated Tregs in PBMC (Table 3, Figure 1A). Additionally, CNI blood levels correlated with CD127+ expression in CD25+FOXP3+ Tregs (Figure 1A). Two groups of CNI patients defined by mean trough CNI levels (< or > 3.6 ng/ml) had differential Treg function, being impaired in those with CNI >3.6 ng/ml. This was observed in all patients, in liver recipients alone, and in CNI monotherapy patients (Figure 1A and not shown), excluding possible additional effects of combined therapies. CNI “low” and “high” groups had no differences in other clinical and demographic data (not shown). In contrast to CNI, rapamycin blood levels were positively correlated with Treg numbers (Figure 1B) and high CTLA4++ expression in isolated Treg cells (Figure 1B). Patients on CNI had significantly higher CD127+ expression in CD4+CD25+FOXP3+ Tregs (8.31±0.85% vs. 4.95±0.47%, p=0.046 for all patients, 7.80±0.74% vs. 4.99±0.54%, p=0.048 for patients on monotherapies). To test effects of high CNI or rapamycin concentrations, we compared patients with CNI blood levels >3.6 ng/ml with patients having rapamycin levels >4.1 ng/ml and found that CNI patients had fewer Tregs in the CD4+ subset (1.73±0.15% in CNI vs. 2.96±0.71% in Rapamycin, p=0.011). These data indicate differential effects of long-term CNI and rapamycin therapy on Treg numbers and suppressive function.
Table 3.
Correlations of CNI blood level (ng/ml) with Treg numbers
| Group | Statistical factors | Treg number calculated as: | |||
|---|---|---|---|---|---|
| CD4+CD25+ cells in PBMC | CD4+CD25+FOXP3+ cells in PBMC | CD4+CD25+ cells in CD4+ subset | TSDR-demethylated CD4+CD25+ cells in PBMC | ||
| All patients on CNI | r, Pearson p (2 tails) No. cases |
−0.371 0.026 36 |
−0.402 0.046 25 |
−0.468 0.003 36 |
−0.437 0.023 27 |
| Liver Tx patients on CNI | r, Pearson p (2 tails) No. cases |
−0.441 0.015 30 |
−0.571 0.009 20 |
−0.498 0.005 30 |
−0.657 0.006 16 |
| Patients on CNI monotherapy | r, Pearson p (2 tails) No. cases |
−0.409 0.028 29 |
−0.505 0.028 19 |
−0.447 0.015 29 |
−0.612 0.012 16 |
Figure 1. Effects of CNI and Rapamycin on Tregs.
(A) Left column: CNI level in blood inversely correlates with CD4+CD25+Tregs number in PBMC (36 patients) and positively correlates with CD127 expression in CD4+CD25+FOXP3+ Tregs (27 patients). Right column: all patients (top) or liver graft patients (bottom) were divided into 2 groups according to mean of CNI blood level, >3.6 ng/ml and <3.6 ng/ml. Patients with higher CNI level showed impaired Treg ability to suppress divisions of CD4+ or CD8+ healthy donor responder cells (20 patients). Suppression was calculated as AUC (see Material and Methods and Figure S1). Compared groups had no differences in FOXP3 expression. (B) Rapamycin therapy positively correlates with CD4+CD25+ Treg number in PBMC (12 patients) and with CTLA4++ expression in Tregs (8 patients). Partial correlation assay with FOXP3 as potential controlling variable showed that rapamycin use still correlated with CTLA4++ expression in Tregs when effects of FOXP3 were removed (r=0.866, p=0.012).
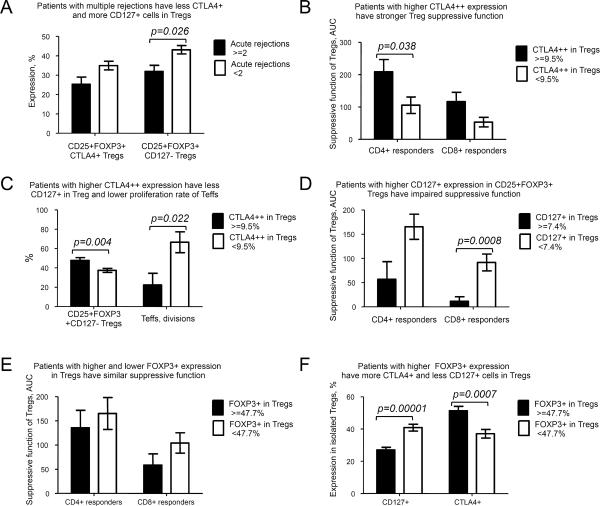
Patients with histories of multiple rejections differ in CTLA4 and CD127 expression in Tregs
In liver Tx patients, tacrolimus blood levels correlated with number of biopsy-proven acute rejection episodes (r=0.327, p=0.042 for all CNI patients; r=0.351, p=0.033 for CNI monotherapy, Spearman tests), but neither rapamycin blood level nor dosing had such correlations. Given CNI-rejection data, we tested if patients with multiple rejections differed from others in any Treg-associated markers. We found CNI patients with multiple rejections (>=2) had more CD127 and tended to have less CTLA4 expression in Tregs compared with 0–1 rejections group (Figure 2A for all patients and not shown for liver recipients). The number of acute rejections correlated with in vitro proliferative ability of patient CD4+CD25− Teffs (r=0,798, p=0.03, Spearman), suggesting a persisting greater capacity for T cell proliferation in patients with histories of multiple rejections.
Figure 2. Correlations of CTLA4, CD127 and FOXP3 with Treg suppression and clinical variables.
(A) 28 patients receiving CNI were divided into 2 groups: those with a history of multiple rejections (≥2) and those with less than 2 acute rejection episodes. Patients who had experienced multiple rejections had less CTLA4+ (p=0.07, not significant) and more CD127 in FOXP3+ Tregs. (B), (C) Patients were divided into 2 groups: with high CTLA++ expression in isolated Tregs (≥9.5%) and decreased CTLA4++ expression (<9.5%). Patients with higher CTLA4++ expression have (B) stronger Treg suppressive function (p=0.038 for CD4+ responders and not significant p=0.073 for CD8+ responders, 21 patients) and (C) lower CD127+ expression in Tregs (37 patients) with lower ex vivo proliferative abilities of patient CD4+CD25− responders (13 patients). (D) Patients with higher CD127+ expression in CD4+CD25+FOXP3+Tregs (≥7.4%) have impaired suppressive function in comparison with those who have less CD127 in Tregs (not significant p=0.066 for CD4+ responders and p=0.0008 for CD8+ responders, 21 patients). Suppression of both CD4+ and CD8+ responders strongly correlated with each other in all tested groups (r=0.8, p<0.0001 for all patients) and CD127+ expression in Tregs inversely correlated with CTLA4 expression in all tested groups (r=−0.552, p<0.001 for CTLA4+, r=−0.381, p=0.019 for CTLA4++ for all patients). (E) Patients with higher (≥47.7%) or lower FOXP3 expression in isolated Tregs did not differ with regard to suppressive function (136 AUC vs. 165 AUC, p=0.55 for CD4+ responders, 58 AUC vs. 104 AUC for CD8+ responders, p=0.16, 21 patients), but (F) differed in CD127 and CTLA4 expression, 37 patients.
CTLA4 and absence of CD127, but not FOXP3 expression are important for Tregs function
To assess whether CTLA4 expression is important for Treg suppression in Tx recipients, we divided patients into 2 groups based upon mean value of CTLA4++ expression (< or > 9.5% CTLA4++ Treg). Tregs in the group with higher CTLA4++ expression had better suppression (Figure 2B), lower CD127 expression, and lower rates of Teff proliferation in vitro (Figure 2C). The same differences were observed in CNI-treated patients, indicating this was not due to rapamycin-CNI differences. The suppressive function of patient Tregs correlated with CTLA4++ expression (r=0.462, p=0.03 for CD4+ responders). Low CD127 expression appeared important for Treg suppression since CD25+FOXP3+ Tregs with higher CD127+ expression were weaker suppressors (Figure 2D), and this was true when CNI-treated liver recipients were analyzed separately (not shown). While FOXP3 expression correlated with CTLA4 and absence of CD127 expression in all tested groups (not shown), and despite variable FOXP3+ expression in Tregs after isolation, FOXP3+ “high” and “low” Tregs (< or > mean 47.7%) had comparable suppressive function (Fig. 2E), despite differences in CTLA4 and CD127 expression (Figure 2F). FOXP3 expression in Tregs did not correlate with suppressive function for any group (all patients, liver grafts, boys, girls, patients on CNI) (Figure S3 and not shown). Collectively, these data suggest that CD127− and CTLA4+ are likely surrogate markers of functional FOXP3 protein and therefore should be used in conjunction with FOXP3 evaluation by flow cytometry.
Analysis of FOXP3 TSDR-demethylation alone in PBMC is not an accurate guide to Treg numbers
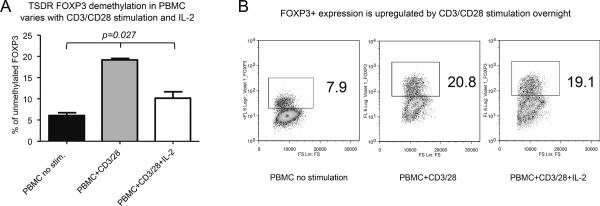
TSDR-demethylation within PBMC or tissue samples is reportedly the most specific indicator of “true” Tregs (13). We therefore tested TSDR-demethylation correlated with the number of Tregs post-Tx, calculated as CD4+CD25+ cells, CD4+CD25+FOXP3+ cells, or as TSDR-demethylated CD4+CD25+ cells. We found no correlations between TSDR-demethylation and any Treg isolate in all patients, or in boys or in girls (Table 4). Moreover, TSDR-demethylation had no correlations with numbers of CD127− or CTLA4+ cells (not shown). To assess whether TSDR-demethylation in PBMC was affected by T cell activation conditions, we stimulated healthy donor PBMC with CD3/CD28 mAbs ± IL-2. CD4+ T cells significantly upregulated FOXP3 expression and increased TSDR-demethylation, and addition of IL-2 reduced TSDR-demethylation without affecting FOXP3 upregulation (Figure 3). Importantly, CD4+FOXP3+ stimulated cells were CD25+ and CD127−, and had comparable CD39, CD31 and Helios markers, despite different TSDR-demethylation. These data indicate monitoring TSDR-demethylation levels in PBMC is not an accurate means to assess Treg numbers post-Tx.
Table 4.
TSDR-demethylation in PBMC does not correlate with numbers of Tregs in PBMC
| Group | Statistical factors | Treg number calculated as: | |||
|---|---|---|---|---|---|
| CD4+CD25+ cells in PBMC | CD4+CD25+FOXP3+ cells in PBMC | TSDR-demethylated CD4+ CD25+ cells in PBMC | CD4+CD25+ cells in CD4+ subset | ||
| All patients | r, Pearson p (2 tails) No. cases |
0.180 0.399 24 |
0.096 0.712 17 |
0.060 0.824 16 |
0.576 0.003 24 |
| Boys | r, Pearson p (2 tails) No. cases |
0.241 0.387 15 |
0.25 0.482 10 |
0.091 0.791 11 |
0.615 0.015 15 |
| Girls | r, Pearson p (2 tails) No. cases |
0.124 0.751 9 |
0.265 0.565 7 |
0.655 0.230 5 |
0.307 0.421 9 |
Figure 3. FOXP3 TSDR-demethylation in PBMC is sensitive to CD3/CD28 stimulation and/or IL-2.
Healthy donor PBMC were stimulated overnight with CD3/CD28 mAb-coated beads (1:1) ± IL-2 (100 U/ml). Cells were split and stained for CD4, CD25 and FOXP3, or used to isolate DNA and test TSDR-demethylation. (A) Compared with untreated controls, CD3/CD28 stimulation increased TSDR-demethylation in PBMC, from 5.3 to 19.7% without IL-2, and to 10.2% with IL-2, p=0.0273, Kruskal-Wallis test. Dunn's multiple comparison post-test showed that untreated control differed from CD3/28 stimulated PBMC, p<0.05. (B) In parallel, CD3/CD28 stimulation led to FOXP3 upregulation from 7.2 to 20.8% (without IL-2) and to 19.1% (with IL-2) in CD4+ cells. Experiment was set up in triplicate, TSDR demethylation was tested separately for each of three wells, and flow cytometry was performed using pooled samples.
Epigenetic status of FOXP3 is an important Treg marker
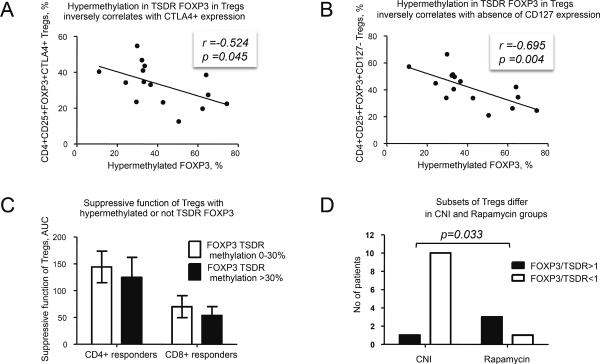
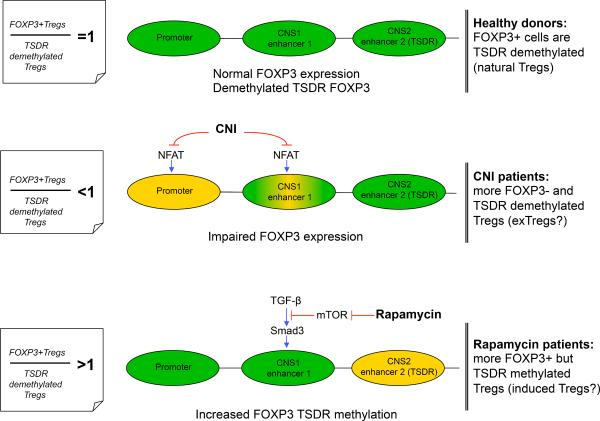
Hypermethylated-TSDR FOXP3 in boys negatively correlated with CTLA4 and with absence of CD127 expression in Tregs (Figure 4 A, B), i.e. with markers linked to enhanced suppressive function. TSDR-hypermethylated Tregs tended to have impaired suppressive function (Figure 4C), but the strongest correlation was found for FOXP3 protein (r=−0.732, p=0.002). In contrast to induced Tregs (iTregs) and FOXP3+ activated T cells, natural Tregs have TSDR-demethylation (13). To characterize patient Treg subsets, we calculated the ratio of %FOXP3+ cells to %TSDR-demethylated cells within isolated suppressive CD4+CD25+ Tregs (FOXP3+/TSDR ratio). In only 12.5% cases was the ratio ≈1, and CNI and rapamycin groups showed interesting differences. Most patients receiving CNI had FOXP3+/TSDR ratios <1, while in rapamycin-treated patients had ratios typically >1 (p=0.033, Fisher's exact test, Figure 4D). FOXP3+/TSDR ratios <1 indicate cells with TSDR-demethylation but not FOXP3 expression, consistent with CNI inhibition of the transcription factor, nuclear factor of activated T cells (NFAT) (14) (Figure 5), whereas FOXP3+/TSDR ratios >1 suggest FOXP3+ cells with methylated-TSDR, e.g. iTregs or activated Teffs. Since the group with FOXP3+/TSDR ratios >1 had more suppressive Tregs and higher CTLA4 expression than the group with FOXP3+/TSDR ratios <1 (Figure S4), it is likely the rapamycin group had increased numbers of FOXP3+ TSDR-methylated suppressive iTregs rather than Teff cells (Figure 5). Collectively, these data suggest TSDR-demethylation is a useful component of Treg (though not PBMC) characterization, and that the associated FOXP3+/TSDR ratio provides insight into the relative contributions of Treg subsets in clinical Tx recipients.
Figure 4. Role of TSDR-methylation in Tregs.
(A) In boys, hypermethylation in TSDR correlates inversely with (A) CTLA4 expression and (B) with absence of CD127 expression in isolated CD4+CD25+FOXP3+ Tregs (15 patients). The same (and also stronger) correlations were observed in boys receiving CNI therapy: r=−0.647, p=0.032 for CD4+CD25+CTLA4+ Tregs and r=−0.838, p=0.001 for CD25+FOXP3+CD127−Tregs (11 patients). (C) Tregs, hypermethylated in TSDR (≥30% methylation adjusted to gender as HMgirls=100-((100-HM)*2) for girls where HM - % of hypermethylated-TSDR) tend to have lower suppressive function than Tregs with <30% methylation: 125 vs. 144 AUC, p=0.68 for CD4+ responders and 55 vs. 70, p=0.54 for CD8+ responders (17 patients). (D) Ratio between FOXP3+ Tregs to TSDR-demethylated Tregs in isolated suppressive CD4+CD25+ cells (FOXP3/TSDR) was calculated in boys in CNI vs. rapamycin groups (15 patients). Most patients in CNI group have FOXP3/TSDR ratios <1, while most patients in the rapamycin group have FOXP3/TSDR ratios >1. An absolute percent of TSDR-demethylation in Tregs was similar in CNI and rapamycin groups (not shown).
Figure 5. Basis for different FOXP3/TSDR ratios in Tregs from CNI and Rapamycin groups.
To initiate FOXP3 gene transcription, the FOXP3 promoter and at least 1 of 2 enhancer elements must be activated. In natural Tregs, enhancer 2 (TSDR) is demethylated, resulting in sustained FOXP3 expression. In peripherally induced Tregs, TGF-β sensitive enhancer is activated, leading to transient FOXP3 expression. Top: in healthy donors, most Tregs are natural thymic-derived cells expressing FOXP3 and demethylated at the TSDR. Middle: CNI exposure disrupts NFAT signaling, leading to impaired FOXP3 promoter activation (14) and, as a result, leads to an increased percent of TSDR-demethylated (in CNS2 region) cells that are unable to activate FOXP3 transcription, within CD4+CD25+ Tregs. The enhancer 1 region may also be sensitive to CNI, since TGF-β activated induction occurs through the cooperation of NFAT and Smad3 and that can lead to further aggravation of impaired FOXP3 expression in Tregs. Bottom: Activation of enhancer 1 through TGF-β-dependent and TGF-β-independent pathways (the latter not shown in schematic) can be disrupted by AKT-mTOR activity and inhibited by rapamycin, resulting in increased numbers of FOXP3+ peripherally induced Tregs with TSDR-methylated FOXP3. Green regions are active, yellow regions are partially inactive.
Discussion
While murine studies showed increased numbers of FOXP3+ Tregs in tolerant allograft recipients (15), the induction of CD25 and FOXP3 upon activation of human Teffs (16) has led to far more variable clinical data. Thus, Treg numbers were increased in tolerant recipients (17, 18), whereas others found no differences (19, 20). Similarly, Treg numbers were decreased in patients with acute (21–23) or chronic (19, 24) rejection in some Centers but not others (25, 26). Despite an ever-expanding list of markers to identify “true” human Tregs, including CD127−/low and CD49d−, CD39+, CTLA4+, GARP+, CD120+, LAP+, CD27+ etc., none have proven appropriately Treg-specific using patient samples. E.g. in healthy donors CD4+CD25+CD127−/low gated cells are 90–95% FOXP3+, whereas that subset is enriched by FOXP3− cells in rheumatoid arthritis (27). In Tx recipients, we found variable FOXP3 expression by CD4+CD25+ cells, many FOXP3+ cells outside the CD25++CD127− gate, and contamination of that gate by FOXP3− cells. Others noted similar findings in renal Tx patients (28). Thus, currently flow cytometry alone cannot satisfactorily enumerate Treg cells post-Tx, and needs to be accompanied by assessment of Treg function.
Treg suppression assays have pitfalls, since isolation of CD4+CD25+ (or CD4+CD25+CD127−) cells from Tx recipient results in a mix of suppressive (Tregs) and activated (Teffs) cells. We are unaware of any literature assessing at least FOXP3 (or better, FOXP3, CD127 and CTLA4) expression within isolated cells and corresponding assessment of Treg suppressive function post-Tx. Moreover, using autologous cells as responders in Treg assays can be misleading, since immunosuppression can decrease their proliferative capacity. Such use may explain why Tregs from patients with rejection had impaired suppressive function in some studies (24, 26) but not others (23, 29, 30). We therefore assessed CD25, CD127, CTLA4 and FOXP3 expression in isolated Tregs, and performed suppression assays under highly standardized conditions, with Tregs as the only variable. We found Treg function post-Tx correlated with CTLA4 and absence of CD127 expression, but not with FOXP3, despite its role as a master regulator in Tregs (2). This likely reflects how activated human T cells can induce FOXP3 without acquiring a Treg phenotype or suppressive function, and is consistent with dominance of CTLA4 over FOXP3 for human Treg suppressive function (11).
The epigenetic status of FOXP3 is useful given the TSDR is demethylated only in natural Tregs with stable FOXP3 expression (13). We found that TSDR-demethylation indeed correlated with a “true” Treg CD127−CTLA4+ phenotype and suppressive function, and described a new way to evaluate Treg subsets within isolated Tregs: the ratio of FOXP3+ cells to TSDR-demethylated cells (FOXP3/TSDR). Almost all Tregs from CNI patients had FOXP3/TSDR ratios <1, showing FOXP3 gene transcription was impaired, while TSDR FOXP3 demethylation was preserved. Conversely, most Tregs in rapamycin-treated patients had FOXP3/TSDR ratios >1, showing the Treg subset was enriched by suppressive, CTLA4+FOXP3+ cells with a methylated-TSDR, and indicative of iTregs (Figure 5). Rapamycin stimulates iTreg conversion through inhibition of the mTOR pathway and enhancing TGF-β production by conventional T cells (2).
Assessment of TSDR-demethylation is proposed as way to determine Treg numbers in PBMC or tissue samples (13), but requires at least two conditions. First, every TDSR-demethylated cell should have normal FOXP3 transcription and hence, FOXP3 protein. Second, cells with methylated-TSDR should be unable to produce FOXP3 or, if FOXP3 is produced, the cells should not be suppressive. This concept does not adequately account for iTregs. TSDR-demethylation in PBMC post-Tx showed no correlations with Treg numbers, calculated as CD4+CD25+, CD4+CD25+FOXP3+ or even as CD4+CD25+TSDR-demethylated cells. We are unaware of any previous comparison of Treg numbers calculated using TSDR-demethylation in PBMC and TSDR-demethylation in Tregs, and note that TSDR-demethylation in PBMC varies with activation.
Our study design has several points to note. We excluded patients undergoing acute or chronic rejection, or those graft failure, allowing us to evaluate Tregs from patients with stable graft function and receiving conventional immunosuppression. However, patients with impaired grafts may provide additional insights. Second, we excluded patients with conditions affecting Treg number or function, such as cancer, autoimmune diseases, hepatitis and serious infections, to minimize their possibly confounding effects. Lastly, our study had a small size and cross-sectional design. Despite this, our composite approach showed that even in patients with stable graft function, CNI use may adversely affect Tregs. Serial studies including collection of samples preand post-Tx will likely produce key additional insights, and are now underway in our lab.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health: 1RC1 DK087270, UL1-RR026314 NCRR/NIH, and NIDDK KO8 1K08DK092282.
Abbreviations
- CNI
calcineurin inhibitors
- GFR
glomerular filtration rate
- NFAT
nuclear factor of activated T cells
- Rapa
rapamycin
- Teffs
conventional CD4+CD25− T-effector cells
- Tregs
T-regulatory cells
- TSDR
Treg-specific demethylated region
- Tx
transplantation
Footnotes
Ethical guidelines Clinical studies were approved by the respective IRB of each Center: CHOP-08-006311_CR1, CCHMC-2009-0018, and HSC-1000014523.
Disclosure The authors declare no conflict of interest.
References
- 1.Srinivas TR, Kaplan B. Transplantation in 2011: New agents, new ideas and new hope. Nature reviews Nephrology. 2012;8:74–75. doi: 10.1038/nrneph.2011.215. [DOI] [PubMed] [Google Scholar]
- 2.Boros P, Bromberg JS. Human FOXP3+ regulatory T cells in transplantation. Am J Transplant. 2009;9:1719–1724. doi: 10.1111/j.1600-6143.2009.02704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penninga L, Wettergren A, Chan AW, Steinbruchel DA, Gluud C. Calcineurin inhibitor minimisation versus continuation of calcineurin inhibitor treatment for liver transplant recipients. Cochrane Database Syst Rev. 2012;3:CD008852. doi: 10.1002/14651858.CD008852.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeiser R, Nguyen VH, Beilhack A, Buess M, Schulz S, Baker J, et al. Inhibition of CD4+CD25+ regulatory T-cell function by calcineurin-dependent interleukin-2 production. Blood. 2006;108:390–399. doi: 10.1182/blood-2006-01-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao W, Lu Y, El Essawy B, Oukka M, Kuchroo VK, Strom TB. Contrasting effects of cyclosporine and rapamycin in de novo generation of alloantigen-specific regulatory T cells. Am J Transplant. 2007;7:1722–1732. doi: 10.1111/j.1600-6143.2007.01842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Zhao L, Sun Z, Sun L, Zhang B, Zhao Y. A potential side effect of cyclosporin A: inhibition of CD4(+)CD25(+) regulatory T cells in mice. Transplantation. 2006;82:1484–1492. doi: 10.1097/01.tp.0000246312.89689.17. [DOI] [PubMed] [Google Scholar]
- 7.Segundo DS, Ruiz JC, Izquierdo M, Fernandez-Fresnedo G, Gomez-Alamillo C, Merino R, et al. Calcineurin inhibitors, but not rapamycin, reduce percentages of CD4+CD25+FOXP3+ regulatory T cells in renal transplant recipients. Transplantation. 2006;82:550–557. doi: 10.1097/01.tp.0000229473.95202.50. [DOI] [PubMed] [Google Scholar]
- 8.Chu Z, Zhang J, Zhao Y, Ji Q, Zhong J, Zhang C, et al. Influence of immunosuppressive drugs on the development of CD4(+)CD25(high) Foxp3(+) T cells in liver transplant recipients. Transplant Proc. 2010;42:2599–2601. doi: 10.1016/j.transproceed.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 9.Ruggenenti P, Perico N, Gotti E, Cravedi P, D'Agati V, Gagliardini E, et al. Sirolimus versus cyclosporine therapy increases circulating regulatory T cells, but does not protect renal transplant patients given alemtuzumab induction from chronic allograft injury. Transplantation. 2007;84:956–964. doi: 10.1097/01.tp.0000284808.28353.2c. [DOI] [PubMed] [Google Scholar]
- 10.Lin WX, Christiansen D, Fu LL, Roberts MA, Sandrin MS, Ierino FL. Foxp3(+) T cells in peripheral blood of renal transplant recipients and clinical correlations. Nephrology (Carlton) 2012 doi: 10.1111/j.1440-1797.2012.01578.x. [DOI] [PubMed] [Google Scholar]
- 11.Akimova T, Ge G, Golovina T, Mikheeva T, Wang L, Riley JL, et al. Histone/protein deacetylase inhibitors increase suppressive functions of human FOXP3+ Tregs. Clin Immunol. 2010;136:348–363. doi: 10.1016/j.clim.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLoS One. 2011;6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wieczorek G, Asemissen A, Model F, Turbachova I, Floess S, Liebenberg V, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69:599–608. doi: 10.1158/0008-5472.CAN-08-2361. [DOI] [PubMed] [Google Scholar]
- 14.Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 15.Joffre O, Santolaria T, Calise D, Al Saati T, Hudrisier D, Romagnoli P, et al. Prevention of acute and chronic allograft rejection with CD4+CD25+Foxp3+ regulatory T lymphocytes. Nat Med. 2008;14:88–92. doi: 10.1038/nm1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 17.Koshiba T, Li Y, Takemura M, Wu Y, Sakaguchi S, Minato N, et al. Clinical, immunological, and pathological aspects of operational tolerance after pediatric living-donor liver transplantation. Transpl Immunol. 2007;17:94–97. doi: 10.1016/j.trim.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Llordella M, Puig-Pey I, Orlando G, Ramoni M, Tisone G, Rimola A, et al. Multiparameter immune profiling of operational tolerance in liver transplantation. Am J Transplant. 2007;7:309–319. doi: 10.1111/j.1600-6143.2006.01621.x. [DOI] [PubMed] [Google Scholar]
- 19.Louis S, Braudeau C, Giral M, Dupont A, Moizant F, Robillard N, et al. Contrasting CD25hiCD4+T cells/FOXP3 patterns in chronic rejection and operational drug-free tolerance. Transplantation. 2006;81:398–407. doi: 10.1097/01.tp.0000203166.44968.86. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Llordella M, Lozano JJ, Puig-Pey I, Orlando G, Tisone G, Lerut J, et al. Using transcriptional profiling to develop a diagnostic test of operational tolerance in liver transplant recipients. J Clin Invest. 2008;118:2845–2857. doi: 10.1172/JCI35342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Q, Fan H, Li JQ, Qi HZ. Decreased circulating CD4+CD25highFoxp3+ T cells during acute rejection in liver transplant patients. Transplant Proc. 2011;43:1696–1700. doi: 10.1016/j.transproceed.2011.03.084. [DOI] [PubMed] [Google Scholar]
- 22.Perrella O, Sbreglia C, Arenga G, Perrella A, Ferrara A, D'Antonio A, et al. Acute rejection after liver transplantation: Is there a specific immunological pattern? Transplant Proc. 2006;38:3594–3596. doi: 10.1016/j.transproceed.2006.10.102. [DOI] [PubMed] [Google Scholar]
- 23.Demirkiran A, Kok A, Kwekkeboom J, Kusters JG, Metselaar HJ, Tilanus HW, et al. Low circulating regulatory T-cell levels after acute rejection in liver transplantation. Liver Transpl. 2006;12:277–284. doi: 10.1002/lt.20612. [DOI] [PubMed] [Google Scholar]
- 24.Akl A, Jones ND, Rogers N, Bakr MA, Mostafa A, El Shehawy el M, et al. An investigation to assess the potential of CD25highCD4+ T cells to regulate responses to donor alloantigens in clinically stable renal transplant recipients. Transpl Int. 2008;21:65–73. doi: 10.1111/j.1432-2277.2007.00560.x. [DOI] [PubMed] [Google Scholar]
- 25.Ashton-Chess J, Dugast E, Colvin RB, Giral M, Foucher Y, Moreau A, et al. Regulatory, effector, and cytotoxic T cell profiles in long-term kidney transplant patients. J Am Soc Nephrol. 2009;20:1113–1122. doi: 10.1681/ASN.2008050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dijke IE, Korevaar SS, Caliskan K, Balk AH, Maat AP, Weimar W, et al. Inadequate immune regulatory function of CD4+CD25bright+FoxP3+ T cells in heart transplant patients who experience acute cellular rejection. Transplantation. 2009;87:1191–1200. doi: 10.1097/TP.0b013e31819ec2fb. [DOI] [PubMed] [Google Scholar]
- 27.Aerts NE, Dombrecht EJ, Ebo DG, Bridts CH, Stevens WJ, De Clerck LS. Activated T cells complicate the identification of regulatory T cells in rheumatoid arthritis. Cell Immunol. 2008;251:109–115. doi: 10.1016/j.cellimm.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Alonso-Arias R, Suarez-Alvarez B, Lopez-Vazquez A, Moro MA, Baltar J, Gomez-Huertas E, et al. CD127(low) expression in CD4+CD25(high) T cells as immune biomarker of renal function in transplant patients. Transplantation. 2009;88:S85–93. doi: 10.1097/TP.0b013e3181afebdb. [DOI] [PubMed] [Google Scholar]
- 29.Braudeau C, Racape M, Giral M, Louis S, Moreau A, Berthelot L, et al. Variation in numbers of CD4+CD25highFOXP3+ T cells with normal immuno-regulatory properties in long-term graft outcome. Transpl Int. 2007;20:845–855. doi: 10.1111/j.1432-2277.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- 30.Kreijveld E, Koenen HJ, van Cranenbroek B, van Rijssen E, Joosten I, Hilbrands LB. Immunological monitoring of renal transplant recipients to predict acute allograft rejection following the discontinuation of tacrolimus. PLoS One. 2008;3:e2711. doi: 10.1371/journal.pone.0002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.