Abstract
Background:
Dysfunctional adipose tissue plays an important role in the etiology of the metabolic syndrome, type 2 diabetes, and dyslipidemia. However, the molecular mechanisms underlying adipocyte dysfunction are incompletely understood.
Aim:
The aim of the study was to identify differentially regulated pathways in sc adipocytes of dyslipidemic subjects.
Methods:
Whole-genome expression profiling was conducted on sc adipocytes from a discovery group of nine marginally overweight subjects with familial combined hyperlipidemia (FCHL) and nine controls of comparable body sizes as well as two independent confirmation groups. In this study, FCHL served as a model of familial insulin resistance and dyslipidemia, in the absence of frank obesity.
Results:
Functional analyses and gene set enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes or a custom pathway database identified the complement system and complement regulators as one of the top up-regulated pathways in FCHL [false discovery rate (FDR) < 1E-30]. Higher adipocyte complement expression in FCHL was confirmed in the appropriate confirmation group. Higher complement gene expression was associated with lower adipocyte insulin receptor substrate-1 expression as marker of adipocyte insulin resistance, independent of age, sex, or disease status, and this association was corroborated in the two confirmation groups. Additionally, complement gene expression was associated with triglycerides in the discovery set and with triglycerides and/or waist circumference in the confirmation groups. Complement pathway up-regulation did not appear to be driven by hypertriglyceridemia because a 40% pharmacological reduction in triglycerides did not affect complement expression.
Conclusions:
These findings point to an up-regulation of a complement-related transcriptome in sc adipocytes under metabolically stressed conditions, even in the absence of overt obesity. Such up-regulation may subsequently influence downstream processes, including macrophage infiltration into adipose tissue and adipocyte insulin resistance.
Dysfunctional adipose tissue plays a central role in the etiology of a large number of metabolic disorders including the metabolic syndrome, type 2 diabetes, and dyslipidemia. However, the molecular mechanisms underlying adipocyte dysfunction are incompletely understood.
Studies investigating transcriptional changes in adipose tissue that are associated with obesity and obesity-related disorders have often relied on the use of whole adipose tissue samples (1, 2). However, the use of total adipose tissue samples does not allow for the exclusive evaluation of gene expression in adipocytes due to the substantial number of nonadipose cells (macrophages, CD4 and CD8 T cells, fibroblasts, endothelial cells, and mesenchymal cells) that also reside in adipose tissue (3). This is particularly relevant when studying metabolic disorders related to obesity-associated insulin resistance because these conditions are generally characterized by an increased influx of inflammatory cells into the adipose tissue. These inflammatory cells can, by themselves, express a large array of genes. The use of the isolated adipocyte fraction, as opposed to whole adipose tissue, allows for the study of gene expression patterns originating directly from the adipocyte without possible confounding from other cell types.
Studies on adipose tissue gene expression have been typically focused on comparisons between obese and lean subjects (2, 4–7), profiling of different fat depots (2, 6, 8–10), or studies on the effects of dietary and/or lifestyle intervention on the adipose transcriptome (1, 11–13). Relatively little is known, however, about systemic changes in the adipocyte transcriptional program in metabolically compromised nonobese subjects, e.g. subjects with insulin resistance, impaired glucose tolerance, or moderate hyperlipidemia, compared with metabolically healthy subjects of comparable body sizes (8)
A metabolic disorder involving adipose tissue dysfunction in the absence of pronounced obesity is familial combined hyperlipidemia (FCHL), which is the most common familial dyslipidemia. FCHL is associated with an increased risk of cardiovascular disease, with prevalence rates as high as 20% in patients with premature coronary artery disease (14). Although FCHL patients are usually nonobese, the complications associated with FCHL are thought to be caused, at least in part, by deregulations in adipose tissue metabolism (15). In addition to hyperlipidemia (14), FCHL is characterized by insulin resistance (16), which predisposes to the development of type 2 diabetes (17). In this study, we have used FCHL as a model of metabolic stress involving familial insulin resistance and dyslipidemia. Specifically, we have conducted whole-genome expression profiling on the sc adipocyte fraction of marginally overweight FCHL patients and unaffected, unrelated controls matched for body mass. We complemented the transcriptome analysis with anthropometric and biochemical determinations to relate adipocyte gene expression changes to functional consequences. Our findings provide novel insights into transcriptional remodeling in the insulin-resistant adipocyte in predominantly nonobese, dyslipidemic subjects.
Materials and Methods
Subjects
FCHL probands and their family members were recruited following previously described inclusion-exclusion criteria (18). Briefly, FCHL probands had primary hyperlipidemia [untreated fasting plasma cholesterol > 6.5 mm (250 mg/dl) and/or fasting plasma triglyceride concentration > 2.3 mm (200 mg/dl)], and a positive family history of premature cardiovascular disease (less than 60 yr).
FCHL patients were ascertained as such when they were hyperlipidemic members of a FCHL family that contained at least two other first-degree relatives with fasting plasma cholesterol and/or triglycerides concentrations exceeding the diagnostic values. FCHL patients who used lipid-lowering medication had stopped their therapy for 14 d to obtain untreated plasma and adipose tissue samples. The control subjects were normolipidemic spouses or relatives of the FCHL patients or volunteers who were recruited via advertisement among hospital personnel. We used a discovery group consisting of nine marginally overweight subjects with FCHL and nine controls of comparable body sizes and two independent confirmation groups (confirmation group 1: 16 FCHL patients, 23 normolipidemic relatives, and seven controls; confirmation group 2: 11 FCHL patients). The Human Investigation Review Committee of the Academic Hospital Maastricht approved the study protocol and all subjects gave informed consent.
Sample preparation, microarray hybridization, and quantitative PCR
Details of the isolation and purification of adipocyte subfractions, total RNA isolation, and hybridization to Affymetrix microarrays (Santa Clara, CA), gene signal extraction, and quantitative PCR (qPCR) are presented in Supplemental Data and Table 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org.
Table 1.
Basic characteristic of the study subjects (discovery set)
| Control (n = 6–9) | HL-FCHL (n = 8–9) | P value | |
|---|---|---|---|
| Age (yr) | 40.3 ± 6.4 | 48.4 ± 8.5 | 0.036 |
| Sex (male/female) | 2/7 | 6/3 | 0.058 |
| BMI (kg/m2) | 25.1 ± 3.6 | 26.1 ± 2.7 | 0.526 |
| Waist (cm) | 89.0 ± 5.9 | 95.4 ± 8.0 | 0.114 |
| Triglycerides (mm) | 0.9 (0.9; 1.4) | 3.1 (2.5; 5.1) | <0.001 |
| Cholesterol (mm) | 5.0 ± 0.9 | 7.0 ± 1.1 | 0.001 |
| Glucose (mm)a | 4.9 ± 0.4 | 5.2 ± 0.5 | 0.167 |
| Insulin (μU/ml)a | 7.5 (4.2; 9.2) | 6.6 (5.0; 12.1) | 0.426 |
| HOMA2ira | 1.0 (0.6; 1.3) | 1.0 (0.8; 1.7) | 0.288 |
Full data are available for age, sex, BMI, waist, and triglyceride and cholesterol concentrations.
Glucose concentration was missing for one HL-FCHL patient; insulin and HOMA2ir were missing for three controls and one HL-FCHL.
Statistical analyses
Variables with a normal distribution are presented as mean ± sd. Variables with skewed distribution are presented as median (interquartile range) and were log transformed before further analyses. Comparisons between different groups were assessed by Student's t test (continuous data) or χ2 test (proportions). Linear regression models were used to generate age- and sex-adjusted unstandardized residuals for correlation analyses and to perform age- and sex-adjusted comparisons between groups. Statistical significance was assessed at the P < 0.05 level. These analyses were performed in the Statistical Package for the Social Sciences, version 15 (SPSS Inc., Chicago, IL).
Functional profiling and transcript interaction mapping
Differential gene expression data were used to identify the significantly overrepresented biological themes between the phenotypes via the FunNet tool (19). Pathway enrichment was ascertained via the Gene Set Enrichment Analysis (GSEA) software package (20). In each case, statistical significance was assessed at a false discovery rate (FDR) of 5%. Details about the methodologies are described in Supplemental Data. An author-developed Custom pathway database, used for the GSEA, is shown in Supplemental Table 2.
To obtain an overall measure of gene expression for a GSEA-derived pathway of interest, genes that significantly contributed to the core enrichment of that pathway were compiled into a generalized pathway expression score, representing the overall expression of that pathway. First, the expression data for each gene of the pathway contributing to core enrichment were converted to z-scores [i.e. (individuals' observed values − population mean)/sd]. Then the average of the z-scores of the genes that contributed to the core enrichment was calculated. This general pathway expression score represents the overall expression of that pathway in a specific dataset and is expressed in units of sd. By using z-scores, the expression of a pathway in the discovery data set (microarray) and in the confirmation data sets (microarray and qPCR) could be directly compared.
Results
Basic characteristics
Demographic and biochemical characteristics of the subjects in the discovery group are summarized in Table 1. FCHL patients had, by definition, higher triglyceride and cholesterol levels than controls. In addition, FCHL patients were slightly older and comprised a higher proportion of male subjects. FCHL patients and controls did not differ significantly with respect to the other characteristics. Demographic characteristics of the confirmation groups are presented in Supplemental Table 3. Expression analysis of adipose tissue macrophage markers showed nondetectable expression for TNF-α and CD68 antigen, which argues against any significant presence of macrophages in the adipocyte fractions.
Differential pathway enrichment in the sc adipocyte fraction
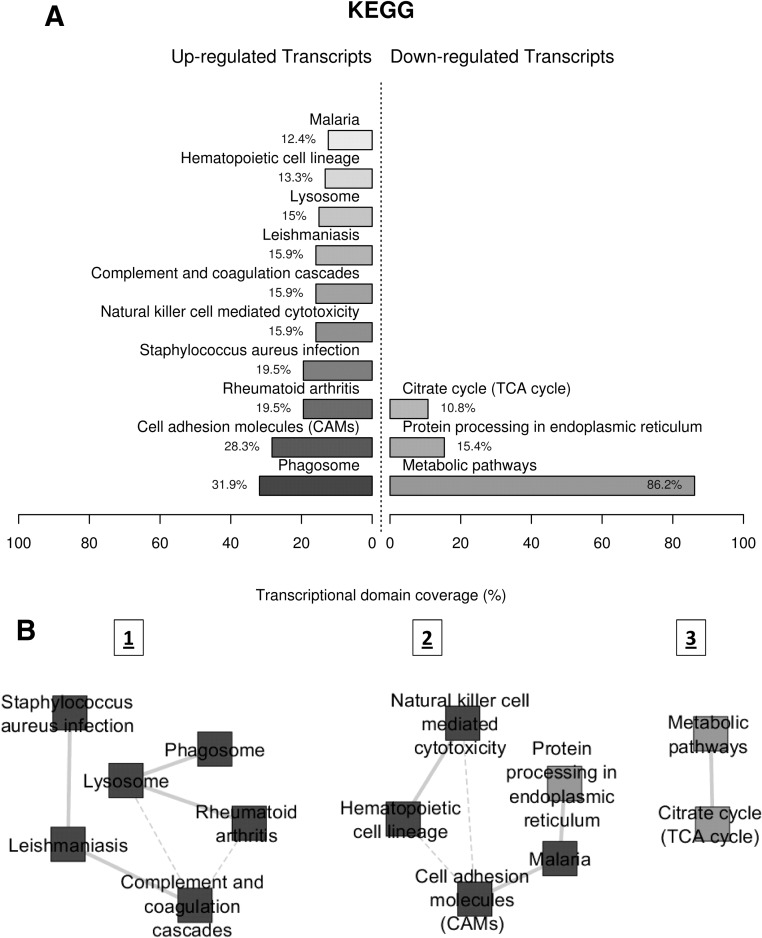
We first carried out a functional profiling of gene expression data to identify overrepresented biological themes among the up- and down-regulated transcripts in FCHL adipocytes, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. Figure 1A displays the major enriched KEGG pathways from among the up- and down-regulated genes in the FCHL adipocytes. Notable overrepresentations of up-regulated and differentially expressed genes were observed for immune/infection related pathways (phagosome, rheumatoid arthritis, Staphylococcus aureus infection, complement and coagulation cascades, etc.) and also for pathways controlling cell adhesion (cell adhesion molecules). Conversely, the most significant overrepresentation of down-regulated genes was observed for the metabolic pathways, followed by protein processing in the endoplasmic reticulum and the citrate (tricarboxylic acid cycle) cycle. We then measured the transcriptional interaction between these overrepresented KEGG pathways, based on the similarity of the expression profiles of their annotated transcripts (21). These measurements led to the generation of biological interaction networks, shown in Fig. 1B. We identified three distinct interaction modules: one consisting entirely of up-regulated pathways (module 1), one containing a mix of up- and down-regulated pathways (module 2), and one consisting of only down-regulated pathways (module 3). Module 1 is enriched for immune/infection related pathways, module 2 showed an enrichment of cell adhesion/hematopoietic cell activity and intracellular protein processing, and module 3 consisted of an interaction between metabolic pathways and the TCA cycle. The three most highly connected nodes among the three modules were the complement and coagulation cascades and lysosome (module 1) and cell adhesion molecules (module 2) with each connecting to three neighboring nodes.
Fig. 1.
KEGG functional profiling and interaction modules, illustrating transcriptomic interactions in FCHL adipocytes. A, Functional profiling depicts significantly overrepresented KEGG pathways on the basis of differentially expressed genes in FCHL vs. control adipocytes. Functional profiling was conducted via the FunNet tool by considering all differentially expressed genes at an FDR of 5%. Transcriptional domain coverage for each pathway corresponds to the percentage of the transcriptional domain covered by the genes identified for that pathway. Up- and down-regulated pathways are shown separately. B, Functional themes, represented by enriched KEGG pathways, were correlated in a proximity interaction network driven by the expression similarity of their annotated genes. KEGG categories are indicated by nodes, whereas transcriptional interactions among the KEGG categories are indicated by edges. Continuous lines indicate stronger, whereas dashed lines depict medium-strength, interactions.
The overrepresentation/transcript interaction analysis described above was complemented with a different pathway analysis approach based on the GSEA methodology. Pathway (gene set) enrichment analysis of adipose tissue gene expression data identified 15 pathways from the KEGG database as significantly up-regulated and six pathways as significantly down-regulated in the FCHL patients (FDR < 5%) (Table 2). Several of the FCHL-up-regulated pathways were again associated with inflammation and immune response-related processes. Three of the 10 up-regulated pathways identified in Fig. 1A were also identified by GSEA at an FDR less than 5%, these included the complement and coagulation cascades, cell adhesion molecules, and natural killer cell-mediated cytotoxicity pathways. The citrate cycle pathway was also identified as a FCHL down-regulated pathway at an FDR less than 5%. The metabolic pathways from Fig. 1A, which contains more than 200 genes, were not identified in GSEA because the analysis was restricted to KEGG pathways with 200 or less genes.
Table 2.
KEGG pathways that were up-regulated and down-regulated in FCHL patients compared with controls
| Name | Size | ES | NES | NOM P value | FDR q value | FWER P value | Rank at max |
|---|---|---|---|---|---|---|---|
| KEGG pathways that are up-regulated in FCHL (FDR q value <0.05) | |||||||
| HSA04612_antigen_processing_and_presentation | 36 | 0.7782 | 2.85797 | 0 | 1 E-30 | 0 | 925 |
| HSA04514_cell_adhesion_molecules | 56 | 0.6689 | 2.67752 | 0 | 1 E-30 | 0 | 1597 |
| HSA04610_complement_and_coagulation_cascades | 19 | 0.7961 | 2.41627 | 0 | 1 E-30 | 0 | 1191 |
| HSA04940_type_1_diabetes_mellitus | 23 | 0.7214 | 2.36695 | 0 | 2.15E-04 | 0.001 | 1480 |
| HSA04640_hematopoietic_cell_lineage | 22 | 0.7116 | 2.30842 | 0 | 1.72E-04 | 0.001 | 1557 |
| HSA04512_ECM_receptor_interaction | 42 | 0.5952 | 2.26881 | 0 | 1.43E-04 | 0.001 | 1017 |
| HSA04060_cytokine_cytokine_receptor_interaction | 63 | 0.5239 | 2.17418 | 0 | 6.86E-04 | 0.005 | 1526 |
| HSA01032_glycan_structures_degradation | 17 | 0.7017 | 2.12834 | 0 | 0.00123 | 0.01 | 2025 |
| HSA01430_cell_communication | 36 | 0.5845 | 2.10512 | 0 | 0.00121 | 0.011 | 1660 |
| HSA04670_leukocyte_transendothelial_migration | 58 | 0.5201 | 2.10461 | 0 | 0.001089 | 0.011 | 1999 |
| HSA04650_natural_killer_cell_mediated_cytotoxicity | 60 | 0.5008 | 2.02132 | 0 | 0.002171 | 0.024 | 1599 |
| HSA00511_N_glycan_degradation | 10 | 0.7608 | 1.96087 | 0.0042 | 0.00408 | 0.048 | 2025 |
| HSA03010_ribosome | 63 | 0.4611 | 1.88859 | 0 | 0.006919 | 0.086 | 3519 |
| HSA04360_axon_guidance | 64 | 0.4502 | 1.84904 | 0.0021 | 0.009319 | 0.123 | 1904 |
| HSA04510_focal_adhesion | 119 | 0.3698 | 1.69301 | 0 | 0.035797 | 0.427 | 1017 |
| KEGG pathways that are down-regulated in FCHL (FDR q value <0.05) | |||||||
| HSA00280_valine_leucine_and_isoleucine_degradation | 34 | −0.6564 | −23340 | 0.0 | 1 E-30 | 0.0 | 1577 |
| HSA00640_propanoate_metabolism | 25 | −0.6194 | −20359 | 0.001879 | 0.00747 | 0.015 | 1577 |
| HSA00650_butanoate_metabolism | 25 | −0.5745 | −18970 | 0.00185 | 0.02188 | 0.061 | 1577 |
| HSA00020_citrate_cycle | 24 | −0.5928 | −18958 | 0.0 | 0.01669 | 0.062 | 3091 |
| HSA01040_polyunsaturated_fatty_acid_biosynthesis | 12 | −0.70587 | −18875 | 0.0 | 0.01459 | 0.068 | 1431 |
| HSA00030_pentose_phosphate_pathway | 14 | −0.6339 | −1.663 | 0.001855 | 0.0462 | 0.235 | 1609 |
Pathways that are significant at an FDR of 5% or less are listed. Column headings are as follows: name, name of the KEGG pathway; size, number of genes contained in the pathways; ES, enrichment score (a measure of the extent to which a pathway is overrepresented at the top or bottom of a ranked list of genes); NES, normalized enrichment score, after adjustment for pathway size; NOM P value, nominal P value obtained from gene-set level permutations; FDR q value, FDR to correct for multiple testing; FWER P value, family-wise error rate correction for multiple testing; rank at max, the position in the ranked gene list at which maximum enrichment of the pathway was observed. Nominal P < 0.001 values and FDR q < 0.0001 values are listed as zeros.
We were particularly interested in the identification of the KEGG complement and coagulation cascade pathway (third most up-regulated pathway in FCHL patients; FDR < 1 × 10−30), given the recent publications on the potential role of C3a/C3a-receptor signaling in the following: 1) macrophage infiltration into adipose tissue and development of insulin resistance (22), 2) the known role of C3a-desarg/ASP in triglyceride metabolism (23), and 3) the identification of central complement component C3 as a risk factor for weight gain (24), type 2 diabetes mellitus (25), and myocardial infarction (26). However, KEGG pathway analysis was not able to discern whether the observed pathway enrichment was due to contributions of genes specifically related to the complement or coagulation pathway or both (Supplemental Fig. 1). To ascertain whether the complement pathway by itself was indeed up-regulated in FCHL, we performed GSEA against a user-defined custom database (Supplemental Table 2) that included the full gene set of complement-related genes represented on the Affymetrix array (Supplemental Table 4). Pathway enrichment analysis on the custom pathway database identified six pathways as up-regulated and one pathway as down-regulated in the FCHL patients (FDR < 5%), with the full complement pathway being the top-ranked, FCHL-up-regulated pathway (FDR < 1 × 10−30, Table 3). These results strongly imply that the complement cascade pathway was indeed significantly up-regulated in the FCHL patients. Further analysis indicated that 16 of 20 expressed genes belonging to the full complement pathway were significantly overexpressed in FCHL patients and made significant contributions to the core enrichment of this pathway (see heat map in Supplemental Fig. 1).
Table 3.
Custom pathways that were up-regulated and down-regulated in FCHL patients compared with controls
| Name | Size | ES | NES | NOM P value | FDR q value | FWER P value | Rank at max |
|---|---|---|---|---|---|---|---|
| Custom pathways that were significantly up-regulated in FCHL (FDR q value <0.05) | |||||||
| Full pathway of complement | 20 | 0.81 | 2.52 | 0.0 | 0.0 | 0.0 | 1332 |
| T_B cell activation | 25 | 0.65 | 2.21 | 0.0 | 0.0 | 0.0 | 594 |
| Prostaglandin and leukotriene biosynthesis | 11 | 0.82 | 2.20 | 0.0 | 0.0 | 0.0 | 862 |
| Lymphocyte_affytechnote | 12 | 0.76 | 2.06 | 0.0 | 0.002 | 0.006 | 1480 |
| Innate and adaptive immunity | 34 | 0.55 | 1.97 | 0.0 | 0.006 | 0.02 | 1142 |
| CXCR4-mediated signaling events | 16 | 0.64 | 1.91 | 0.002 | 0.013 | 0.056 | 1557 |
| Custom pathways that were significantly down-regulated in FCHL (FDR q value <0.05) | |||||||
| Mitochondria | 47 | −0.5293 | −20.273 | 0.0 | 0.004 | 0.007 | 1628 |
Column headings are as follows: name, name of the KEGG pathway; size, number of genes contained in the pathways; ES, enrichment score (a measure of the extent to which a pathway is overrepresented at the top or bottom of a ranked list of genes); NES, normalized enrichment score, after adjustment for pathway size; NOM P value, nominal P value obtained from gene-set level permutations; FDR q value, FDR to correct for multiple testing; FWER P value, family-wise error rate correction for multiple testing; rank at max, the position in the ranked gene list at which maximum enrichment of the pathway was observed. Nominal P < 0.001 values and FDR q < 0.0001 values are listed as zeros.
Because there was a difference in the proportions of female and male subjects between the FCHL and control groups (Table 1), we further tested whether the observed results could be attributed to sex instead of disease status. Using either KEGG or the custom database, we did not observe evidence for significant differential expression of the previously identified pathways between the male and female subjects (FDR > 15%). Thus, the observed pathway differences between FCHL and control subjects were unlikely to be confounded by sex but were rather a consequence of disease status.
The complement genes that were responsible for the core enrichment of the complement pathway were compiled into a complement expression score to represent the overall expression of the complement pathway (described in Materials and Methods). In the discovery set, this overall complement expression score was 1.02. sd [95% confidence interval (CI) 0.25; 1.80] higher in hyperlipidemic-FCHL patients than in the control subjects (age and sex adjusted, P = 0.014). The complement score (compiled from the expression of C1S, DCN, C1R, C3aR, C3, and CD59 obtained in qPCR analyses) in independent confirmation data set 1 was 0.51 sd (95% CI 0.09; 0.92) higher in the HL-FCHL patients than in the control subjects and normolipidemic (NL) relatives combined (age and sex adjusted, P = 0.018) and also 0.52 sd (95% CI 0.06; 0.98) higher in the HL-FCHL patients than in their NL relatives (age and sex adjusted, P = 0.028). These findings confirmed our observation of an overall higher complement gene expression in the adipocyte fractions of HL-FCHL patients. When five subjects with body mass index (BMI) greater than 30 kg/m2 were excluded from the confirmation data set, the results did not materially change, i.e. complement expression remained higher in HL-FCHL patients than either control and NL-relatives combined or in NL-FCHL relatives only [differences were 0.46 (95% CI −0.02; 0.93), P = 0.059 and 0.48 (95% CI −0.06; 1.01), P = 0.079, respectively].
Association of adipocyte-expressed complement pathway gene expression with quantitative phenotype traits
We next investigated whether the complement pathway was significantly related to quantitative traits of insulin resistance and dyslipidemia. To control for potential confounding, age- and sex-adjusted values for homeostasis model assessment 2 insulin resistance index (HOMA2ir), BMI, waist circumference, triglycerides, and cholesterol were used. Gene sets (pathways) that were significantly correlated with these traits were identified via GSEA (Supplemental Table 5). We observed a positive and highly significant association of the KEGG complement and coagulation cascade pathway with HOMA2ir (FDR < 0.003), waist (FDR < 0.005), plasma triglyceride (FDR < 0.0001), and cholesterol (FDR < 0.0001) but not BMI (FDR < 0.69). When GSEA analysis was conducted against the custom pathway database, the full complement gene set displayed positive and highly significant relations to HOMA2ir (FDR < 1 × 10−30), waist (FDR < 1 × 10−30), and plasma triglycerides (FDR < 1 × 10−30). In comparison, a weaker correlation was observed for BMI (FDR < 0.006), and no correlation was observed for cholesterol (FDR 0.43). The associations of complement with waist and plasma triglycerides were also apparent in the discovery and the confirmation groups analyses with overall expression of the complement system represented by the complement score (Table 4), whereas the association with HOMA2ir was not (r = 0.460, P = 0.213). Thus, the associations observed in the discovery set were supported by the two independent confirmation sets, although in the discovery set, the association with plasma triglycerides appeared to be the most prominent one, whereas in the confirmation sets, this was true for waist.
Table 4.
Age- and sex-adjusted association of the complement score with waist, TG, and IRS1 expression in the discovery and in the two confirmation data sets
| Age- and sex-adjusted Pearson's r |
||||||
|---|---|---|---|---|---|---|
| Waist | P value | TG | P value | IRS1a | P value | |
| Controls and HL subjects | ||||||
| Complement scoreb | ||||||
| Discovery set (array; n = 15) | 0.479 | 0.097 | 0.771 | 0.002 | −0.751 | 0.003 |
| Confirmation set 1 (qPCR; n = 46) | 0.554 | <0.001 | 0.295 | 0.055 | −0.320 | 0.037 |
| Confirmation set 1; FCHL family members only (qPCR; n = 39) | 0.592 | <0.001 | 0.358 | 0.032 | −0.401 | 0.015 |
| Confirmation set 1; FCHL family members with BMI < 30 kg/m2 only (qPCR; n = 34) | 0.571 | 0.001 | 0.329 | 0.066 | −0.366 | 0.040 |
| HL subjects only | ||||||
| Complement scoreb | ||||||
| Discovery set (array; n = 9) | 0.619 | 0.139 | 0.340 | 0.455 | −0.737 | 0.059 |
| Confirmation set 1 (qPCR; n = 16) | 0.595 | 0.032c | 0.118 | 0.701 | −0.372 | 0.211 |
| Confirmation set 2 (array; n = 11) | 0.751 | 0.020C | 0.364 | 0.336 | −0.768 | 0.016 |
TG, Triglycerides.
For the arrays data, IRS1 expression is represented by the average z-scores of the expression levels of IRS-1 (two probesets); for the qPCR data, IRS1 is the z-value of IRS1 expression. For details on confirmation sets 1 and 2, see Supplemental Data.
For the array data, the complement score is the average z-score of the expression levels of C1S, DCN (five probe sets), C1R, ITGAX, C3AR1, C5R1, C1QG, C1QB, C3, ITGAM, CD59 (four probe sets), DAF (four probe sets), CFH (two probe sets), C7, SERPING, and CLU (three probe sets); for the qPCR data, the complement score is the average z-score of the expression levels of C1R, C1S, C3, C3AR, CD59, and DCN.
The positive associations did not remian statistically sugnificant after exclusion of subjects with BMI greater than 30 kg/m2 (confirmation set 1, r = 0.511, P = 0.131, n = 12; confirmation set 2, r = 0.567, P = 0.271, n = 8).
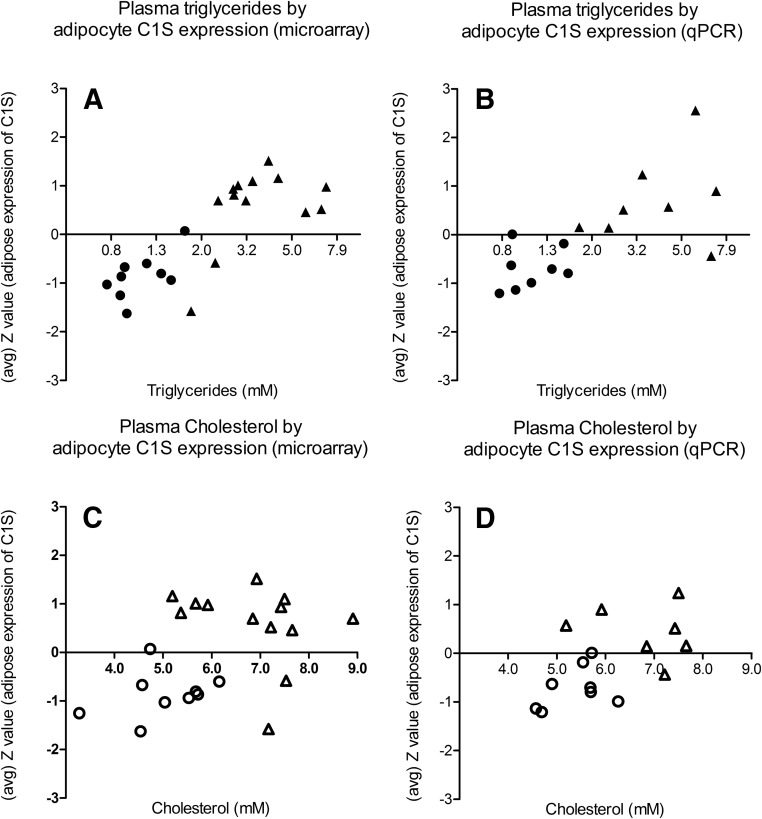
FCHL patients are defined by higher levels of triglycerides and/or cholesterol compared with controls. Therefore, the observed correlation of the complement genes with these plasma traits could simply be a reflection of the subjects' disease status (i.e. FCHL or control). To evaluate this possibility, we repeated the GSEA analysis with additional adjustment for disease status. The association of both the KEGG complement and coagulation cascade pathway and the custom full complement pathway with triglycerides (age, sex, and disease status adjusted) remained statistically significant (FDR < 0.002 and FDR < 1 × 10−30, respectively) and was therefore not entirely explained by disease status. Additionally, we observed a positive correlation of complement factor C1S expression with plasma triglyceride levels, regardless of the FCHL or control status of the subjects (Fig. 2). In contrast, the relationship between cholesterol and the complement and coagulation cascade gene set was no longer significant when the analyses were adjusted for age, sex, and disease status (FDR > 0.99). Likewise, in the custom database analysis, the full complement pathway was not significantly associated with cholesterol levels in these adjusted analyses (FDR > 0.99). This finding suggests that the apparent association observed with cholesterol was most likely due to the FCHL and control status, rather than to a true association with cholesterol levels. In agreement, the correlation between C1S gene expression and cholesterol levels in FCHL and control subjects was also substantially weaker than what was observed for triglycerides (Fig. 2).
Fig. 2.
Complement C1S expression in the sc adipocyte fraction is associated with plasma triglyceride levels, rather than with plasma cholesterol levels. Circles represent the control subjects, and triangles are the hyperlipidemic subjects. Upper panels, The regression coefficients r were 0.807 (P < 0.001) and 0.720 (P = 0.002) for the correlations of plasma triglycerides (log transformed) with C1s on microarray (A) and qPCR (B) data, respectively. Lower panels, The regression coefficients r were 0.437 (P = 0.042) and 0.561 (P = 0.030) for the correlations of plasma cholesterol with C1S on microarray (C) and qPCR (D) data, respectively.
Association of complement gene expression with local adipocyte insulin resistance markers
In addition to the identification of pathways significantly associated with systemic insulin resistance (represented by HOMA2ir), we further evaluated whether genes from the complement pathway were also associated with insulin resistance of the adipocytes. The expression of the insulin receptor substrate-1 (IRS1) was used as a proxy measure of adipocyte insulin resistance, as reported earlier (27, 28). Although the HL-FCHL and control subjects in the discovery set did not differ with respect to HOMA2ir, IRS-1 expression was 1.24 sd (95%CI 0.39; 2.09) lower in the HL-FCHL patients than in the controls (age and sex adjusted, P = 0.008), consistent with the presence of adipocyte insulin resistance. This was corroborated in confirmation group 1 in which age- and sex-adjusted IRS1 expression was 0.77 sd (95% CI 0.24; 1.30) lower in the HL-FCHL patients than in control subjects and NL relatives combined (P = 0.005) and was 0.74 sd (0.17; 1.31) lower in the HL-FCHL patients than in their NL relatives (P = 0.013). These findings did not change when subjects with BMI less than 30 kg/m2 were excluded.
The complement score was also used to evaluate the relation of adipocyte complement and IRS1 expression (Table 4). There was a clear inverse association between complement and IRS1 expression in the adipocytes in the discovery set, and this was corroborated by data obtained in both confirmation sets (Table 4).
Effect of acute lipid lowering on complement gene expression
The subjects in confirmation set 2 were part of an intervention study in which HL-FCHL patients were treated for 8 wk with 40 mg atorvastatin. Fat biopsies were taken before and after the intervention and processed for microarray analysis. Atorvastatin treatment resulted in an approximately 40% decrease in plasma triglycerides (P < 0.006) and plasma cholesterol (P < 0.001) with no effect on HOMA2ir (3% increase, P = 0.898). However, the transcriptome profiling in the fat biopsies showed that the lipid-lowering treatment affected neither the expression of complement genes nor the expression of IRS1 in the adipose fraction (data not shown).
Discussion
The primary purpose of this investigation was to query the transcriptomic remodeling of human adipocytes under conditions of increased cardiometabolic risk but without confounding effects from overt adiposity. We used primarily nonobese FCHL patients as a disease model and contrasted the gene expression patterns in sc adipocytes to those from unrelated, unaffected controls with comparable body mass. We further applied bioinformatic analysis to generate a systems-level view of alterations in the adipocyte transcriptome. Functional profiling and transcript interaction analysis, using FunNet (19), identified multiple KEGG pathways as significantly overrepresented among the differentially expressed genes. These pathways could be further organized into three separate modules representing immune/infection-related processes (module 1); hematopoietic cell function, cell adhesion, and intracellular protein processing (module 2); and metabolic process (module 3). Gene-set enrichment analysis on two different pathway databases also identified several pathways related to cell-cell communication, immune activation, and tissue integrity as being up-regulated in the FCHL subjects (at < 5% FDR). Conversely, pathways related to metabolism and fuel use were down-regulated in FCHL. The complement pathway was up-regulated in FCHL subjects and was highly ranked and highly significant in GSEA analyses in both the KEGG and custom databases. We subsequently focused our investigation on the expression of genes related to the complement system. In addition to its significant association with FCHL disease status, the complement pathway was also significantly correlated to plasma triglyceride levels and waist circumference (adjusted for age and sex). These findings were subsequently confirmed in an independent cohort of hyperlipidemic FCHL subjects. Moreover, expression of several complement pathway genes was strongly associated with proxy measures of local insulin resistance in the adipocytes and was largely maintained, even after adjustments for disease status.
With respect to the expression of individual complement genes, our current findings are consistent with, but substantially extend, previously published data. For example, Gabrielsson et al. (29) showed that a large range of complement factors is expressed in the adipose tissue of obese men but did not report on differences between groups. Koistinen et al. (30) demonstrated higher C3 expression in sc adipose tissue of nondiabetic obese and type 2 diabetic patients compared with controls. In another study, C3 expression did not differ between FCHL patients and controls but was associated insulin resistance (31). Activation of the alternative complement pathway in adipocytes was first shown by Choy et al. (32), whereas more recently Zhang et al. (33) reported that expression of some proximal components of the classical pathway was altered in sc adipose cells of insulin-resistant individuals. Insulin resistance has also been reported to be associated with higher expression of factors H and B in the omental, but not sc, fat depot (34). Additionally, C3, C1S, C7, and CD59 were also reported to be higher in omental fat from obese nondiabetic and diabetic patients than in lean subjects (35). Some apparent discrepancies also exist between our results and those previously published. Notably, some complement factors (e.g. complement factor B and C5L2/GPR77) were below the detection limit in our study but have been reported elsewhere (35, 36). A possible explanation is that these studies used total adipose tissue, including nonadipocyte cells, that are capable of independently producing various complement components (37).
Our data add important new information to current knowledge. For example, through the use of gene-set enrichment analysis, we showed that proximal factors of the classical pathway as well as most components of the alternative pathway and a substantial number of regulators of complement activation contributed to higher expression of the complement system in FCHL patients. This association was also extended to plasma triglyceride levels and waist circumference measures, even after adjustment for age, sex, and disease status. Moreover, we also identified a strong correlation between adipocyte insulin resistance and expression of the complement system in sc adipocytes that was, at least partly, independent of the disease status of the subjects.
Our findings have several implications. First, overall dysregulation of complement activation may trigger the innate immune system and thereby contribute to the overall inflammatory response that is observed in insulin resistance in fat tissue. For instance, increased production and activation of C3 by adipocytes may lead to increased infiltration of macrophages in adipose tissue in response to signaling via the C3-C3a-C3aR pathway and may thereby aggravate insulin resistance (22). Second, altered signaling via the C3a/ASP-C5L2 pathway may, in turn, alter triglyceride synthesis in the adipocyte (36). Third, activation of the classical pathway (C1q in particular) may lead to effective apoptotic cell removal (38), similar to what has been observed in aortic lesions (39). Fourth, the observed changes in complement expression in insulin-resistant individuals may not be confined to (sc) adipocytes alone but could also occur also in ectopic fat depots such as perivascular and peri/epicardial fat (40, 41). As recently reported elsewhere (42), the induction of complement C3 from perivascular fat may adversely affect adventitial fibroblast function. The multifaceted relations between complement and adipose tissue (also reviewed in Ref. 43) support the concept that altered expression of complement in insulin-resistant adipocytes is a highly relevant finding. Further investigations are needed to determine whether changes in complement expression lead to altered insulin signaling in adipocytes (or vice versa) or whether they may both be regulated by a common predecessor, e.g. low-grade inflammation. These actions may all contribute to the various vicious cycles that are involved in the disturbed adipose tissue homeostasis that underlies the metabolic derangements in insulin resistance-associated diseases.
Our current study has some limitations. First, we cannot draw conclusions on causal relations between the differentially expressed pathways in adipose cells and the metabolic derangements as seen in FCHL. Likewise, we cannot draw conclusions on causal relations between adipocyte insulin resistance and expression of the complement pathway because we did not actively intervene in insulin resistance. The fact that we did not observe any change in complement expression after a 40% lowering of triglyceride levels does, however, suggest that hypertriglyceridemia per se was not the driving force behind the observed up-regulation of complement in adipose cells. Other limitations lie in the fact that we studied gene expression, which may not fully reflect protein expression for all genes. Also, our study was restricted to the sc adipocyte fraction and does not illuminate transcriptomic changes in visceral adipocytes, which might play distinct roles in the pathophysiology of insulin resistance-associated metabolic diseases. Studies comparing complement expression in sc and omental (visceral) fat generally show higher levels of complement expression (29) and secretion (37) in visceral than in sc adipose tissue. However, recent data by Moreno-Navarrette et al. (34) suggest that these differences may be due, at least for a substantial part, to the nonadipocyte fraction of these fat depots, rather than to the adipocytes themselves. Additionally, there have been various publications that suggest an independent role for sc abdominal fat in insulin resistance-related disorders (44, 45).
In conclusion, our study provides evidence for dysregulation of the complement system in purified adipocytes from nonobese, hypertriglyceridemic subjects and further demonstrates a statistically significant positive association of complement pathway expression with markers of adipocyte insulin resistance. Dysregulation of complement activation in adipocytes are expected to significantly impact adipose tissue homeostasis, local inflammation, and lipid handling. Combined with previously published reports, our current findings suggests that expression of complement system in the adipose faction of human adipose tissue in vivo may play a role in the control of insulin resistance and low-grade inflammation with adverse metabolic sequelae, even in the absence of overt obesity.
Supplementary Material
Acknowledgments
We thank Yingnian Shen for RNA processing and microarray hybridization and Joan Stuart for data management.
This work was partially supported by National Institutes of Health Grants DK088319-01, MD000175, and HL059868, and American Heart Association Grant AHA10SDG4230068 (to S.G.).
Disclosure Summary: The authors report there are no conflicts of interest to disclose.
Footnotes
- BMI
- Body mass index
- CI
- confidence interval
- FCHL
- familial combined hyperlipidemia
- FDR
- false discovery rate
- GSEA
- Gene Set Enrichment Analysis
- HL
- hyperlipidemic
- HOMA2ir
- homeostasis model assessment 2 insulin resistance index
- IRS1
- insulin receptor substrate-1
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- NL
- normolipidemic
- qPCR
- quantitative PCR.
References
- 1. Márquez-Quiñones A, Mutch DM, Debard C, Wang P, Combes M, Roussel B, Holst C, Martinez JA, Handjieva-Darlenska T, Kalouskova P, Jebb S, Babalis D, Pfeiffer AF, Larsen TM, Astrup A, Saris WH, Mariman E, Clément K, Vidal H, Langin D, Viguerie N. 2010. Adipose tissue transcriptome reflects variations between subjects with continued weight loss and subjects regaining weight 6 mo after caloric restriction independent of energy intake. Am J Clin Nutr 92:975–984 [DOI] [PubMed] [Google Scholar]
- 2. Linder K, Arner P, Flores-Morales A, Tollet-Egnell P, Norstedt G. 2004. Differentially expressed genes in visceral or subcutaneous adipose tissue of obese men and women. J Lipid Res 45:148–154 [DOI] [PubMed] [Google Scholar]
- 3. Dixit VD. 2008. Adipose-immune interactions during obesity and caloric restriction: reciprocal mechanisms regulating immunity and health span. J Leukoc Biol 84:882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. von Eyben FE, Kroustrup JP, Larsen JF, Celis J. 2004. Comparison of gene expression in intra-abdominal and subcutaneous fat: a study of men with morbid obesity and nonobese men using microarray and proteomics. Ann NY Acad Sci 1030:508–536 [DOI] [PubMed] [Google Scholar]
- 5. Lee YH, Nair S, Rousseau E, Allison DB, Page GP, Tataranni PA, Bogardus C, Permana PA. 2005. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia 48:1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dolinková M, Dostálová I, Lacinová Z, Michalský D, Haluziková D, Mráz M, Kasalický M, Haluzik M. 2008. The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol 291:63–70 [DOI] [PubMed] [Google Scholar]
- 7. Ghosh S, Dent R, Harper ME, Gorman SA, Stuart JS, McPherson R. 2010. Gene expression profiling in whole blood identifies distinct biological pathways associated with obesity. BMC Med Genomics 3:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacLaren R, Cui W, Simard S, Cianflone K. 2008. Influence of obesity and insulin sensitivity on insulin signaling genes in human omental and subcutaneous adipose tissue. J Lipid Res 49:308–323 [DOI] [PubMed] [Google Scholar]
- 9. Bujalska IJ, Quinkler M, Tomlinson JW, Montague CT, Smith DM, Stewart PM. 2006. Expression profiling of 11beta-hydroxysteroid dehydrogenase type-1 and glucocorticoid-target genes in subcutaneous and omental human preadipocytes. J Mol Endocrinol 37:327–340 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y, Bossé Y, Marceau P, Biron S, Lebel S, Richard D, Vohl MC, Tchernof A. 2007. Gene expression variability in subcutaneous and omental adipose tissue of obese men. Gene Expr 14:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, Pérusse L, Vohl MC. 2010. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr 91:309–320 [DOI] [PubMed] [Google Scholar]
- 12. Kolehmainen M, Salopuro T, Schwab US, Kekäläinen J, Kallio P, Laaksonen DE, Pulkkinen L, Lindi VI, Sivenius K, Mager U, Siitonen N, Niskanen L, Gylling H, Rauramaa R, Uusitupa M. 2008. Weight reduction modulates expression of genes involved in extracellular matrix and cell death: the GENOBIN study. Int J Obes (Lond) 32:292–303 [DOI] [PubMed] [Google Scholar]
- 13. Clément K, Viguerie N, Poitou C, Carette C, Pelloux V, Curat CA, Sicard A, Rome S, Benis A, Zucker JD, Vidal H, Laville M, Barsh GS, Basdevant A, Stich V, Cancello R, Langin D. 2004. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J 18:1657–1669 [DOI] [PubMed] [Google Scholar]
- 14. Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG. 1973. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest 52:1544–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van der Kallen CJ, Voors-Pette C, Bouwman FG, Keizer HA, Lu JY, van de Hulst RR, Bianchi R, Janssen MJ, Keulen ET, Boeckx WD, Rotter JI, de Bruin TW. 2002. Evidence of insulin resistant lipid metabolism in adipose tissue in familial combined hyperlipidemia, but not type 2 diabetes mellitus. Atherosclerosis 164:337–346 [DOI] [PubMed] [Google Scholar]
- 16. Bredie SJ, Tack CJ, Smits P, Stalenhoef AF. 1997. Nonobese patients with familial combined hyperlipidemia are insulin resistant compared with their nonaffected relatives. Arterioscler Thromb Vasc Biol 17:1465–1471 [PubMed] [Google Scholar]
- 17. Brouwers MC, van der Kallen CJ, Schaper NC, van Greevenbroek MM, Stehouwer CD. 2010. Five-year incidence of type 2 diabetes mellitus in patients with familial combined hyperlipidaemia. Neth J Med 68:163–167 [PubMed] [Google Scholar]
- 18. Keulen ET, Kruijshoop M, Schaper NC, Hoeks AP, de Bruin TW. 2002. Increased intima-media thickness in familial combined hyperlipidemia associated with apolipoprotein B. Arterioscler Thromb Vasc Biol 22:283–288 [DOI] [PubMed] [Google Scholar]
- 19. Prifti E, Zucker JD, Clement K, Henegar C. 2008. FunNet: an integrative tool for exploring transcriptional interactions. Bioinformatics 24:2636–2638 [DOI] [PubMed] [Google Scholar]
- 20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henegar C, Tordjman J, Achard V, Lacasa D, Cremer I, Guerre-Millo M, Poitou C, Basdevant A, Stich V, Viguerie N, Langin D, Bedossa P, Zucker JD, Clement K. 2008. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol 9:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mamane Y, Chung Chan C, Lavallee G, Morin N, Xu LJ, Huang J, Gordon R, Thomas W, Lamb J, Schadt EE, Kennedy BP, Mancini JA. 2009. The C3a anaphylatoxin receptor is a key mediator of insulin resistance and functions by modulating adipose tissue macrophage infiltration and activation. Diabetes 58:2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baldo A, Sniderman AD, St. Luce S, Avramoglu RK, Maslowska M, Hoang B, Monge JC, Bell A, Mulay S, Cianflone K. 1993. The adipsin-acylation stimulating protein system and regulation of intracellular triglyceride synthesis. J Clin Invest 92:1543–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engström G, Hedblad B, Janzon L, Lindgärde F. 2005. Weight gain in relation to plasma levels of complement factor 3: results from a population-based cohort study. Diabetologia 48:2525–2531 [DOI] [PubMed] [Google Scholar]
- 25. Engström G, Hedblad B, Eriksson KF, Janzon L, Lindgärde F. 2005. Complement C3 is a risk factor for the development of diabetes: a population-based cohort study. Diabetes 54:570–575 [DOI] [PubMed] [Google Scholar]
- 26. Muscari A, Bozzoli C, Puddu GM, Sangiorgi Z, Dormi A, Rovinetti C, Descovich GC, Puddu P. 1995. Association of serum C3 levels with the risk of myocardial infarction. Am J Med 98:357–364 [DOI] [PubMed] [Google Scholar]
- 27. Jager J, Grémeaux T, Cormont M, Le Marchand-Brustel Y, Tanti JF. 2007. Interleukin-1β-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology 148:241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Serrano R, Villar M, Martínez C, Carrascosa JM, Gallardo N, Andrés A. 2005. Differential gene expression of insulin receptor isoforms A and B and insulin receptor substrates 1, 2 and 3 in rat tissues: modulation by aging and differentiation in rat adipose tissue. J Mol Endocrinol 34:153–161 [DOI] [PubMed] [Google Scholar]
- 29. Gabrielsson BG, Johansson JM, Lönn M, Jernås M, Olbers T, Peltonen M, Larsson I, Lönn L, Sjöström L, Carlsson B, Carlsson LM. 2003. High expression of complement components in omental adipose tissue in obese men. Obes Res 11:699–708 [DOI] [PubMed] [Google Scholar]
- 30. Koistinen HA, Vidal H, Karonen SL, Dusserre E, Vallier P, Koivisto VA, Ebeling P. 2001. Plasma acylation stimulating protein concentration and subcutaneous adipose tissue C3 mRNA expression in nondiabetic and type 2 diabetic men. Arterioscler Thromb Vasc Biol 21:1034–1039 [DOI] [PubMed] [Google Scholar]
- 31. Ylitalo K, Pajukanta P, Meri S, Cantor RM, Mero-Matikainen N, Vakkilainen J, Nuotio I, Taskinen MR. 2001. Serum C3 but not plasma acylation-stimulating protein is elevated in Finnish patients with familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol 21:838–843 [DOI] [PubMed] [Google Scholar]
- 32. Choy LN, Rosen BS, Spiegelman BM. 1992. Adipsin and an endogenous pathway of complement from adipose cells. J Biol Chem 267:12736–12741 [PubMed] [Google Scholar]
- 33. Zhang J, Wright W, Bernlohr DA, Cushman SW, Chen X. 2007. Alterations of the classic pathway of complement in adipose tissue of obesity and insulin resistance. Am J Physiol Endocrinol Metab 292:E1433–E1440 [DOI] [PubMed] [Google Scholar]
- 34. Moreno-Navarrete JM, Martínez-Barricarte R, Catalán V, Sabater M, Gómez-Ambrosi J, Ortega FJ, Ricart W, Blüher M, Frühbeck G, Rodríguez de Cordoba S, Fernández-Real JM. 2010. Complement factor H is expressed in adipose tissue in association with insulin resistance. Diabetes 59:200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Corominola H, Conner LJ, Beavers LS, Gadski RA, Johnson D, Caro JF, Rafaeloff-Phail R. 2001. Identification of novel genes differentially expressed in omental fat of obese subjects and obese type 2 diabetic patients. Diabetes 50:2822–2830 [DOI] [PubMed] [Google Scholar]
- 36. Kalant D, MacLaren R, Cui W, Samanta R, Monk PN, Laporte SA, Cianflone K. 2005. C5L2 is a functional receptor for acylation-stimulating protein. J Biol Chem 280:23936–23944 [DOI] [PubMed] [Google Scholar]
- 37. Hocking SL, Wu LE, Guilhaus M, Chisholm DJ, James DE. 2010. Intrinsic depot-specific differences in the secretome of adipose tissue, preadipocytes, and adipose tissue-derived microvascular endothelial cells. Diabetes 59:3008–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fraser DA, Laust AK, Nelson EL, Tenner AJ. 2009. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J Immunol 183:6175–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhatia VK, Yun S, Leung V, Grimsditch DC, Benson GM, Botto MB, Boyle JJ, Haskard DO. 2007. Complement C1q reduces early atherosclerosis in low-density lipoprotein receptor-deficient mice. Am J Pathol 170:416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eringa EC, Bakker W, Smulders YM, Serné EH, Yudkin JS, Stehouwer CD. 2007. Regulation of vascular function and insulin sensitivity by adipose tissue: focus on perivascular adipose tissue. Microcirculation 14:389–402 [DOI] [PubMed] [Google Scholar]
- 41. Mazurek T, Zhang L, Zalewski A, Mannion JD, Diehl JT, Arafat H, Sarov-Blat L, O'Brien S, Keiper EA, Johnson AG, Martin J, Goldstein BJ, Shi Y. 2003. Human epicardial adipose tissue is a source of inflammatory mediators. Circulation 108:2460–2466 [DOI] [PubMed] [Google Scholar]
- 42. Ruan CC, Zhu DL, Chen QZ, Chen J, Guo SJ, Li XD, Gao PJ. 2010. Perivascular adipose tissue-derived complement 3 is required for adventitial fibroblast functions and adventitial remodeling in deoxycorticosterone acetate-salt hypertensive rats. Arterioscler Thromb Vasc Biol 30:2568–2574 [DOI] [PubMed] [Google Scholar]
- 43. Pattrick M, Luckett J, Yue L, Stover C. 2009. Dual role of complement in adipose tissue. Mol Immunol 46:755–760 [DOI] [PubMed] [Google Scholar]
- 44. Frederiksen L, Nielsen TL, Wraae K, Hagen C, Frystyk J, Flyvbjerg A, Brixen K, Andersen M. 2009. Subcutaneous rather than visceral adipose tissue is associated with adiponectin levels and insulin resistance in young men. J Clin Endocrinol Metab 94:4010–4015 [DOI] [PubMed] [Google Scholar]
- 45. Goodpaster BH, Thaete FL, Simoneau JA, Kelley DE. 1997. Subcutaneous abdominal fat and thigh muscle composition predict insulin sensitivity independently of visceral fat. Diabetes 46:1579–1585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.