Abstract
During the last couple decades, we have significantly advanced our understanding of mechanisms underlying the development of pancreatic ductural adenocarcinoma (PDAC). In the late 1990s into the early 2000s, a model of PDAC development and progression was developed as a multi-step process associated with the accumulation of somatic mutations. The correlation and association of these particular genetic aberrations with the establishment and progression of PDAC has revolutionized our understanding of this process. However, this model leaves out other molecular events involved in PDAC pathogenesis that contribute to its development and maintenance, specifically those being epigenetic events. Thus, a new model considering the new scientific paradigms of epigenetics will provide a more comprehensive and useful framework for understanding the pathophysiological mechanisms underlying this disease. Epigenetics is defined as the type of inheritance not based on a particular DNA sequence but rather traits that are passed to the next generation via DNA and histone modifications as well as microRNA-dependent mechanisms. Key tumor suppressors that are well established to play a role in PDAC may be altered through hypermethylation, and oncogenes can be upregulated secondary to permissive histone modifications. Factors involved in tumor invasiveness can be aberrantly expressed through dysregulated microRNAs. A noteworthy characteristic of epigenetic-based inheritance is its reversibility, which is in contrast to the stable nature of DNA sequence-based alterations. Given this nature of epigenetic alterations, it becomes imperative that our understanding of epigenetic-based events promoting and maintain PDAC continues to grow.
1. Introduction
Genetics refers to the expression and heredity of the DNA sequence. The transfer of sequence information from DNA to RNA to protein via the genetic code became known as the central dogma of molecular biology. The DNA sequence is static throughout life, with mechanisms in existence to maintain this stability as much as possible. However, changes to this sequence do occur, whether they are more subtle stochastic changes that occur during replication and development or more imminently detrimental changes that may occur due to endogenous or environmental stimuli[1; 2; 3]. While the DNA sequences themselves are relatively static throughout life, other aspects of what constitutes the DNA are highly dynamic. The DNA sequence itself is rarely seen naked in cells. Rather, DNA exists in cells as a complex structure of DNA and proteins that constitute what is known as chromatin. As these characteristics have been identified, the term epigenetics has arisen[4]. Epigenetics is any heritable genomic mechanism mediated through changes in chromatin structure and DNA methylation that are unrelated to changes in the DNA sequence. Being a relatively new field of molecular biology, the definition of epigenetics was highly debated among scientists at the 2004 69th Cold Spring Harbor Symposium[5; 6]. For every scientist, the idea of epigenetics as a mode of inheritance which works in parallel to genetics was understood, but given the various mechanisms that comprised the field there was controversy over how exactly to define it. Subsequently, the term epigenetics, first used by Waddington in 1942, was defined as any heritable trait not involving the DNA sequence that influences the phenotype of a developing organism[7]. Even more simply, epigenetics can be defined as the idea that all cells have the same genotype but have different phenotypes that persist through many generations[5; 8].
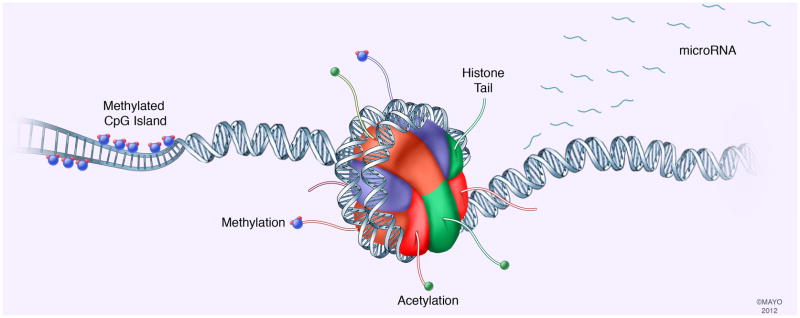
Although the modern definition of epigenetics was not coined until 2004, epigenetics has been engrained in scientific research for the last century. Before the 1950’s, the word epigenetics was used to describe all the developmental effects leading from the fertilized egg to a fully developed organism[7]. This concept actually traces its origin back to the 19th century. After the development of the mature organism, it was not clear to scientists at the time if each cell retained the full complement of DNA that was present in the fertilized egg. With the elucidation of DNA structure by Watson and Crick in 1953, the field of science began to slowly shift from the idea of DNA, not protein, being the carrier of genetic information due to experiments performed by Avery, MacLeod and McCarty, which showed DNA to be the “transformative factor”[9]. In 1970, it was demonstrated that the somatic cells possess genes other than those necessary for development by introducing a somatic cell nucleus into an enucleated egg, which lead to embryogenesis[7]. Once it became clear that DNA was not deleted from somatic cells, the question remained as to exactly how these cells expressed such different phenotypes if they were retaining the same DNA. If the DNA sequence is strongly conserved and stable, there must be other features linked to the DNA sequence that are more dynamic and can account for these alterations. Epigenetics and the mechanisms that constitute this field are indeed the links that account for the dynamic variation not explained by the DNA sequence itself. There are several well-known epigenetic mechanisms, including DNA methylation, histone modification, and microRNAs (fig. 1). These epigenetic modifications affect the genome by either inducing or suppressing gene expression, thus resulting in changing phenotypes. Most importantly, the epigenetic mechanisms regulating gene expression are not only applicable to normal cellular development and maintenance but they can be responsible for deregulation of gene expression that is associated with diseased cellular phenotypes. In particular, deregulation of epigenetic mechanisms can contribute to cancer development[10], in particular PDAC, is an important example of epigenetic-based influence on development and progression. While genetic-based drivers of PDAC have been well studied, they do not account for all of the phenotypic and molecular alterations demonstrated by PDAC cells[11]. Some tumor suppressor and oncogenes involved with PDAC pathogenesis have aberrant expression and function not due to genetic causes within these genes but rather due to the effects of epigenetic mechanisms regulating their expression. Tumor suppressors, such as p16, can be silenced through the epigenetic hypermethylation of its promoter, but not a change in sequence[12]. The oncogenes C-MYC can be upregulated secondary to histone modifications[13; 14]. Tumor invasiveness, metastatic potential, and maintenance of stem cell phenotypes are at least in part regulated by microRNAs and not from genetic aberrations in their genes. While this review provides a mechanistic view of each of the example epigenetic processes, the processes do not necessarily operate independently. For example, as will be described further, the methylase component of the PRC2 silencing complex is known to interact with DNA methyltransferases which may further aid in gene silencing[15]. Thus, PDAC initiation and progression is the result of a heterogenous and dynamic combination of both genetic and epigenetic mechanisms. With the identification of epigenetic alterations seen in early PanIN lesions through the development of PDAC, there implies an inherent complexity in epigenetic changes that occurs in parallel to genetic changes. This brings into question, as do the progression of genetic changes that are seen, what changes occur first. At this time, much more research is necessary to advance our understanding of these mechanisms and their potential for manipulation. This review will focus on three specific epigenetic mechanisms, DNA methylation, histone modifications and miRNAs, and their involvement in PDAC pathogenesis.
Fig. 1. Summary of major epigenetic mechanisms mediating PDAC pathogenesis.
This review focuses on three specific epigenetic mechanisms to alter gene expression: DNA methylation, histone-based epigenetics, and microRNAs.
2. DNA-based epigenetics
DNA methylation is one of the best understood epigenetic modifications for transcriptional regulation. DNA methylation is often associated with gene repression. DNA methylation commonly occurs on dinucleotide CpGs, where cytosines precede guanines. The process of DNA methylation entails the addition of a methyl group to the number 5 carbon of the cytosine pyrimidine ring to form 5-methylcytosine, which ultimately silences gene expression. This modification can be inherited and passed through generations to daughter cells or it may be a de novo modification. The addition of the methyl group is often found in CpG-islands in the promoter regulatory regions of many genes. This methylation interferes with the binding of transcription factors and simultaneously attracts methyl-CpG- binding domain proteins (MBDs) to initiate chromatin compaction and gene silencing[16]. A variety of enzymes called DNA methyltransferases (DNMTs) are responsible for maintenance and addition of these methylation patterns. However, not all DNMTs have the exact same function. DNMT1 is thought to function as a maintenance methyltransferase in that it is responsible for maintaining methylation patterns from the parent strand of DNA to the newly synthesized daughter strand[17; 18]. Alternatively, DNMT3a and DNMT3b are thought to be responsible for applying de novo methyl groups to DNA[19]. These enzymes do require the aid of a co-factor to function, namely the DNA methyltransferase 3-like protein (DNMT3L), and the roles of this co-factor continue to be elucidated[20; 21; 22]. Demethylation of DNA is not as straightforward as methyl groups are not easily removed with only speculation regarding the existence and role of DNA demethylases. Loss of DNA methylation patterns appears to occur through the loss of DNMT activity or through DNA repair mechanisms such as the use of base excision repair machinery[23; 24]. More recently, an additional modification, 5-hydroxymethylcytosine has been proposed to serve as an intermediate in the demethylation process. Certain members of the TET family of proteins have been shown to mediate the conversion of 5-mehtylcytosine to 5-hydroxymethylcytosine in mammalian cells[25]. It has also been shown that some 5-methylcytosine-binding proteins do not bind to 5-hydroxymethylcytosines, and that DNMT1 does not recognize the hydroxy-group resulting in a loss of maintenance of methylation patterns[26; 27; 28]. Additionally, inhibitors of DNMTs such as 5-aza-2′-deoxycytidine have been used in the research setting to further understand the process and mechanism of DNA methylation. Studies performed by Sato and colleagues used this inhibitor to evaluate genes that are aberrantly methylated in PDAC cell lines and found many genes that were greatly induced (5-fold or more) with the addition of this inhibitor[29]. Some of these genes, such as NPTX2, SARP2 and CLDN5, were confirmed to have aberrant (increased) methylation in the PDAC samples evaluated. Yet it is important to note that in cancers, hypermethylation of promoters is not the only mechanism of gene dysregulation. Hypomethylation occurring in the promoters as well as other genomic maintenance regions can occur to result in chromosomal and genomic changes. In a separate study, Sato and colleagues also looked at the importance of hypomethylation[30]. They found that 7 genes that were overexpressed in PDAC samples and cell lines but not in normal pancreatic duct samples had a high degree of hypomethylation[30]. These findings show the importance of not only hypermethylation in the disease process but also hypomethylation with is associated with the overexpression of affected genes. Understanding the role of DNA methylation is noteworthy as it normally has a significant physiological importance. These normal examples of DNA methylation include genomic imprinting to ensure monoallelic expression and hypermethylation of repetitive genomic sequences to prevent chromosomal instability, translocations, and gene disruption caused by the reactivation of transposable DNA sequences[31]. However, during tumorigenesis, aberrant DNA methylation can modulate the cancer phenotype.
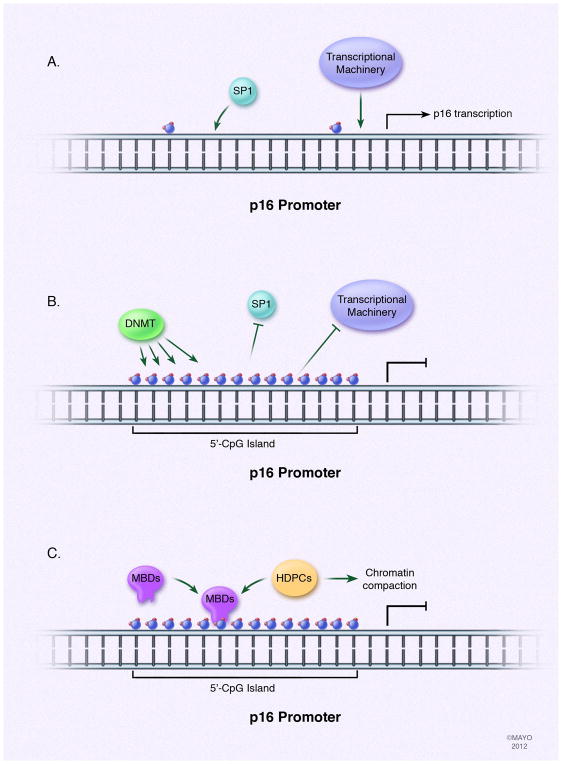
In PDAC, DNA methylation has been known for a long time as a mechanism to inactivate tumor suppressor genes. Specifically, the inactivation of the p16 tumor suppressor gene promoter via methylation in PDAC has been well established. The importance of this process is significant as greater than 95% of PDACs have a loss of p16[12]. The p16 protein inhibits the binding of the D-family cyclins to their cyclin-dependent kinase (CDK) partners, and the loss of p16 protein can result in increased phosphorylation of Retinoblastoma [12] and a subsequent progression through G1 phase into S phase of the cell cycle. Methylation of the promoter of the 5′-CpG island in the p16 gene is one of the most common mechanisms of p16 inactivation in PDAC, emphasizing the importance of an epigenetic mechanism in modulating tumor suppressor function in this disease[12]. The result of this silencing is an interference of binding of permissive transcription factors, such as SP1, and the recruitment of MBDs that can recruit other proteins like HDACs and lead to chromatin compaction [33](fig. 2). The identification of genes affected in this manner had been slow to identify as initial methodologies only provided insights at the single gene level. However, recent developments in methodologies have advanced enough to allow for genome-wide scale methylation analysis. Advantage can derive by combining both methodologies since methylation analysis of a single gene is a specific candidate gene approach, while the genome-wide analysis possesses power in its unbiased approach. More recently, the use of large-scale methylation analysis has allowed the identification of other genes affected by aberrant methylation, such as the PDAC candidate tumor suppressor KLF11[34; 35]. However, silencing of a gene having a tumor suppressor function is not the only example of the significance of aberrant methylation in PDAC. The loss of methylation of a normally silenced promoter in pancreatic cells can lead to its misexpression. An example of this is the gene encoding the hematopoietic-specific guanine nucleotide exchange factor, VAV1. The promoter of VAV1 is demethylated in PDACs, rendering it active and responsible for promoting the activity of oncogenic KRAS to aid in cellular proliferation in PDACs[36]. These examples of aberrant methylation of genes in PDAC are important to understand because they may contribute to the initiation and progression of the disease, combined with the genetic alterations associated with the progression model of PDAC.
Fig. 2. Aberrant DNA methylation contributes to PDAC pathogenesis.
DNA methylation occurs on dinucleotide CpGs, primarily in islands located within promoter regions of genes. DNA methyltransferases are responsible for applying the methyl groups. The consequence of this process is the interference with binding of transcription factors to the DNA, such as SP-1 in the case of p16, and the recruitment of methyl-CpG-binding domain proteins (MBDs) that participate in recruitment of other proteins, such as HDACs, to facilitate chromatin compaction leading to gene repression. In the case of PDAC, the tumor suppressor gene p16 is often found silenced through hypermethylation of the 5′-CpG island of its promoter.
Current evidence supports the idea that aberrant methylation occurs very early during the histopathological progression of PDAC. Using a specific gene candidate approach, Rosty and colleagues demonstrated that PanIN lesions in patients with chronic pancreatitis show loss of p16 expression, suggesting that this alteration may contribute to the predisposition of patients with chronic pancreatitis to develop PDAC[37]. Interestingly, in a large-scale methylation analysis with subsequent validation via methylation-specific PCR, Sato and colleagues analyzed DNA samples from 65 PanIN lesions for methylation status of eight genes identified prior by a microarray approach as aberrantly hypermethylated in invasive PDAC[29]. Of the PanIN lesions examined in this study, methylation of any of these genes was identified in 68%, and the earliest lesions, PanIN-1A lesions, aberrant methylation was present in approximately 70% of the cases[29]. Among the genes analyzed, methylation prevalence increased from PanIN-1 to PanIN-2 for NPTX2, and from PanIN-2 to PanIN-3 for SARP2, Reprimo, and LHX1[31]. The most striking result from both of these studies is that aberrant CpG island hypermethylation begins in early stages of PanINs and its prevalence progressively increases during neoplastic progression including the above mentioned genes [31]. A more recent study by Goggins’ group using methylated CpG island amplification followed by microarray analysis identified 1,658 known loci that were differentially methylated in PDAC compared with normal pancreas. A subset of these aberrantly methylated loci show differentially gene expression, thus further supporting the functional role of these DNA modifications in the regulation of gene expression [32].
It is clear that aberrant DNA methylation is involved in various cancers, and the importance of this process has been well established for PDAC. Evidence for methylation in tumor suppressor gene silencing continues to reconfirm its clear role in the progression of PDAC. Initially, some genes, such as p16, were believed to be methylated solely in the malignant pancreatic lesions. However, current and ever-growing evidence indicates that methylation also occurs earlier at the preneoplastic stage. Furthermore, DNA methylation can act in concert with other epigenetic mechanisms mediating the pathogenesis of this disease, such as histone-based modifications.
3. Histone-based epigenetics
Histone-based alterations serve as one of the mechanisms of epigenetics, which determines the epigenetic inheritance of either a phenotypic trait from the germ line (imprinting) or from one somatic cell to its daughter. In terms of transcription, it was thought that histones and nucleosomes were rich solely in heterochromatin, which is transcriptionally silent, and relatively poor in euchromatin, which is transcriptionally active. However, these states can be interchanged, meaning that chromatin is more dynamic than initially speculated. Chromatin dynamics is regulated by several factors and signaling events that form the basis of the histone code. To understand the role histone-based epigenetics plays in normal cellular function as well as in the diseased state, particularly in the context of PDAC, insights into histone architecture is necessary.
Histones are comprised of an octamer of proteins that include dimers of H2A, H2B, H3, and H4[38; 39]. While each of these core proteins has a role in histone and chromatin dynamics, including the potential for variant protein integration, it is histone H3 that is one of the more studied and modified of the core subunits. The first 24 amino acids of histone H3, known as the histone tail, are almost identical in most organisms[38; 39]. The histone H3 tail contains several serine (S), threonine (T), and tyrosine (Y) residues that can undergo phosphorylation in addition to other residues, such as lysine (K) and arginine (R) that can be extensively modified by methylation, acetylation, ubiquitination, and sumoylation[40]. In fact, the lysines of the histone H3 tail even have the potential to be in different states of methylation, namely mono-, di-, and tri-methylated, and each of these states of methylation codes for different transcriptional states in a certain genomic region. Although the knowledge about modifications made to the histone tails is much more extensive, it is important to note that modifications to the histones away from the tail and in the nucleosomal regions associated with DNA are also found. These modifications participate in histone mobility and stability, which can also contribute to the cancer phenotype[41; 42]. These histone modifications have come to be known as “marks” because in many cases they are utilized as clues for epigenetics. This has led to the formulation of the histone code hypothesis, which utilizes the type, location, and combination of histone marks to predict whether a gene may be expressed or silenced under a particular set of circumstances[40; 43]. Included within the histone code hypothesis are not only the marks themselves, but the proteins that serve to apply the marks, or writers, those that interpret the marks, or readers, and those that remove the marks, or erasers[40; 43]. Each of these players coordinates all facets of the histone code hypothesis and contributes to the dynamic properties dictating histone regulation. The writers, readers and erasers have been found to have defective function in cancerous cellular states, including PDAC [46; 47; 48; 49; 50]. In fact, aberrant expression and genetic mutations in these chromatin remodelers including p300, HDACs, Brg1, PBRM1, and others have been found in PDAC as well as other pancreatic tumors like IPMN [46; 47; 48; 49; 50]. As there are numerous modifications that may occur to these histones, the remainder of this section will focus on two major mechanisms that have been studied in regards to PDAC: histone acetylation and histone methylation.
In the case of histone acetylation, this process occurs via histone acetyltransferases (HATs), such as CREB binding protein (CBP), p300, and p300/CBP-associated factor (P/CAF), to result in gene expression activation. In contrast to the activity of HATs, deacetylation is mediated by histone deacetylases (HDACs) to induce gene silencing. Together, these enzymes provide a fine-tuned, highly dynamic mechanism, which upon alteration, has the possibility to cause the activation of oncogenic pathways and the silencing of tumor suppressors. However, differently from other epigenetic regulators, such as the histone methylation associated polycomb complexes and HP1 described further below, HATs and HDACs mediate short-term responses [51; 52; 53; 54].
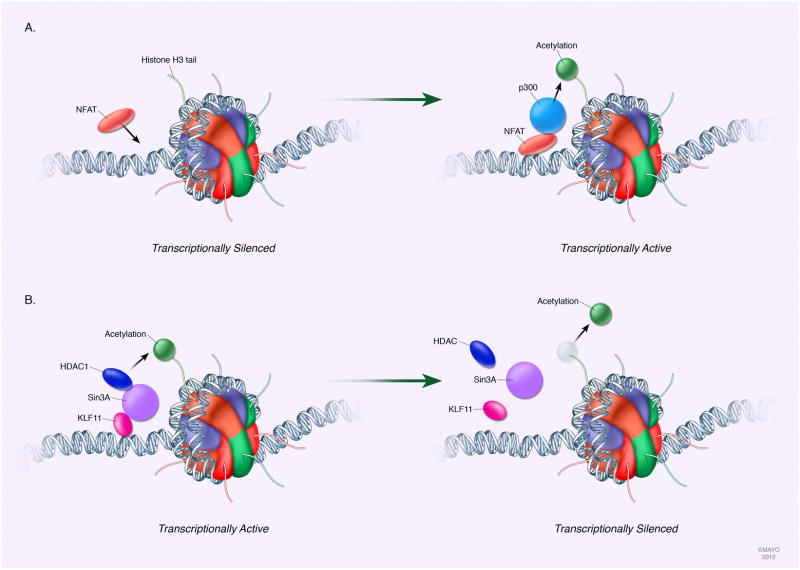
Although HATs have been implicated in contributing to PDAC carcinogenesis, sparse knowledge about the function of individual HATs currently exists, which might be attributed to the redundancy within this enzyme family. The role of p300 in the NFAT- and GLI-pathways has been clearly described. Often, the aberrant activity of p300 in PDAC is found to be influenced by specific transcription factors, such as nuclear factor of activated T cells (NFAT) and GLI3[14; 54; 55]. NFAT, a calcium/calcineurin responsive transcription factor, is able to bind to the promoter of target genes with other factors such as chromatin remodeling proteins. In PDAC cells for instance, NFAT promotes cell growth in vitro and in vivo through the transcriptional induction of C-MYC[13]. Mechanistically, NFATc1 binds to the serum responsive element of the C-MYC promoter where it then recruits p300 to induce local hyperacetylation of the chromatin rendering it transcriptionally activated (fig. 3a)[14]. Importantly, this event is required for K-Ras-dependent recruitment of the oncogenic transcription factor Elk-1 and the resulting induction of C-MYC expression following stimulation of the Ca2+/calcineurin pathway[14]. Disruption of this transcription complex through inhibition of the calcineurin/NFAT signaling prevents both C-MYC promoter transactivation and PDAC growth stimulation in vitro and in vivo[14]. These findings are clinically relevant to humans with PDAC. It has been shown, for instance, that NFATc1 and NFATc2 are induced and transcriptionally active in early PanIN-2 precursor lesions and are highly expressed in the majority of advanced PDAC[13; 55]. In addition to the calcineurin/NFAT pathway, p300 is also recruited to target promoters by the transcription factor GLI3, an effector component of the Hedgehog pathway that was generally thought to function as a transcriptional repressor. In a model of KRAS-induced autophagy in PDAC cell lines, GLI3 and p300 bound to the promoter of the autophagy gene vacuole membrane protein 1 (VMP1)[56]. This interaction was found to be necessary to allow complete transcriptional activation of VMP1 and thereby mediate a subsequent cancer autophagy phenotype[56]. However, histone acetylation status in PDAC is not affected solely by HATs but also by HDACs.
Fig. 3. Histone-based modifications affect gene expression in PDAC.
A particular set of histone modifications cooperate to mark transcriptionally active or silent chromatin. While histone marks are extensive and highly dynamic with numerous associated writers, readers, and erasers, the most studied alterations related to PDAC include those of histone H3 lysine acetylation or methylation. Recruitment of modulating enzymes is often aberrant in PDACs, such as with HATs and HDACs. A specific example of faulty HAT recruitment in PDAC is p300. Induction of the transcription factor NFAT can bind to target gene promoters where it recruits p300 to hyperacetylate the promoter and render the gene transcriptionally active. Similarly, HDAC recruitment to a target gene promoter can be disrupted. An example of this is the loss of KLF11 in PDAC, a transcription factor that functions to recruit a Sin3a-HDAC complex to promoters targeted for silencing. Loss of normal silencing of target genes in this manner contributes to PDAC pathogenesis.
One example of HDAC involvement with PDAC is the Sin3a–HDAC system. Pancreatic cells express three different Sin3 proteins that are recruited by tumor suppressors proteins and require binding to the Sin3a–HDAC complex to perform their function (fig. 3b)[57; 58]. For example, KLF11, a ppel-like family protein member, was found to function as a suppressor of cell growth and to be downregulated in various cancers, including PDAC[34; 35; 58]. Furthermore, KLF11 contains a SID domain that interacts with Sin3a to mediate its repressive activity through the Sin3a-HDAC complex. Thus, this system is both active and important for antagonizing PDAC carcinogenesis, and the finding that decreased KLF11 in PDAC may contribute to dysregulated cell growth emphasizes the importance of this HDAC complex. Although many other gene-silencing complexes exist in mammalian cells and have been shown to participate in several cancer-associated functions in other organs, studies on these proteins in the pancreas remain underrepresented. Yet it is clear that HDAC activity plays a clinically relevant role in PDAC.
HDACs1, 2, 3, and 7 are overexpressed in PDAC[32; 59; 60; 61; 62]. Consistently, HDAC activity is higher in PDAC tissue samples compared to chronic pancreatitis and normal pancreas. One study has shown that high HDAC7 expression can discriminate PDACs from other pancreatic tumors, such as serous cystadenoma and intraductal papillar mucinous tumor (IMPN) or chronic pancreatitis[59]. Furthermore, HDAC activity and prognosis associated with PDAC has been shown[32; 54]. High HDAC2 expression correlates with poorly differentiated PDACs[32]. This correlation is has functional relevance as well. The HDAC2 gene was demonstrated to be induced by C-MYC, linking HDAC2 to oncogenic programs driven by C-MYC[63]. Other relevant tumor processes ranging from proliferation, apoptosis and tumor maintenance to epithelial to mesenchymal transition (EMT). Downregulation of E-cadherin, an adherens junction protein, is an important mechanism of EMT. In models of PDAC, HDAC1 and/or HDAC2 containing co-repressor complexes are involved with the repression of the E-cadherin gene. EMT-inducing transcription factors, such as SNAIL and ZEB1, are able to interact with and recruit HDAC1 and HDAC2 to the proximal E-cadherin promoter to repress gene activity. Importantly, the recruitment of HDAC1 and HDAC2 to this gene has been detected in human tissue samples of PDAC, further emphasizing the importance of the functionality of HDAC dysregulation[64; 65]. Thus, it is clear the HDACs play an important role in the maintenance of the proper balance of chromatin marks on a given promoter. However, other aspects of the histone code also are likely players in PDAC development. In particular, evidence directs attention to changes in histone methylation marks.
As mentioned at the beginning of this section, histone methylation is an important component of the histone code that is mediated by histone methylases and histone methyltransferases. Additionally, it is important to note that is appears there is a greater specificity of the methylation machinery for certain histone lysines than for the acetylation machinery. Polycomb proteins silence gene expression by specifically methylating histone H3, on K27 [15]. At the core of this pathway, polycomb group (PcG) proteins act via the stepwise recruitment of the histone H3 K27 methylase containing polycomb repressive complex 2 (PRC2) to chromatin. Subsequently, the trimethyl-K27-H3 mark deposited by PRC2 recruits the PRC1 complex, thereby completing the gene silencing complex formation. The methylase activity of the PCR2 complex involves the enzyme enhancer of zeste homolog 2 (EZH2)[15; 66; 67; 68]. The role of polycomb proteins in PDAC is an emerging area of research. One of the outcomes of aberrant polycomb regulation may be related to the silencing of the p16 gene, which could occur prior to DNA methylation via altered recruitment of members of this family to the p16 promoter sequence[69]. In recent studies, EZH2 was found to physically and functionally interact with all three human DNMTs with co-dependency of certain target gene silencing from both EZH2 and DNMTs[68]. Therefore, the presence of polycomb proteins on the p16 promoter could recruit DNA methylases, which then further inactivate the expression of p16 via DNA methylation[69]. Whether histone H3-K27 methylation and recruitment of DNMT leads to DNA methylation and the permanent silencing of the gene or if these mechanisms of p16 inactivation are independent of each other remains to be discovered, but the interaction between EZH2 and DNMTs is an example of the cross- talk that can exist between two epigenetic mechanisms. Adding to its functional importance, EZH2 has been specifically implicated in PDAC clinically.
Studies have demonstrated that loss of trimethylation at K27 of histone H3, which is a mark applied by EZH2, is a predictor of poor outcome in PDACs[70]. In fact, together with tumor size and lymph node status, the level of trimethyl-K27-H3 was found to have a strong and independent prognostic influence in PDAC[70]. In another recent study, nuclear accumulation of EZH2 was identified as a hallmark of poorly differentiated PDAC, and this nuclear overexpression of EZH2 contributes to PDAC cell proliferation, suggesting EZH2 as a potential therapeutic target for the treatment of PDAC[66; 71]. Thus, although these initial studies inspire much more to learn about the composition and function of polycomb complexes in PDAC, the association of this pathway with poor survival of patients affected by this disease makes this area of research one of paramount importance. Despite the emerging importance of EZH2, other proteins involved with histone methylation marks are also being found to have relevance.
Another example of the importance of histone methylation in relation to PDAC is the formation of heterochromatin through the protein HP1[55; 72]. HP1 binds methylated K9 of histone H3, causing transcriptional repression. This occurs through the N-terminal chromodomain of HP1, while the highly related C-terminal chromoshadow domain allows for dimerization of these HP1 molecules and subsequently serves as a docking site for various factors[53]. To mediate gene silencing via the formation of heterochromatin, HP1 must interact with two different H3-K9 histone methylases, G9a (EuHMTase-2) and Suv39h1. These methylases work in concert with HP1 in a circular manner to form silenced chromatin. When either of the methylases adds methyl groups to K9, this, in turn, forms an HP1 docking site on chromatin. Since HP1 also recruits the methylases, this cycle repeats, and the HP1– methylase pair can spread the formation of silenced chromatin to adjacent nucleosomes, causing long-term silencing of entire genes[53]. While the function of HP1 proteins in both normal and cancerous cell states is emerging, there is evidence that HP1 is involved in the pathogenesis of PDAC.
One specific example of how the methyl-K9 H3-HP1 type of chromatin dynamics can impact on the field of PDAC is the regulation of mucin 1 (MUC1) expression[73]. In PDAC tumors, the MUC1 protein has been detected in >90% of samples examined via immunohistochemistry, as well as in the pancreatic secretions of PDAC patients by comprehensive proteomic analysis and in many PDAC cell lines[74]. The sialylated form of MUC1 is overexpressed in invading and metastatic PDAC cells, but absent in normal pancreas, cases of chronic pancreatitis, and pancreatic ductal hyperplasia[75]. Studies have recently demonstrated that a mechanism responsible for changes in the expression of MUC1 is regulated by DNA methylation and histone H3 lysine 9 modification, which is bound by HP1, on the MUC1 promoter[73]. MUC1-negative cancer cell lines correlated with high H3 methyl-K9 levels, while MUC1-positive cell lines had low levels of this epigenetic mark[73]. Overall, all of these findings mentioned in this section clearly indicate the importance of histone-based mechanisms of epigenetics in PDAC. This also supports the argument that continued investigations into these and other related mechanisms are necessary to further help the development of therapeutic options for PDAC.
4. MicroRNA-based epigenetics
MicroRNAs (miRNAs) are a class of small non-protein coding RNAs which participate in post-transcriptional control of gene expression in eukaryotic organisms. Currently more than 1500 Homo sapiens miRNAs (miRBase, Release 18) have been identified which are believed to influence and control the expression of a large part of the cellular proteome. In fact, miRNAs are currently predicted to control the activity of approximately 30% of all protein-coding genes in mammals. They obstruct the synthesis of proteins via a process involving their pairing to the mRNAs of protein-coding genes to direct posttranscriptional repression, but miRNAs are typically generated by transcription of long precursors and the processing into their final effector form involves many steps[76].
In the nucleus primary miRNA transcripts (pri-miRNAs) are cleaved by the enzymes DROSHA and PASHA to form pre-miRNA precursors with a characteristic hairpin structure. Following transport into the cytoplasm, miRNAs associate with a multi-protein complex containing DICER and ARGONAUT proteins called the RNA Inducing Silencing Complex (RISC). In this RISC complex, the loop of the hairpin structure is cleaved by DICER and the mature miRNA strand guides the RISC complex to its complementary target sequence mainly (but not exclusively) in the 3′-UTR of a target messenger RNA. Bound miRNAs execute a negative control on gene expression by one of three different mechanisms: 1) inhibition of translation initiation, 2) inhibition of translation elongation, 3) deadenylation of mRNA by recruitment of a deadenylase complex that destabilizes the mRNA and leads to degradation[76; 77; 78]. It has been also shown that miRNAs can induce the expression of a target mRNA by binding to it 5′-UTR region, however this mechanism does not seem to play a major role in miRNA biology[79]. Although the importance of miRNAs remains to be fully understood, dysregulated protein expression resulting from dysfunctional miRNA-based gene regulation has been reported to play critical roles in many key biological processes, such as cell growth and proliferation, differentiation, and apoptosis. Hence, mutation of miRNAs, dysfunction of miRNA biogenesis, and dysregulation of miRNAs target interactions may represent key etiologic factors in various diseases including PDAC.
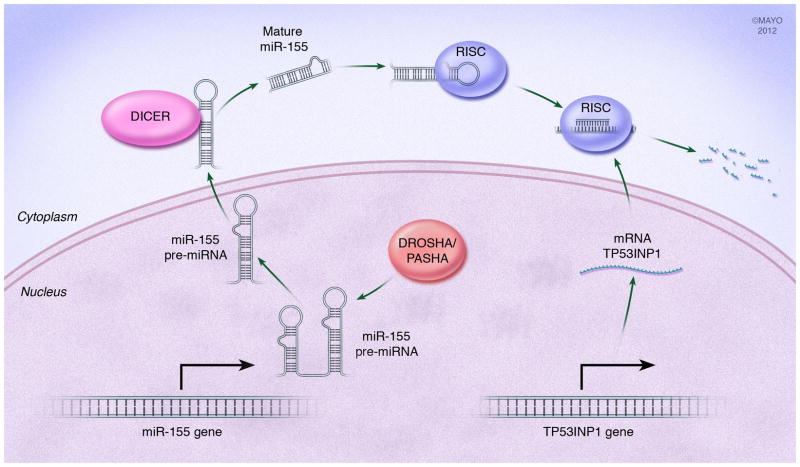
Advanced global screening technologies in the last years have enabled large scale analysis of miRNA profiles in diverse tissue samples, indicating that miRNAs can function as either oncogenes or tumor suppressors in the development of various human cancer types including PDAC[80; 81; 82; 83]. The analysis of miRNA expression patterns has led to completely new insights into cancer biology. Specific miRNAs, such as the miR-200 family, miR10a, miR-34a and miR-155 are involved in PDAC biology by regulating genes associated with metastatic phenotype and cell stemness. Members of the miR-200 family were identified as modulators of the epithelial-to-mesenchymal transition (EMT) by having negative activity on zinc finger E-box-binding homeobox 1 and 2 (ZEB1 and ZEB2) that function as repressors of EMT-opposing genes[84; 85; 86]. The miR-200 family has a putative tumor suppressor activity by silencing ZEB1 and ZEB2 thereby allowing expression of genes mitigating EMT such as E-cadherin[86]. Interestingly, in most PDAC cell lines and primary tumors, miR-200 family members are expressed but do not have a negative effect on ZEB2 expression because its promoter is already silenced by hypermethylation[86]. MiRNA are also involved in the expression of stem cell phenotypes. In a double negative feedback loop, ZEB1 also represses the expression of stemness-inhibiting miR-203, whose targets may include the stem cell factors Sox2 and KLF4[84]. Another group of miRNA that plays a role in cell stemness is the miR-34 family (composed of miRNA-34a, 34b and 34c), whose targets include the stemness inducing Notch1/2 and the anti-apoptotic Bcl-2[87; 88]. MiR-34 family is upregulated in a p53-dependent manner, and this is of striking importance in cancers where p53 function is lost as miR-34 family is thereby downregulated[88]. However, miR-34s have been found to be primarily inactivated by aberrant CpG methylation in PDAC, independent of the p53 status, revealing an interesting example of one epigenetic mechanism influencing the function of another[87]. Conversely, it has been demonstrated that the miRNA-29 is able to directly regulates DNA methylation by regulating the expression family of In a study evaluating PDAC stem cells and cell lines, treatment with the a demethylating agent or an HDAC inhibitor resulted in re-expression of miR-34 and resulted in significantly inhibited clonogenic cell growth and invasion, induced apoptosis and G2/M arrest in cell cycle, and inhibited the NOTCH pathway in PDAC stem cells[87]. An additional miRNA that functions to repress expression of a pro-apoptotic gene is miR-155 (fig. 4)[83]. One of the targets of miR-155 is TP53INP1[89]. The TP53INP1 gene functions as a pro- apoptotic gene regulated under induction of p53. Expression of TP53INP1 has been found to be significantly repressed in PDACs. While normal expression is found in benign pancreatic and early PanIN lesions, by PanIN3 lesions TP53INP1 expression is lost. The mechanism of this loss is through the induction of miR-155, as loss of this miRNA results in return of TP53INP1 expression[89]. Furthermore, new signaling pathways linking miRNAs to alteration in specific gene expression are being identified. A recent report suggested that miR-10a is a retinoid acid target and that retinoic acid receptor antagonists effectively repress miR-10a expression to completely block metastasis of PDAC cells[90]. This antimetastatic activity was prevented by specific knockdown of Hox genes HOXB1 and HOXB3, which are suspected targets of miR-10a. Thus, regulatory networks will continue to emerge that link the post-transcriptional control of miRNAs to inducers of EMT. In combination with stemness- maintenance by suppressing stemness-inhibiting miRNAs this control mechanism may promote mobile, migrating cells[90].
Fig. 4. microRNA influence PDAC pathogenesis.
miRNAs are a class of non-protein coding RNAs regulating gene expression post-transcriptionally in eukaryotes. After processing to a final effector form, miRNAs pair to their target protein-coding mRNAs in the cytoplasm to mediate post-transcriptional repression. An example of this in PDAC is the miR-155. Pri-miR-155 is processed in the nucleus by DROSHA to its pre-miRNA form where it is then translocated to the cytoplasm. In the cytoplasm, its hairpin loop is cleaved by DICER to form the mature miR-155 duplex. This mature form associated with the RISC complex where the single strand miR-155 is shuttled to its target mRNA. miR-155 represses expression of the TP53INP1 gene that is involved in promoting apoptosis under stress-activation of p53. Loss of TP53INP1 protein occurs early in PDAC pathogenesis and contributes to tumor formation.
In PDAC specifically, microarray data and multiplex analysis have revealed specific miRNA profiles that clearly define malignant phenotypes and may potentially aid to differentiate PDAC from benign pancreatic tissue or chronic pancreatitis. In one study, miR-196a was found to predict long-term vs. short-term survival, and consequently their prognostic potential could be helpful for therapeutic treatment options[80]. Several miRNAs, including miR-205, -18a, -31, -93, -221, and -224, were demonstrated to be over-expressed in primary neoplastic ductal cells and PDAC cell lines. Furthermore, 26 miRNAs were identified as the most significantly dysregulated miRNAs in PDAC, as compared to normal pancreas or pancreatitis[81]. The analysis of merely two, miR-217 and -196a, allowed discrimination between normal and neoplastic tissues, further supporting the potential use of miRNAs for the diagnosis of PDAC[81]. However, another important distinction should be made between PDAC and the precursor PanIN lesions. One study evaluated miRNA differences between various PanIN lesions and normal pancreatic ducts using laser capture microdissection for cell isolation followed by microarray and validation with FISH and qRT- PCR[91]. While several miRNAs identified in this study confirmed previous study results and identified new candidates, only miR-196b was selectively differentially expressed in PanIN-3 lesions, thereby providing another candidate for diagnostic differentiation of disease[91].
A major challenge still remains in the elucidation of biological pathways or signaling networks underlying cancer development and how specific miRNAs interact or contribute to the malignant transformation. There is increasing evidence suggesting miRNA-disease associations and numerous reports improve our knowledge about how miRNAs influence established cancer signaling pathways or connect previously unknown signaling routes or targets, which may open new roads to therapy. The identification of disease-related miRNAs is essential for understanding the pathogenesis of diseases at the molecular level.
5. Conclusion
As reviewed in this paper, the epigenetic concept integrates genomic methylation, histone modifications, and the regulatory effects of microRNAs on gene expression. It is evident that multiple epigenetic mechanisms are indeed crucial in the development and progression of PDAC. In addition to genetic changes, epigenetic alterations add another layer of complexity and contribute to the heterogeneity of PDAC. Continued studies on chromatin dynamics alone are unveiling the existence of robust machineries that can mediate epigenetic changes in pancreatic cells. These findings highlight the need to further our insight into how epigenetic mechanisms are able to independently and cooperatively influence gene regulation and thereby PDAC development. While a significant portion of our research into PDAC continues to focus on somatic genetic alterations, we must also continue to expand our research into the epigenetic mechanisms to complete the full story of altered gene expression in PDAC. This important observation is what has driven the focus of this review and highlights the need for the design of a more comprehensive model that can accommodate the emerging epigenetic data. The era of epigenetics has emerged strongly and will continue into a frontier area for PDAC research. Furthermore, it is important to emphasize one of the characteristics of epigenetic mechanisms of gene regulation – their reversibility. This feature provides a unique target for the introduction of specific therapeutic interventions for PDAC.
Acknowledgments
We thank Michael A. King for their help with the illustrations and Emily Porcher for the secretarial assistance. This work was supported by the Mayo Clinic Cancer Center, NCI CA136526, Mayo Clinic Pancreatic SPORE P50 CA102701 and Mayo Clinic Center for Cell Signaling in Gastroenterology P30 DK084567 to MEFZ and NIDDK DK52913 to RU. FUW was supported by grants from the Deutsche Forschungsgemeinschaft (DFG GRK840-E4) and the University Medicine Greifswald (FV Molekulare Medizin).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 3.Lomberk G, Mathison AJ, Grzenda A, Urrutia R. The sunset of somatic genetics and the dawn of epigenetics: a new frontier in pancreatic cancer research. Curr Opin Gastroenterol. 2008;24:597–602. doi: 10.1097/MOG.0b013e32830b111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Haig D. The (dual) origin of epigenetics. Cold Spring Harb Symp Quant Biol. 2004;69:67–70. doi: 10.1101/sqb.2004.69.67. [DOI] [PubMed] [Google Scholar]
- 6.Gottschling DE. Summary: epigenetics--from phenomenon to field. Cold Spring Harb Symp Quant Biol. 2004;69:507–519. doi: 10.1101/sqb.2004.69.507. [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH, Pantelouris EM. Transplantation of nuclei in newt’s eggs. Nature. 1953;172:1050–1051. doi: 10.1038/1721050a0. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1979;149:297–326. doi: 10.1084/jem.149.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature Reviews in Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clinical Can Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 12.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, Baylin SB, Kern SE, Herman JG. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 13.Buchholz M, Schatz A, Wagner M, Michl P, Linhart T, Adler G, Gress TM, Ellenrieder V. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006;25:3714–3724. doi: 10.1038/sj.emboj.7601246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G, Singh G, Hofmann L, Kunsch S, Buch T, Schafer E, Gress TM, Fernandez-Zapico ME, Ellenrieder V. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology. 2010;138:1189–1199. e1181–1182. doi: 10.1053/j.gastro.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzenda A, Ordog T, Urrutia R. Polycomb and the emerging epigenetics of pancreatic cancer. J Gastrointest Cancer. 2011;42:100–111. doi: 10.1007/s12029-011-9262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 17.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 18.Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 19.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 20.Hata K, Okano M, Lei H, Li E. Dnmt3L. cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 21.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 22.Wienholz BL, Kareta MS, Moarefi AH, Gordon CA, Ginno PA, Chedin F. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010;6:e1001106. doi: 10.1371/journal.pgen.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vairapandi M, Duker NJ. Enzymic removal of 5-methylcytosine from DNA by a human DNA-glycosylase. Nucleic Acids Res. 1993;21:5323–5327. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 25.Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-Methylcytosine to 5- Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: A mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67:5583–5586. doi: 10.1158/0008-5472.CAN-07-0846. [DOI] [PubMed] [Google Scholar]
- 27.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 28.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–3742. [PubMed] [Google Scholar]
- 30.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 31.Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2008;21:238–44. doi: 10.1038/modpathol.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res. 2011;17:4341–54. doi: 10.1158/1078-0432.CCR-10-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Feng Y, Pan L, Wang Y, Xu X, Lu J, Huang B. The proximal GC-rich region of p16(INK4a) gene promoter plays a role in its transcriptional regulation. Mol and Cell Biochem. 2007;301:259–266. doi: 10.1007/s11010-007-9427-4. [DOI] [PubMed] [Google Scholar]
- 34.Navarro A, Yin P, Monsivais D, Lin SM, Du P, Wei JJ, Bulun SE. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS One. 2012;7:e33284. doi: 10.1371/journal.pone.0033284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potapova A, Hasemeier B, Romermann D, Metzig K, Gohring G, Schlegelberger B, Langer F, Kreipe H, Lehmann U. Epigenetic inactivation of tumour suppressor gene KLF11 in myelodysplastic syndromes. Eur J Haematol. 2010;84:298–303. doi: 10.1111/j.1600-0609.2009.01389.x. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, Savoy DN, Molina JR, Fonseca R, Smyrk TC, Chari ST, Urrutia R, Billadeau DD. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Rosty C, Geradts J, Sato N, Wilentz RE, Roberts H, Sohn T, Cameron JL, Yeo CJ, Hruban RH, Goggins M. p16 Inactivation in pancreatic intraepithelial neoplasias (PanINs) arising in patients with chronic pancreatitis. Am J Surg Pathol. 2003;27:1495–1501. doi: 10.1097/00000478-200312000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin in Genet & Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 39.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 40.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 41.Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 43.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 44.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Ann Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 45.Lehmann A, Denkert C, Budczies J, Buckendahl AC, Darb-Esfahani S, Noske A, Muller BM, Bahra M, Neuhaus P, Dietel M, Kristiansen G, Weichert W. High class I HDAC activity and expression are associated with RelA/p65 activation in pancreatic cancer in vitro and in vivo. BMC Cancer. 2009;9:395. doi: 10.1186/1471-2407-9-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dal Molin M, Hong SM, Hebbar S, Sharma R, Scrimieri F, de Wilde RF, Mayo SC, Goggins M, Wolfgang CL, Schulick RD, Lin MT, Eshleman JR, Hruban RH, Maitra A, Matthaei H. Loss of expression of the SWI/SNF chromatin remodeling subunit BRG1/SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum Pathol. 2012;43:585–91. doi: 10.1016/j.humpath.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, Delhanty JD, Ponder BA, Kouzarides T, Caldas C. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300–3. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 48.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, Maitra A, Pollack JR. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109:E252–9. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosson GB, Bartlett C, Reed W, Weissman BE. BRG1 loss in MiaPaCa2 cells induces an altered cellular morphology and disruption in the organization of the actin cytoskeleton. J Cell Physiol. 2005;205:286–94. doi: 10.1002/jcp.20397. [DOI] [PubMed] [Google Scholar]
- 51.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 52.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 53.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider G, Kramer OH, Schmid RM, Saur D. Acetylation as a transcriptional control mechanism-HDACs and HATs in pancreatic ductal adenocarcinoma. J Gastrointest Cancer. 2011;42:85–92. doi: 10.1007/s12029-011-9257-1. [DOI] [PubMed] [Google Scholar]
- 55.Baumgart S, Glesel E, Singh G, Chen NM, Reutlinger K, Zhang J, Billadeau DD, Fernandez-Zapico ME, Gress TM, Singh SK, Ellenrieder V. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology. 2012;142:388–398. doi: 10.1053/j.gastro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lo Re AE, Fernandez-Barrena MG, Almada LL, Mills L, Elsawa SF, Lund G, Ropolo A, Molejon MI, Vaccaro MI, Fernandez-Zapico ME. A novel AKT1-GLI3-VMP1 pathway mediates KRAS-induced autophagy in cancer cells. J Biol Chem. 2012;287:25325–34. doi: 10.1074/jbc.M112.370809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathison A, Liebl A, Bharucha J, Mukhopadhyay D, Lomberk G, Shah V, Urrutia R. Pancreatic stellate cell models for transcriptional studies of desmoplasia-associated genes. Pancreatology. 2010;10:505–516. doi: 10.1159/000320540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez-Zapico ME, Mladek A, Ellenrieder V, Folch-Puy E, Miller L, Urrutia R. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 2003;22:4748–4758. doi: 10.1093/emboj/cdg470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouaissi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, Payan MJ, Dahan L, Pirro N, Seitz JF, Mas E, Lombardo D, Ouaissi A. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann of Surg Oncol. 2008;15:2318–2328. doi: 10.1245/s10434-008-9940-z. [DOI] [PubMed] [Google Scholar]
- 60.Fritsche P, Seidler B, Schuler S, Schnieke A, Gottlicher M, Schmid RM, Saur D, Schneider G. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3- only protein NOXA. Gut. 2009;58:1399–1409. doi: 10.1136/gut.2009.180711. [DOI] [PubMed] [Google Scholar]
- 61.Ouaissi M, Cabral S, Tavares J, da Silva AC, Mathieu Daude F, Mas E, Bernard J, Sastre B, Lombardo D, Ouaissi A. Histone deacetylase (HDAC) encoding gene expression in pancreatic cancer cell lines and cell sensitivity to HDAC inhibitors. Cancer Biol Ther. 2008;7:523–531. doi: 10.4161/cbt.7.4.5480. [DOI] [PubMed] [Google Scholar]
- 62.Schneider G, Kramer OH, Fritsche P, Schuler S, Schmid RM, Saur D. Targeting histone deacetylases in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2010;14:1255–1263. doi: 10.1111/j.1582-4934.2009.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall GM, Gherardi S, Xu N, Neiron Z, Trahair T, Scarlett CJ, Chang DK, Liu PY, Jankowski K, Iraci N, Haber M, Norris MD, Keating J, Sekyere E, Jonquieres G, Stossi F, Katzenellenbogen BS, Biankin AV, Perini G, Liu T. Transcriptional upregulation of histone deacetylase 2 promotes Myc-induced oncogenic effects. Oncogene. 2010;29:5957–5968. doi: 10.1038/onc.2010.332. [DOI] [PubMed] [Google Scholar]
- 64.Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, Friess H, Buchler M, Evert M, Lerch MM, Weiss FU. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439–448. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- 65.von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, von Werder A, Schmidt A, Mages J, Pagel P, Schnieke A, Schmid RM, Schneider G, Saur D. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361–371. 371 e361–365. doi: 10.1053/j.gastro.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Grzenda AL, Lomberk G, Urrutia R. Different EZH2 isoforms are expressed in pancreatic cells: Evidence for a polycomb-mediated subcode within the context of the histone code. Pancreas. 2007;35:404. [Google Scholar]
- 67.Cao R, Wang LJ, Wang HB, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 68.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 69.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes & Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, Albarracin C, Yu D, Abbruzzese JL, Mills GB, Bast RC, Jr, Hortobagyi GN, Hung MC. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47:701–706. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res. 2008;14:6790–6796. doi: 10.1158/1078-0432.CCR-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lomberk G, Mathison AJ, Grzenda A, Seo S, DeMars CJ, Rizvi S, Bonilla-Velez J, Calvo E, Fernandez-Zapico ME, Iovanna J, Buttar NS, Urrutia R. Sequence-specific recruitment of heterochromatin protein 1 via interaction with Kruppel-like factor 11, a human transcription factor involved in tumor suppression and metabolic diseases. J Biol Chem. 2012;287:13026–13039. doi: 10.1074/jbc.M112.342634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008;68:2708–2716. doi: 10.1158/0008-5472.CAN-07-6844. [DOI] [PubMed] [Google Scholar]
- 74.Hollingsworth MA, Strawhecker JM, Caffrey TC, Mack DR. Expression of Muc1, Muc2, Muc3 and Muc4 Mucin Messenger-Rnas in Human Pancreatic and Intestinal Tumor- Cell Lines. Int J Cancer. 1994;57:198–203. doi: 10.1002/ijc.2910570212. [DOI] [PubMed] [Google Scholar]
- 75.Masaki Y, Oka M, Ogura Y, Ueno T, Nishihara K, Tangoku A, Takahashi M, Yamamoto M, Irimura T. Sialylated MUC1 mucin expression in normal pancreas, benign pancreatic lesions, and pancreatic ductal adenocarcinoma. Hepatogastroenterology. 1999;46:2240–2245. [PubMed] [Google Scholar]
- 76.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schier AF, Giraldez AJ. MicroRNA function and mechanism: insights from zebra fish. Cold Spring Harb Symp Quant Biol. 2006;71:195–203. doi: 10.1101/sqb.2006.71.055. [DOI] [PubMed] [Google Scholar]
- 78.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 79.Vasuden Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 80.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 81.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 82.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–346. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 85.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J, Brabletz T. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, Chaix A, Fazli L, Motoo Y, Wang Q, Rocchi P, Russo A, Gleave M, Dagorn JC, Iovanna JL, Carrier A, Pebusque MJ, Dusetti NJ. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2145. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 91.Yu J, Li A, Hong SM, Hruban RH, Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18:981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]