Abstract
The ribosome undergoes numerous large-scale conformational changes during protein synthesis, but the molecular bases for these changes has been unclear. Recent cryo-electron microscopic and x-ray crystallographic structures of both the bacterial and eukaryotic ribosome now provide snapshots of the wide range of motions that occur within the ribosome. X-ray crystallographic structures of the ribosome have also pinpointed local deformations in ribosomal RNA that occur when the two ribosomal subunits rotate with respect to each other. These structural results establish the foundation for unraveling the mechanics of the ribosome that are universal, and those that differ in bacteria and eukaryotes.
Introduction
Translation of messenger RNAs (mRNAs) into proteins by the ribosome requires its movement along the mRNA in a 5′ to 3′ direction. To accomplish this feat, the ribosome is constructed as a highly dynamic machine with several independently moving elements, assembled into two ribosomal subunits. In bacteria, the ribosome consists of small and large ribosomal subunits (30S and 50S, respectively) that sum to a mass of about 2.5 MDa for the intact 70S ribosome. In eukaryotes, the ribosome is much larger, and is composed of 40S and 60S subunits that associate to form a 3.3 MDa or larger 80S ribosome. Structural and biophysical results from many labs have revealed the extent of the ribosome’s dynamics and flexibility, and have started to reveal the complex energy landscape that the ribosome traverses as each amino acid is added to the growing peptide chain [1]. Remarkably, the energy landscape of ribosome conformations is rather “flat” in the absence of bound substrates (mRNA and transfer RNAs (tRNAs)) and the myriad of translation factors required for protein synthesis. The binding and release of these substrates and factors is therefore an integral component of the ribosome’s unidirectional movement along mRNA [2], and requires a structural framework to understand the many transitions that occur.
Our knowledge of the molecular basis of conformational changes in the ribosome as well as the contacts between the two ribosomal subunits, called inter-subunit bridges [3], has been building for more than a decade due to rapid advances in electron microscopy (EM) and x-ray crystallography, and their application to ribosome structure determination. Low resolution structures of the ribosome determined by cryo-EM revealed many of the global changes in ribosome conformation [4-15](Figure 1), and high resolution x-ray crystallographic studies revealed the molecular details of the inter-subunit bridges [16]. However until recently, all of the high resolution structures of the ribosome had clustered in conformations closely related to the so-called “unrotated” state, in which tRNAs are bound in the classical acceptor-tRNA, peptidyl-tRNA, or exit-tRNA sites (A/A, P/P or E/E sites in the small/large subunits, respectively)[17-19]. Only apo-70S ribosome structures had revealed the molecular details of dynamics in the small subunit head domain [16], dynamics that were previously observed in lower-resolution cryo-EM reconstructions (Figure 1) [9]. Furthermore, the critical rotated state of the ribosome–the rotation of the small subunit relative to the large–that is essential for translocation, the movement of mRNA and tRNA after peptide bond formation, and for aspects of initiation, termination and ribosome recycling, had only been observed by cryo-EM [4,6,9-11,14,20] and biophysical experiments [21-25].
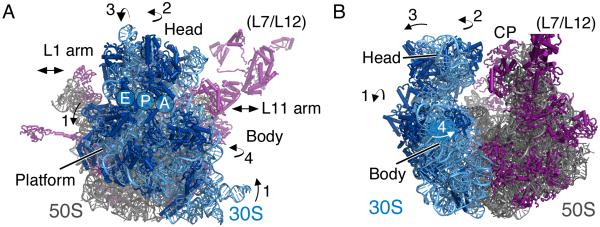
Figure 1. Global changes in ribosome conformation during translation.
A) During translation the small ribosomal subunit undergoes four large-scale motions: rotation of the body/platform (1), swiveling of the head (2), tilting of the head (3), and body closure (4). In the large ribosomal subunit, movement of the L1 arm and L11 arm are also observed during translation. The stalk proteins in the large subunit (L7/L12 in bacteria) are highly dynamic. The tRNA A, P, and E sites, which occur at the subunit interface, are indicated on the solvent side of the small subunit. B) The bacterial ribosome as in A) but rotated by 90° around the vertical axis so that the ribosome is viewed from the side of the L11 arm, into the A site. Movements are indicated by arrows. In both panels, the small subunit rRNA and proteins are shown in light blue and dark blue, respectively. Large subunit rRNA and proteins are shown in gray and magenta, respectively.
This review summarizes recent x-ray crystal structures and cryo-EM reconstructions that have revealed many new ribosomal conformational states, as well as the molecular interactions in the ribosome in its fully rotated states stabilized with different translation factors. These structures reveal the inter-subunit bridges in fully rotated states, and also shed light on differences between the bacterial and eukaryotic ribosome. Furthermore, these recent structures reveal that the conformational landscape of the ribosome is more extensive than was previously known. These structural findings pave the way for a far deeper understanding of ribosome function in the years ahead.
Ribosome dynamics revealed
The large scale conformational changes in the ribosome that accompany movement of tRNAs from the classical state (tRNAs in the A/A and P/P configuration) to hybrid site binding (tRNAs in the A/P and P/E configuration) were originally described as a rigid body rotation of the small ribosomal subunit with respect to the large by up to 10° around an axis perpendicular to the subunit interface (Figure 1) [4]. Higher resolution cryo-EM reconstructions further elucidated additional degrees of freedom during translocation, including swiveling of the small subunit head domain [9] and inward movement of the L1 stalk on the large subunit towards the E site on the large subunit (Figure 1) [6]. Movement of the L1 stalk was proposed to stabilize the P/E tRNA in its hybrid binding site. Swiveling of the small subunit head domain in the direction of the E site results in displacements of up to 20 Å at the ribosomal subunit interface, which is equivalent to the width of a tRNA molecule. This movement, coupled with opening of a “gate” between the P and the E sites in the small subunit, was proposed to enable the anticodon stem-loop (ASL) of P-site tRNA to move into the E site, resulting in tRNAs bound in the post-translocation state, or P/P and E/E configurations [9,16]. After the tRNAs have moved into the P/P and E/E configurations, these motions would reverse to prepare the ribosome for a new elongation cycle (Figure 2). Similar but not identical dynamics are thought to occur during termination and ribosome recycling [26,27].
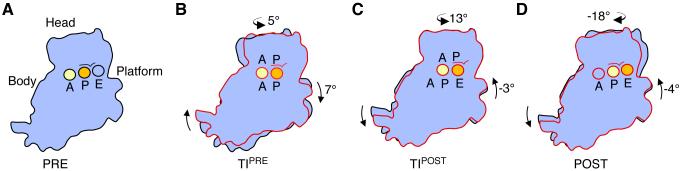
Figure 2. Structural intermediates of the small ribosomal subunit during mRNA and tRNA translocation.
A) Immediately after peptide bond formation the ribosome assumes a pre-translocation state (PRE) in which a peptidyl-tRNA occupies the A site (yellow) and a deacylated tRNA occupies the P site (orange). In this unrotated state of the ribosome, tRNAs are bound in their classical conformation (A/A, P/P, and E/E configuration). The “gate” component of the small subunit head domain is marked above the P site. B) Binding of the GTPase EF-G in complex with GTP stabilizes a pre-translocational intermediate state (TIPRE) in which the small subunit has rotated as a rigid body by ~7° with respect to the large subunit, the head of the small subunit swivels by ~5°, and the acceptor arms of the tRNAs move into the P and E site on the large subunit to assume a hybrid state of binding (A/P and P/E configuration in the small/large subunit, respectively). The A and P sites in the small subunit head and body/platform are marked. C) Hydrolysis of GTP and phosphate release is accompanied by reverse rotation of the small subunit body by −3° and additional head swiveling of +13° compared to TIPRE. In this “post-translocational intermediate” state (TIPOST), the anticodon stem-loops (ASL) of the tRNAs bind in small-subunit hybrid sites in which the ASL of the peptidyl-tRNA contacts the A and the P site in the small subunit head domain and the deacylated tRNA contacts the P and the E site in the small subunit body/platform domains, yielding ap/P and and pe/E tRNA configurations on the small/large subunits, respectively. D) Dissociation of EF-G-GDP from the ribosome leads to complete reversal of the small subunit rotation and back-swiveling of the small subunit head domain, yielding a post translocation state (POST) in which the peptidyl-tRNA and deacylated tRNA are bound in P/P (yellow) and E/E (orange) configurations, respectively. The above model for translocation is adapted from [30].
Structures of ribosomes in intermediate states of rotation indicate that transitions between unrotated and rotated states involve multiple steps [28,29]. Notably, cryo-EM studies of 70S ribosomes with a P site tRNA stabilized by EF-G in its GTP state revealed a conformation of the ribosome that probably occurs in a late step of mRNA and tRNA translocation, when the P-site tRNA ASL transits to the E site of the 30S subunit. In this proposed post-translocational intermediate state (termed TIPOST), the 30S subunit body partially reverses its rotation (to 4°), and the 30S subunit head domain swivels by 18° (Figure 2) [30]. This leads to a 8-10 Å movement of the tRNA ASL, resulting in tRNA ASL contacts to parts of both the P and E site in the small subunit, and tRNA acceptor arm contacts to the E-site in the large subunit (named the pe/E tRNA hybrid site). As head swiveling is thought to guide translocation of the tRNA/mRNA complex [9,14,16], this structure is expected to closely match a post-translocation intermediate. Crystal structures of the E. coli 70S ribosome in intermediate states of rotation [29] provide some clues into the molecular rearrangements that likely occur in states like those in the TIPOST state [30].
Recently, additional structures underlined the range of motions the ribosome can sample. The crystal structure of the ribosome bound to the stationary-phase factors such as ribosome modulation factor (RMF) or hibernation promoting factor (HPF) results in a movement of the 30S head domain away from the central protuberance in a way that promotes 100S ribosome dimer formation [31]. Further, ribosome rescue in bacteria by transfer-messenger RNA (tmRNA) and the protein SmpB induces an unusual 12° tilt movement of the head domain around an axis that is almost parallel to the mRNA in addition to moderate body rotation of 5° and a large head swiveling of 19° [32] (Figure 1).
The first high-resolution view of the ribosome in a fully rotated state was revealed in a crystal structure of the ribosome recycling complex with tRNA and ribosome recycling factor (RRF) [26]. In the structure, the 30S subunit is rotated relative to the large by 9°, and the 30S subunit domain is swiveled by 4° (Figure 3). This structure revealed for the first time the molecular details of how inter-subunit contacts are rearranged to stabilize the ribosome in the fully rotated state and how the ribosome accommodates P/E hybrid state tRNA. Two additional crystal structures of the ribosome with release factor 3 (RF3) [33,34], one containing P/E tRNA, also revealed molecular details of rearrangements in inter-subunit bridges, which will be described below.
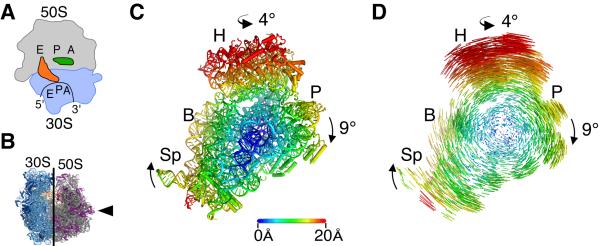
Figure 3. The bacterial ribosome recycling complex in the fully rotated state.
A) During ribosome recycling, ribosome recycling factor (RRF, green) stabilizes a fully rotated state of the ribosome in which the deacylated tRNA (orange) assumes a P/E hybrid configuration in the 30S/50S subunits, respectively. B) Direction of view of the ribosome in C) and D) is indicated by an arrow, which shows the 30S subunit from the subunit interface with the 50S subunit removed. C) Movements of the 30S subunit during inter-subunit rotation. The 30S subunit body and platform rotate as a rigid body by 9° and the head domain of the 30S subunit swivels by 4°. Displacements of rRNA phosphorus and ribosomal protein Cα atoms between the unrotated and the fully rotated state are color-coded by distance in Å, as indicated. D) Difference vectors between equivalent phosphorus and Cα atoms in the unrotated and the fully-rotated state are color-coded as in C). H head, B body, P platform, Sp spur. Reproduced with permission from [26].
The eukaryotic ribosome also exhibits large-scale dynamics during translation, including inter-subunit rotation [9,14,15]. However, the rotated states adopted by the eukaryotic ribosome may differ in some details from those observed with bacterial ribosomes [35]. The eukaryotic ribosome is much larger than the bacterial ribosome, due to additional rRNA domains called expansion segments (ES), as well as extensions in conserved ribosomal proteins and eukaryotic specific ribosomal proteins. Elements of these additional rRNA and protein components nearly double the interaction surface between the two ribosomal subunits compared to their bacterial counterparts (Figure 4) [36-39]. The increased interaction surface between the subunits may explain why the eukaryotic ribosome favors the rotated state in the absence of ligands whereas the bacterial ribosome favors the unrotated state in its vacant form [9].
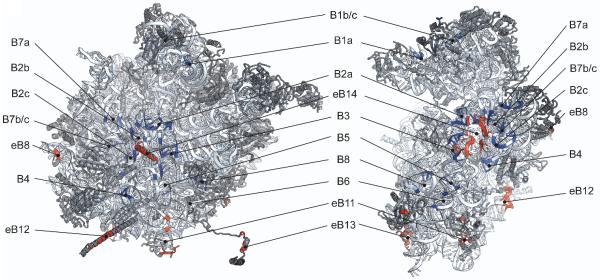
Figure 4. Inter-subunit contacts of the eukaryotic ribosome in the rotated state.
Global view of the inter-subunit contacts in the rotated state of the yeast ribosome. The 60S subunit (left) and 40S subunit (right) are shown from their interface sides. Bridges that are conserved between bacteria and eukaryotes are shown in blue (B1-B8) whereas eukaryotic-specific bridges are shown in red (eB8-eB14). Reproduced with permission from [39].
In recent cryo-EM reconstructions of pre-translocation complexes of the rabbit 80S ribosome, spontaneous rotation of the small subunit body seems to occur without the coupled swiveling of the 40S head domain [35]. Notably, differences in inter-subunit rotation are reflected in slightly different positioning of tRNAs relative to the ribosomal subunits. Structures of the yeast 80S ribosome trapped with the stress protein Stm1p reveal the first high-resolution views of eukaryotic ribosome conformational states [39]. These structures will be highly useful in understanding the elements of ribosome dynamics that are conserved in bacteria, as well as those aspects that are unique to eukaryotes. For example, one of the yeast 80S structures is close in conformation to that of the bacterial translocation intermediate with pe/E tRNA [30]. The slight differences in the position of the small subunit head domain in the two structures may be related to differences in how bacterial and eukaryotic ribosomes control tRNA movement [35].
Ribosomal RNA dynamics in inter-subunit bridges
The two ribosomal subunits are held together by several inter-subunit bridges that are composed of both rRNA and ribosomal proteins (Figure 4). These inter-subunit contacts need to rearrange in order to accommodate the transition from unrotated to rotated states of the ribosome. The molecular details of inter-subunit contacts in the unrotated bacterial ribosome [16-19] can now be compared to those in structures with different extents of inter-subunit rotation. Structures of the E. coli ribosome in intermediate states of rotation, which bear similarities to those seen in cryo-EM reconstructions [28,30], revealed how contacts between the ribosomal subunits rearrange in a stepwise manner [29]. As noted above, one of the structures of the yeast 80S ribosome also adopts an intermediate state of rotation [36,39]. The molecular details of how the fully rotated state is stabilized have now been revealed in structures of a functional ribosome recycling complex [26]. Crystal structures of the ribosome in complexes with RF3, and a structure of the yeast 80S ribosome also shed light on how the inter-subunit contacts are rearranged to accommodate ribosomal subunit rotation [33,34,39].
Bridge B3 serves as the “pivot” point between the two ribosomal subunits during inter-subunit rotation, and remains essentially unchanged during rotation of the small subunit [26]. Bridge B3 is composed of two sheared G-A base pairs in ribosomal RNA helix h44 of the small subunit that form A-minor interactions with two G-C base pairs in H71 of the large subunit [16]. This kind of A-minor interaction is widespread in the ribosome and other large, folded RNAs [40,41]. In contrast to the pivot point contact between the ribosomal subunits, other key rRNA inter-subunit bridges (B2a and B4), as well as the rRNA helix h27 that connects the head domain of the small subunit to the rest of the small subunit, undergo significant deformations to allow rotation of the small subunit relative to the large, and to allow tRNA movement between the three tRNA binding sites [16,26,29,33,34,39]. Bridge B2a, in particular, is notable in that it is located immediately adjacent to the tRNA A and P sites (Figure 4), and had been observed to change conformation in different steps of translocation and ribosome recycling [8,11,20,42,43]. This bridge involves an rRNA hairpin including helix H69 in the large subunit and rRNA helix h44 in the mRNA decoding site and P site of the small subunit. During inter-subunit rotation, h44 moves laterally towards the E-site by ~6 Å. To maintain all of the RNA-RNA contacts between the subunits, helix H69 compresses by ~5Å in the rotated state (Figure 5). To varying degrees, this compression is also seen in the structures of the ribosome with RF3 and in one of the yeast 80S ribosomes [26,33,34,36,39]. The RNA hairpin in H69 is buttressed by a sharp U-turn motif, which unstacks to different extents in the available structures. The striking conformational changes in this short hairpin/U-turn motif that contribute to inter-subunit rotation help to explain its nearly universal conservation at the sequence level [44].
Figure 5. Bridge B2a during subunit rotation.
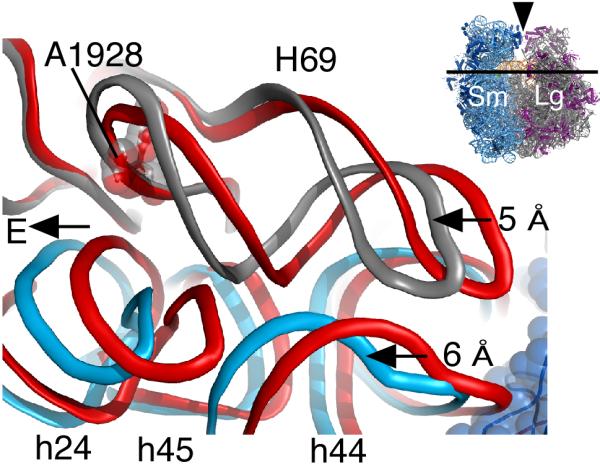
Rotation of the small subunit during translation leads to a lateral displacement of h44 in the small subunit by 6° towards the E site. Upon rotation H69 in the large subunit compresses by 5° in order to maintain its inter-subunit contacts with h44. Nucleotide A1928 is nearly invariant in position and is shown for reference. Ribosomal RNA of the fully rotated ribosome is shown in blue (30S) and gray (50S) whereas rRNA of the unrotated ribosome is shown in red. Inset, icon of the ribosome indicating the direction of view. Reproduced with permission from [26].
Conclusion
The last few years have yielded an abundance of new structures of the ribosome, both bacterial and eukaryotic in origin, determined by cryo-EM and x-ray crystallography. Many of these structures reveal large-scale rearrangements in the ribosome that are likely essential for the fidelity of many steps of the translation cycle. Efforts to parameterize these global motions are still at an early stage, and range from simple descriptions of rigid-body rotations [26,28-30,35,39] to more complex tensor analyses [45]. However, these efforts are challenging due to the variable resolutions of the available structural models and the different degrees of flexibility at the periphery of the ribosome. For example, it is still not clear whether the fully rotated state in translocation is equivalent to those in termination or ribosome recycling. Furthermore, the conformational plasticity of RNA and protein elements connecting rigid domains within the small and large ribosomal subunits is still coming into focus. Future efforts that combine the new structures of the ribosome with approaches such as molecular dynamics and other biophysical methods will be required to fully elucidate how structural changes in the ribosome contribute to the speed and fidelity of translation.
Acknowledgments
This work was funded by a grant from the National Institutes of Health (R01-GM65050) to J.H.D.C. and by a Human Frontiers in Science Program Postdoctoral Fellowship to J.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spirin AS. The ribosome as a conveying thermal ratchet machine. J Biol Chem. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank J, Verschoor A, Li Y, Zhu J, Lata RK, Radermacher M, Penczek P, Grassucci R, Agrawal RK, Srivastava S. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem Cell Biol. 1995;73:757–765. doi: 10.1139/o95-084. [DOI] [PubMed] [Google Scholar]
- 4.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 5.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 6.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 7.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc Natl Acad Sci U S A. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- 11.Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 14.Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 17.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 18.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci U S A. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 21.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 22.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 23.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. The first structure of the ribosome in the fully rotated state, with tRNA bound in the hybrid P/E site stabilized by ribosome release factor (RRF), is presented at 3.2 Å. The structure reveals intermolecular contacts in the fully rotated state and also provides details of how tRNA adopts a highly bent conformation in its hybrid P/E configuration. The structure helps to explain how inter-subunit rotation contributes to translocation, termination, and ribosome recycling.
- **27.Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK. Structural insights into initial and intermediate steps of the ribosome-recycling process. EMBO J. 2012;31:1836–1846. doi: 10.1038/emboj.2012.22. Cryo-EM reconstructions of ribosome recycling complexes reveal insight into different steps of the recycling process. The authors show that the conformation of EF-G bound to the ribosome with RRF differs from previously seen EF-G conformations indicating that the mode of action of EF-G is different during elongation and recycling.
- **28.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. This paper describes cryo-EM reconstructions of the E. coli 70S ribosome trapped after different time points in a reverse translocation reaction, in which several intermediate states of rotation were identified.
- **29.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. The first crystal structure of the bacterial ribosome in an intermediate state of rotation reveals how contacts at the subunit interface are rearranged in a step-wise manner, and provides insight into how tRNAs are moved into the hybrid state upon subunit rotation.
- **30.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. Cryo-EM reconstructions of the ribosome in complexes with tRNA and EF-G in intermediate pre-translocation (TIPRE) and post-translocation (TIPOST) states are presented. The post-translocation intermediate contains a novel pe/E state of tRNA binding, in which the ribosome adopts an intermediate state of inter-subunit rotation. This intermediate state likely follows the fully-rotated state in the trajectory of the translocation process.
- *31.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. Structures of ribosome modulation factor (RMF) and hibernation promoting factor (HPF) bound to the 70S ribosome reveal additional conformational states the ribosome adopts when stored as 100S dimers.
- **32.Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, et al. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature. 2012;485:526–529. doi: 10.1038/nature11006. Cryo-EM reconstructions of the bacterial ribosome in an aberrant termination complex requiring rescue by transfer-messenger RNA (tmRNA) and the protein SmpB reveal dramatic rearrangements in the conformation of the 30S subunit. These rearrangements are likely needed to clear the ribosome complex of the damaged mRNA and allow tmRNA to transit the space between the ribosomal subunits.
- *33.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci U S A. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. The crystal structure of the ribosome bound to release factor 3 (RF3) in the presence of nonhydrolyzable GTP analogue is presented. This structure, refined to 3.8 Å, reveals the ribosome in the rotated conformation with hybrid P/E tRNA bound, and suggests how RF3 facilitates dissociation of class 1 release factors from the ribosome by stabilizing the rotated state.
- *34.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18:230–240. doi: 10.1261/rna.031187.111. This work presents the crystal structure of a ribosome complex with RF3 in a GTP-bound state at 3.3 Å. In the complex, the ribosome adopts a rotated state similar to the fully rotated state. The structure also reveals conformational rearrangements of RF3 upon binding to the ribosome, with implications for the mechanism of elongation factor G (EF-G) in translocation.
- *35.Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. Cryo-EM reconstructions of translocation intermediates on the rabbit ribosome reveal some variation in the conformation of the fully rotated state, when compared to the bacterial 70S ribosome.
- *36.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. The first crystal structure of the eukaryotic ribosome is presented at 4.15 Å resolution. This structure reveals the higher complexity of the eukaryotic ribosome relative to the bacterial ribosome and shows how additional elements expand interactions between the two ribosomal subunits.
- 37.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc Natl Acad Sci U S A. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci U S A. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **39.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. This paper presents structures of the yeast 80S ribosome refined to 3.0 Å resolution, yielding more complete models of the ribosome than those from 2010. The structures contain models of the eukaryotic ribosome in two states of inter-subunit rotation, with nearly complete descriptions of the molecular interactions involving eukaryotic specific elements.
- 40.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 41.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 43.Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK. Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci U S A. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. The authors describe the conformation of the ribosome in different steps of the translocation process by using tensor descriptions of domain motions within the small ribosomal subunit, and compare these to the positioning of tRNAs and the L1 stalk of the large subunit.