Abstract
Hepatitis C virus (HCV) NS5B polymerase is a key target for the development of anti-HCV drugs. Here we report on the identification of novel allosteric inhibitors of HCV NS5B through a combination of structure-based virtual screening and in vitro NS5B inhibition assays. One hundred and sixty thousand compounds from the Otava database were virtually screened against the thiazolone inhibitor binding site on NS5B (thumb pocket-2, TP-2), resulting in a sequential down-sizing of the library by 2.7 orders of magnitude to yield 59 NS5B non-nucleoside inhibitor (NNI) candidates. In vitro evaluation of the NS5B inhibitory activity of the 59 selected compounds resulted in a 14% hit rate, yielding 8 novel structural scaffolds. Of these, compound 1 bearing a 4-hydrazinoquinazoline scaffold was the most active (IC50 = 16.0 µM). The binding site of all 8 NNIs was mapped to TP-2 of NS5B as inferred by a decrease in their inhibition potency against the M423T NS5B mutant, employed as a screen for TP-2 site binders. At 100 µM concentration, none of the eight compounds exhibited any cytotoxicity, and all except compound 8 exhibited between 40–60% inhibition of intracellular NS5B polymerase activity in BHK-NS5B-FRLuc reporter cells. These inhibitor scaffolds will form the basis for future optimization and development of more potent NS5B inhibitors.
Keywords: Hepatitis C virus, NS5B polymerase, virtual screening, NS5B inhibitors, thumb pocket
1. Introduction
Hepatitis C virus (HCV) is a blood-borne pathogen of global public health significance, associated with the development of chronic liver disease. Approximately 3% of the world population is chronically infected with HCV and at risk of developing liver cirrhosis and hepatocellular carcinoma [1, 2]. There is no prophylactic anti-HCV vaccine and the combination therapy of pegylated interferon and ribavirin is expensive, associated with severe adverse effects and lacks uniform efficacy against the various HCV genotypes [3–8]. The recent clinical approval of the HCV protease inhibitors has changed the standard of care to triple drug therapy and dramatically improved the sustained virological response in patients with genotype 1 HCV infection [9–11]. However, this therapy has a complicated dosing regimen, in addition to drawbacks of adverse side effects and the challenge of drug-resistant variants remain. Therefore, the quest for new directly acting antivirals with high therapeutic index, reduced side-effects, and a convenient route of administration is an area of high priority.
HCV is an enveloped positive-sense single stranded RNA virus belonging to the Flaviviridae family [12]. It’s 9.6 kb RNA genome encodes a single large polyprotein of ~ 3000 amino acids, which is co- and post-translationally processed by cellular and viral proteases into three structural (core, E1, and E2) and seven nonstructural proteins ( p7, NS2, -3, -4A, -4B, -5A, and -5B) [13, 14]. Currently, several HCV proteins and its RNA are being explored as candidate targets for anti-HCV therapeutic development. Of these, nonstructural proteins NS3 and NS5B are the most promising and remain in the forefront of anti-HCV therapeutic approaches [9–11, 15].
HCV NS5B is a pivotal component of the viral replication machinery as it encodes the viral RNA-dependent RNA polymerase (RdRp) activity essential for replicating the viral RNA genome [16, 17]. This unique and distinctive ability of NS5B to utilize the RNA template, a property which the host mammalian cell lacks, has resulted in its emergence as an attractive and validated drug target [3, 4, 18]. Thus, NS5B has been widely investigated for its biochemical properties and structural parameters. The latter has revealed that NS5B exhibits a classical “right hand” topology of the polymerase family, with the characteristic fingers, palm, and thumb domains [19–22]. This insight has provided a valuable platform for developing NS5B inhibitors. Based on their mode of action, NS5B inhibitors can be broadly categorized into nucleoside and non-nucleoside inhibitors (NIs and NNIs, respectively). The former functions as rNTP substrate mimics and blocks the elongation of new viral RNA strands whereas the latter bind at one of the five distinct allosteric pockets (AP) of NS5B, preventing a conformational transition needed for initiation of RNA synthesis [4, 15, 18].
Previously, we reported on the utility of three-dimensional quantitative structure-activity relationship methodologies and virtual screening approach to identify new HCV NS5B polymerase inhibitors. These investigations resulted in the identification and optimization of two new chemotypes bearing the rhodanine [23] and S-trityl-L-cysteine (STLC) [24, 25] scaffolds. In this study, we have employed structure-based virtual screening to explore the Otava library database for new NNI leads that bind NS5B thumb pocket-2 (TP-2). Here, we report on the identification of 8 novel hit molecules comprised of unique scaffolds in the field of NS5B inhibitor discovery. We further describe molecular modeling studies to explore the potential optimization of these inhibitors.
2. Results and discussion
2.1. Virtual screening of compounds against TP-2 of HCV NS5B
Crystal structure of NS5B polymerase in complex with thiazolone inhibitor (PDB ID: 2O5D), representing TP-2 at the thumb domain of NS5B was chosen for virtual screening [26]. This site is about 30 Ǻ away from the active site that includes a conserved catalytic GDD motif located in the palm domain. This choice was driven by the fact that a number of NS5B inhibitors bound to this allosteric pocket have been reported during the last decade [26–28]. Further, this site is characterized by high affinity binding to NNIs, an important parameter for selectivity of an inhibitor.
To identify new inhibitor scaffolds which potentially bind NS5B TP-2, we utilized a structure-based virtual screening approach as described in the Experimental section. Towards this objective, the Otava library database (Otava Ltd) comprising of 160,000 organic compounds was first subjected to Lipinski’s “Rule of Five” filtering criteria [29]. This eliminated 25% of compounds resulting in the generation of a new database of 120,000 compounds. At the second step, we applied the docking score cutoff value of ≤-35 kcal/mol to further down-size the 120,000 compounds database by ~66%, thus resulting in 41,000 remaining compounds. Next, we computationally investigated the binding modes of several known TP-2 inhibitors such as thiazolone (PDB ID: 2HWH, 2HWI, 2O5D), thiophene-2-carboxylic acid (PDB ID: 2D3Z), dihydropyranone (PDB ID: 1OS5) and benzoisoquinoline (PDB ID: 2WHO) within TP-2 of NS5B [26, 27, 30–32]. Based on this computational analysis, we observed that these inhibitors form either direct or water-mediated hydrogen bonding interactions with the guanidine and ε-NH2 groups, respectively, of Arg501 and Lys533 and the backbone –NH groups of Ser476 and Tyr477. Further, their hydrophobic moiety establishes contacts with the side chains of Leu419, Met423, Leu497 and Trp528.
Based on this pharmacophoric feature, the ensuing 41,000 compounds were subjected to structure-based virtual screening with filtering criteria such that the hit molecules should exhibit hydrogen bonding and/or ionic interactions with Ser476, Tyr477, Arg501 and Lys533 and their hydrophobic moiety form contacts with Leu419, Met423, Leu497 and Trp528. Compounds that lacked these interactions were removed, thus yielding 984 compounds for further investigation. These rigorous in-silico screening criteria along with visual inspection of the ligand-enzyme complexes ultimately filtered out 99.9% of the remaining compounds, thus yielding a set of 59 candidates bearing structural diversity and acceptable docking scores for biological evaluation.
2.2. Screening and identification of NS5B inhibitors
The anti-NS5B activity of the 59 in-silico selected candidates was experimentally investigated employing an in vitro NS5B RdRp inhibition assay [33–35]. In this assay, we employed poly rA/U12 template-primer (TP) and recombinant HCV NS5BCΔ21 (1b). Wedelolactone, a validated NS5B inhibitor was included as an internal reference standard for monitoring NS5B inhibitory activity and yielded IC50 value of 34 µM, consistent with previously reported value [35]. To identify a wider range of candidates, all compounds were initially screened at 100 µM concentration. This analysis yielded eight compounds exhibiting ≥50% inhibition of NS5B RdRp activity at 100 µM concentration (Table 1), accounting for a 13.6% hit rate. All eight compounds bore structurally distinct scaffolds from previously reported NS5B inhibitors and exhibited IC50 values between 16 µM to 57 µM (Table 2). Of these, compound 1 bearing a 4-hydrazinoquinazoline scaffold was the most active (IC50=16.0 µM), while compound 8, a bis-pyranone scaffold was the least active (IC50=56.8 µM).
Table 1.
In-silico identified NS5B inhibitors.
| Compound | Otava Code | Structure | Compound Name |
|---|---|---|---|
| 1 | P7117160121 |

|
2-(Quinazolin-4-yl- hydrazono)-propionic acid |
| 2 | P7020351141 |  |
2-[N'-(3-Bromo-4- hydroxy-5-methoxy- benzylidene)-hydrazino]- benzoic acid |
| 3 | P1588489 |  |
3-[1-(2-Carboxy-ethyl)-3- (4-fluoro-phenyl)-1H- pyrazol-4-yl]-2-cyano- acrylic acid ethyl ester |
| 4 | P1888291 |  |
3-Benzenesulfonyl-1-[4-(3- benzenesulfonyl- propionyl)-piperazin-1-yl]- propan-1-one |
| 5 | P7020431675 |  |
4-(4-Methyl-1,1,3-trioxo- 1λ6-isothiazolidin-2-yl)-N- quinolin-8-yl- benzenesulfonamide |
| 6 | P1297082 |  |
7-[4-(2,4-Difluoro- benzoyl)-piperazine-1- carbonyl]-5-methyl-2- phenyl-2,5-dihydro- pyrazolo[4,3-c]pyridin-3- one |
| 7 | P7020527349 |  |
2-Hydroxy-4-(5-oxo-6- phenyl-4,5-dihydro- [1,2,4]triazin-3-ylamino)- benzoic acid |
| 8 | P7215300002 |  |
3,3'-[(3,4- Dichlorophenyl)methylene ]bis(4-hydroxy-6-methyl- 2H-pyran-2-one) |
Otava codes, structures and names of the compounds are as indicated. All 8 compounds exhibited ≥50% inhibition of NS5B RdRp activity at 100 µM concentration.
Table 2.
Activity of NS5B inhibitors
| Compound | Docking score (kcal/mol) |
NS5B- WT IC50 (µM) |
NS5B- P495L IC50 (µM) |
NS5B- M423T IC50 (µM) |
NS5B- M414T IC50 (µM) |
|---|---|---|---|---|---|
| 1 | −51.27 | 16.0±1.2 | 14.2±2.8 | 126.9±7.4 | 22.9±5.5 |
| 2 | −52.05 | 20.3±1.2 | 19.2±0.4 | 171.8±4.7 | 23.7±1.1 |
| 3 | −38.78 | 33.0±1.3 | 34.3±4.6 | >250 | 28.6±1.8 |
| 4 | −51.28 | 33.1±1.2 | 33.5±1.4 | >250 | 37.8±1.7 |
| 5 | −44.68 | 36.7±1.1 | 43.0±3.8 | >250 | 36.1±3.7 |
| 6 | −44.75 | 39.5±1.3 | 41.0±3.1 | >250 | 44.8±1.2 |
| 7 | −45.45 | 46.2±1.2 | 40.6±2.6 | >250 | 52.1±2.2 |
| 8 | −43.67 | 56.8±1.3 | 63.8±4.4 | >250 | 62.4±2.1 |
Docking scores were determined by docking the compounds into NS5B NNI TP-2 binding site for thiazolone inhibitor (PDB ID: 2O5D). A more negative docking score indicates a better fit at the binding site. The IC50 values of the compounds were determined against wild-type NS5B and its indicated mutant derivatives, from dose-response curves employing 8–12 concentrations for each compound in duplicate in two independent experiments. Curves were fitted to data points using nonlinear regression analysis and IC50 values were interpolated from the resulting curves using GraphPad Prism 3.03 software. The values represent an average from at least two independent experiments in duplicate.
2.3. Correlation between the IC50 values and binding scores of the NS5B inhibitors
Employing Dock package [36–38], we investigated the interactions of the 8 compounds at the NS5B NNI binding site for thiazolone inhibitor (TP-2) and analyzed the relationship between their calculated binding energies (docking score) and IC50 values. This analysis revealed that the docking scores though not-completely in agreement with the compound’s activity (Table 2), served to provide an overall trend. The docking score of compound 3; however, deviated from this trend.
2.4. Mapping the inhibitor binding site on NS5B
With the objective of mapping the binding sites of the identified leads on NS5B, we analyzed their inhibitory activities against NS5B mutants P495L, M423T and M414T as screens for TP-1, TP-2 and Palm pocket-1 (PP-1) site binders, respectively [39, 40]. These residues define the aforementioned allosteric pockets on NS5B and consequently mutations at these sites decrease inhibitor binding, thus resulting in loss of sensitivity to the inhibitor [39–41]. Consistent with our screening strategy selection for TP-2 binders, the inhibitory potency of all 8 compounds was impacted by mutation at M423 residue of NS5B, but not with P495L and M414T mutations (Table 2). This was evident from the 7.9 and 8.5-fold higher IC50 values for compounds 1 and 2 against M423T NS5B relative to wild-type NS5B. The other 6 compounds exhibited >250 µM IC50 value with M423T NS5B, thus amounting to >4.5-fold increase versus wild-type NS5B. By contrast, the IC50 values of the compounds against NS5B mutants P495L and M414T exhibited ≤1.4-fold change relative to their corresponding wild-type NS5B values. Together, this data suggests that the inhibitors bind at TP-2 of NS5B, thus validating our screening strategy.
2.5. Cell-based screening of hit compounds
The BHK-NS5B-FRLuc reporter cells were employed for cellular screening of the 8 hit compounds against HCV NS5B [42]. This reporter cell line carries stably transfected NS5B and a bicistronic reporter gene, (+)FLuc-(−)UTR-RLuc. This enables the simultaneous measurement of intracellular HCV NS5B RdRp activity, derived from the ratio of Renilla to firefly luciferase luminescence, and cellular viability, reflected by the firefly luciferase luminescence, thus facilitating the identification of potent nontoxic inhibitors. All eight compounds displayed no cytotoxicity at 100 µM concentration. Of these, compounds 1 and 2 exhibited between 57–62% inhibition, whereas the remaining 6 compounds with the exception of compound 8 exhibited ~40- 45% inhibition of intracellular NS5B RdRp activity at 100 µM concentration. Compound 8 did not inhibit NS5B at this concentration. While, the overall trend in cell culture seems to be consistent with in vitro inhibition data, confirmation of true antiviral activity in this cell-based assay must await the design of more potent compounds to ensure that the activity is completely devoid of cytotoxicity artifacts.
2.6. Molecular modeling studies
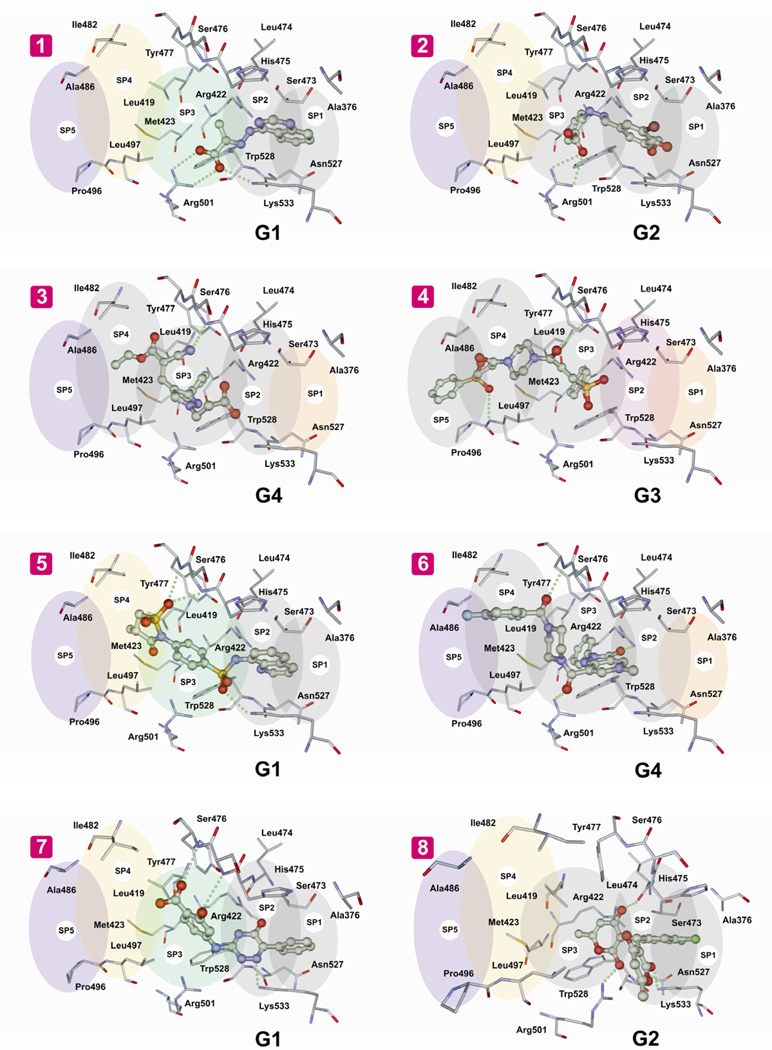
To analyze the binding mode of selected compounds, TP-2 of NS5B was conditionally divided into five subpockets termed SP1 to SP5 (Fig. 1). Each subpocket was defined as a cavity between two flanked residues which describe subpocket’s borders most precisely; other residues potentially involved in the subpocket were neglected. The following residue pairs were attributed to each subpocket: SP1 (Ser473, Asn527), SP2 (His475, Lys533), SP3 (Leu419, Trp528), SP4 (Ile482, Leu497), and SP5 (Ala486, Pro496). According to this simple mapping of NS5B allosteric pocket, the inhibitors were placed into four groups G1-G4, characterized by the inhibitor’s occupancy of one or more distinct subpockets (Fig. 1). Thus, compounds 1, 5 and 7 were placed in group 1, compounds 2 and 8 in group 2, compound 4 in group 3 and compounds 3 and 6 in group 4.
Fig. 1.
Binding mode of NS5B inhibitors and schematic representation of the binding site occupancy. SP1 to SP5 are the five subpockets mapped in the TP-2 of NS5B. G1 to G4 represent the four groups in which the inhibitors were placed, based on their occupancy of one or more distinct pockets. The compounds are represented by their respective numbers 1 to 8. Coloured ovals indicate subpockets appropriate for structural optimization of the inhibitor. Grey ovals reflect the area occupied by the inhibitors. Intermolecular hydrogen bonds/ionic bonds are shown as green dotted lines.
In groups G1 and G2, π-π interaction of the aromatic rings of the inhibitor and residues Trp528 and His475 in SP2 and SP3 are emphasized. These residues are predicted to form key non-polar interactions in NS5B TP-2. Representative inhibitor 4 from group G3 occupies subpockets SP3, SP4 and SP5, while inhibitors 3 and 6 from group G4 occupy opposite part of the allosteric pocket represented by subpockets SP1, SP2 and SP3. All four binding groups G1-G4 display a number of intermolecular hydrogen bonds/ionic interactions with residues Arg501, Lys533, Leu474, Ser476, Tyr477, Leu497, Leu474, and Arg422.
Since the inhibitors identified are structurally diverse, it is hard to extract precisely their key structural components which may be essential for NS5B inhibition. Nonetheless, as per docking analysis, presence of aromatic ring in the structures of inhibitors in close proximity of the indole ring of Trp528, seems to be important for binding. This is common to all the inhibitors except 1 and 5, although in the case of 5 this kind of interaction may also be speculated. There are two main amino acid residue clusters at TP-2, namely Ser476, Tyr477 and Arg501, Lys533, which can form hydrogen bonds and/or ionic interactions with the inhibitors. The corresponding partners for these interactions in the structure of inhibitors 1, 2, 4, 5, 6, 8 are carboxyl, sulfonyl and keto groups. Other H-bond forming substituents are also possible here such as nitrile in 3 and nitrogen atom of 4,5-dihydro-1,2,4-triazin-5-one ring in 7. Taken together, compound 5, a benzenesulfonamide derivative, seems to be druggable and amenable for future structure-based optimization.
3. Conclusions
This study describes our efforts to identify novel HCV NS5B polymerase inhibitor scaffolds. Starting with the virtual screening of 160,000 drug-like organic compounds, followed with post-screening filtering, selection and biochemical investigation of 59 compounds against NS5B, we have identified 8 new structural scaffolds with micromolar inhibitory activity. Further, we have mapped the inhibitor binding site to TP-2 of NS5B by counter screening against TP-2 site mutant, M423T NS5B. Future studies focusing on chemical optimization of druggable scaffolds as potent NS5B inhibitors both in vitro and in cell-based assays are in progress and will be communicated separately.
4. Experimental Section
4.1. Virtual Screening
For virtual screening, drug-like compound library from Otava, Ltd. comprising of 120,000 chemical entities was utilized. This drug-like compound library has been created using Lipinski’s “Rule of Five” [29], and represent Otava’s stock collection of distinct scaffolds of heterocyclic compounds and their fused analogs, produced at the company's synthetic facilities. Purity of compounds tested in biological assays was verified by NMR and/or GC/LC/MS and was found to be greater than 95%. Compounds with reactive groups, biologically unstable compounds, or those containing atoms other than O, N, C, H, Br, I, Cl, F or S, were removed from the library. To predict the NS5B inhibitory activity of these compounds, an in-house screening system was applied. Ligand molecules were processed with SCREENER software [43] which included converting 2D structures to 3D, calculation of ligand geometry with YFF force field [44] and assignment of ligand’s partial atomic charges with Kirchhoff method [45]. Tautomerism and chirality were treated as represented in the compound library without changes. All organic acids and bases were considered in their ionized form. At the next step, molecular docking was performed in the TP-2 of NS5B using DOCK package [36–38]. Crystal structure of NS5B in complex with thiazolone-acylsulfonamide inhibitor (PDB ID 2O5D) was employed for molecular modeling studies. We chose this structure because it is determined at high resolution and converged with the structure-based drug design principle by Yan and colleagues [26]. In addition, the co-crystallized thiazolone derivative occupies major part of the TP-2 site thus producing a favorable binding site conformation suitable for covering wide structural space of docked ligands and opening more directions for further structural optimization. All non-protein molecules were removed from the crystal structure, thus water molecules were not considered in further docking procedure. Minimization of the NS5B crystal structure was performed with GROMACS package (steepest descent algorithm, GROMACS force field) [46]. Partial atomic charges of the protein were calculated using AMBER force field. Allosteric site spheres were calculated with DOCK sphgen software. The spheres with the positions outside the allosteric site were deleted manually. Connolly MS and Grid programs from DOCK package were used to generate allosteric site Connolly surface and energy grids. The grid spacing was set to 0.3 Å. “Multiple anchors” parameter was turned on. The minimum of heavy atoms in ligand molecule anchor was set to 6, the maximum number of orientations was set to 1000, and the “all atoms” model was chosen.
4.2. Biological Studies
4.2.1. HCV NS5B polymerase plasmids and enzyme purification
Wild-type NS5BCΔ21 (1b) and its three mutant derivatives P495L, M423T and M414T, employed in this investigation were a kind gift of Dr. H.-C. Huang [39, 47]. The recombinant NS5BCΔ21 expression plasmids carrying a His6 tag at the C terminus, were expressed and purified from Escherichia coli JM109(DE3) cells by Ni-NTA column chromatography as described previously [33–35].
4.2.2. NS5B inhibition assay
The anti-NS5B activity of the virtually screened candidate compounds was evaluated in vitro by the NS5B RdRp inhibition assay as described previously [33–35]. This reaction utilized poly rA/U12 as the template-primer (TP) and wild-type recombinant NS5BCΔ21 protein as the enzyme source. All compounds were dissolved in dimethylsulfoxide (DMSO) as a 10 mM stock solution and stored at −20°C. Serial dilutions were made in DMSO immediately prior to the assay. To identify candidates belonging to a wider range of structural scaffolds, preliminary screening was conducted at 100 µM compound concentration. Reactions were initiated by the addition of 1.0 mM MnCl2 and terminated by the addition of ice-cold 10% (v/v) trichloroacetic acid (TCA) containing 0.5 mM pyrophosphate. The amount of radioactive UTP incorporated into nascent RNA was subjected to trichloroacetic acid (TCA) precipitation, spotted on a GF-B filter and quantified on a liquid scintillation counter (Packard). Activity of NS5B in the presence of equivalent amounts of DMSO was set at 100% and that in the presence of the inhibitor was calculated relative to this control. Compounds inhibiting NS5B RdRp activity by greater than 50% at 100 µM, were further investigated for their IC50 values.
4.2.3. Mutant counter screen assay
The binding site of the inhibitors on NS5B was investigated employing a mutant counter-screen assay as described previously [39, 40]. Recombinant NS5BCΔ21 mutant proteins P495L, M423T and M414T served as screens for TP-1, TP-2 and PP-1 site binders, respectively. NS5B inhibition assay with the mutants was essentially carried out as described for the wild-type NS5B.
4.2.4. Cell culture
BHK-NS5B-FRLuc reporter cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated fetal bovine serum, 5% antibiotic-antimycotic, 5% nonessential amino acid, 1 mg/mL G418 and 10 µg/mL blasticidin. The reporter cell lines were culured at 37°C in the presence of 5% CO2 supplement.
4.2.5. BHK-NS5B-FRLuc reporter assay
The effect of the compounds on the intracellular HCV NS5B polymerase activity was investigated employing the BHK-NS5B-FRLuc reporter system as previously described [42]. Briefly, BHK-NS5B-FRLuc reporter cells were treated with 100 µM compounds or equivalent concentration of DMSO (1%). After incubation at 37°C for 42 hr, cell lysates were harvested and reporter genes expressions were measured using the Dual-LuciferseR reporter kit (Promega Corporation) in accordance with the manufacturer’s instructions. Effect of the compounds on cell viability was estimated as the relative levels of Firefly luciferase expression in compound treated cells versus DMSO controls. The inhibitory effect of the compounds on intracellular NS5B RdRp activity was evaluated as a percent reduction in Renilla luminescence in the presence of the compound versus DMSO controls after exclusion of any cellular effects of the compounds through normalization of RLuc/FLuc ratio.
Table 3.
Effect of Otava compounds on intracellular NS5B activity in cell based reporter assay.
| Compounds | Cytotoxicity (CC50, µM) | % Inhibition |
|---|---|---|
| 1 | >100 | 62.4±13.0 |
| 2 | >100 | 57.8±16.7 |
| 3 | >100 | 44.2±5.6 |
| 4 | >100 | 42.1±1.6 |
| 5 | >100 | 40.5±17.6 |
| 6 | >100 | 43.3±16.3 |
| 7 | >100 | 45.1±7.3 |
| 8 | >100 | No inhibition |
BHK-NS5B-FRLuc reporter cells were treated with the indicated compounds at 100 µM concentration for 42 hours. Cytotoxicity was estimated as the relative levels of Firefly luciferase in compound treated cells versus DMSO controls, while percent inhibition of intracellular NS5B RdRp activity was evaluated from the percent reduction in RLuc to FLuc luminescence signal in compound treated cells versus DMSO controls. Data represents an average of at least three independent experiments in duplicate.
Research Highlights.
HCV NS5B inhibitors were screened by structure-based virtual screening protocol.
NS5B inhibition and mutant counter screen assay validated HCV NS5B inhibitor leads.
Molecular modeling provided insight for inhibitor scaffold optimization.
Acknowledgements
This work was supported by the National Institute of Health Research Grants DK066837 and CA153147 to N.K.-B. We thank Dr. H.-C. Huang (Merck Research Laboratories, New Jersey, United States), for the wild-type NS5BCΔ21 (1b) and its P495L, M423T and M414T mutant derivative plasmids.
Abbreviations
- AP
allosteric pocket
- HCV
hepatitis C virus
- NI
nucleoside inhibitor
- NNI
non-nucleoside inhibitor
- PP-1
palm pocket-1
- RdRp
RNA-dependent RNA polymerase
- SP
subpocket
- TP
thumb pocket
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seeff LB. Dilemma of the natural history of hepatitis C. J. Gastroenterol Hepatol. 1999;14:199–201. doi: 10.1046/j.1440-1746.1999.01837.x. [DOI] [PubMed] [Google Scholar]
- 2.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 3.Dillon JF. Hepatitis C: What is the best treatment? J. Viral Hepat. 2004;11(Suppl 1):23–27. doi: 10.1111/j.1365-2893.2004.00573.x. [DOI] [PubMed] [Google Scholar]
- 4.Ni ZJ, Wagman AS. Progress and development of small molecule HCV antivirals. Curr. Opin. Drug Discov. Devel. 2004;7:446–459. [PubMed] [Google Scholar]
- 5.Cornberg M, Wedemeyer H, Manns MP. Treatment of chronic hepatitis C with PEGylated interferon and ribavirin. Curr. Gastroenterol. Rep. 2002;4:23–30. doi: 10.1007/s11894-002-0034-y. [DOI] [PubMed] [Google Scholar]
- 6.Reichard O, Norkrans G, Fryden A, Braconier JH, Sonnerborg A, Weiland O. Randomised, double-blind, placebo-controlled trial of interferon alpha-2b with and without ribavirin for chronic hepatitis C. The Swedish Study Group. Lancet. 1998;351:83–87. doi: 10.1016/s0140-6736(97)06088-1. [DOI] [PubMed] [Google Scholar]
- 7.Farci P, Quinti I, Farci S, Alter HJ, Strazzera R, Palomba E, Coiana A, Cao D, Casadei AM, Ledda R, Iorio R, Vegnente A, Diaz G, Tovo PA. Evolution of hepatitis C viral quasispecies and hepatic injury in perinatally infected children followed prospectively. Proc. Natl. Acad. Sci. USA. 2006;103:8475–8480. doi: 10.1073/pnas.0602546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferenci P. Pegylated interferon plus ribavirin for chronic hepatitis C: the role of combination therapy today, tomorrow and in the future. Minerva gastroenterologica e dietologica. 2006;52:157–174. [PubMed] [Google Scholar]
- 9.Rice C. Perspective: miles to go before we sleep. Nature. 2011;474:S8. doi: 10.1038/474S8a. [DOI] [PubMed] [Google Scholar]
- 10.Sheridan C. New Merck and Vertex drugs raise standard of care in hepatitis C. Nat. Biotechnol. 2011;29:553–554. doi: 10.1038/nbt0711-553. [DOI] [PubMed] [Google Scholar]
- 11.Garber K. Hepatitis C: move over interferon. Nat. Biotechnol. 2011;29:963–966. doi: 10.1038/nbt.2031. [DOI] [PubMed] [Google Scholar]
- 12.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 13.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 14.Tanji Y, Hijikata M, Hirowatari Y, Shimotohno K. Hepatitis C virus polyprotein processing: kinetics and mutagenic analysis of serine proteinase-dependent cleavage. J. Virol. 1994;68:8418–8422. doi: 10.1128/jvi.68.12.8418-8422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermehren J, Sarrazin C. New HCV therapies on the horizon. Clin. Microbiol. Infect. 2011;17:122–134. doi: 10.1111/j.1469-0691.2010.03430.x. [DOI] [PubMed] [Google Scholar]
- 16.Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNAdependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 17.Moradpour D, Brass V, Bieck E, Friebe P, Gosert R, Blum HE, Bartenschlager R, Penin F, Lohmann V. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 2004;78:13278–13284. doi: 10.1128/JVI.78.23.13278-13284.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barreca ML, Iraci N, Manfroni G, Cecchetti V. Allosteric inhibition of the hepatitis C virus NS5B polymerase: in silico strategies for drug discovery and development. Future Med. Chem. 2011;3:1027–1055. doi: 10.4155/fmc.11.53. [DOI] [PubMed] [Google Scholar]
- 19.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 20.Bressanelli S, Tomei L, Rey FA, De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 23.Talele TT, Arora P, Kulkarni SS, Patel MR, Singh S, Chudayeu M, Kaushik-Basu N. Structure-based virtual screening, synthesis and SAR of novel inhibitors of hepatitis C virus NS5B polymerase. Bioorg. Med. Chem. 2010;18:4630–4638. doi: 10.1016/j.bmc.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols DB, Fournet G, Gurukumar KR, Basu A, Lee JC, Sakamoto N, Kozielskie F, Musmuca I, Joseph B, Ragno R, Kaushik-Basua N. Inhibition of Hepatitis C Virus NS5B Polymerase by S-Trityl-L-Cysteine Derivatives. Eur. J. Med. Chem. 2012;49:191–199. doi: 10.1016/j.ejmech.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musmuca I, Caroli A, Mai A, Kaushik-Basu N, Arora P, Ragno R. Combining 3-D quantitative structure-activity relationship with ligand based and structure based alignment procedures for in silico screening of new hepatitis C virus NS5B polymerase inhibitors. J. Chem. Inf. Model. 2010;50:662–676. doi: 10.1021/ci9004749. [DOI] [PubMed] [Google Scholar]
- 26.Yan S, Appleby T, Larson G, Wu JZ, Hamatake RK, Hong Z, Yao N. Thiazoloneacylsulfonamides as novel HCV NS5B polymerase allosteric inhibitors: convergence of structure-based drug design and X-ray crystallographic study. Bioorg. Med. Chem. Lett. 2007;17:1991–1995. doi: 10.1016/j.bmcl.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 27.Love RA, Parge HE, Yu X, Hickey MJ, Diehl W, Gao J, Wriggers H, Ekker A, Wang L, Thomson JA, Dragovich PS, Fuhrman SA. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 2003;77:7575–7581. doi: 10.1128/JVI.77.13.7575-7581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Ng KK, Cherney MM, Chan L, Yannopoulos CG, Bedard J, Morin N, Nguyen-Ba N, Alaoui-Ismaili MH, Bethell RC, James MN. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 2003;278:9489–9495. doi: 10.1074/jbc.M209397200. [DOI] [PubMed] [Google Scholar]
- 29.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 30.Yan S, Appleby T, Larson G, Wu JZ, Hamatake R, Hong Z, Yao N. Structure-based design of a novel thiazolone scaffold as HCV NS5B polymerase allosteric inhibitors. Bioorg. Med. Chem. Lett. 2006;16:5888–5891. doi: 10.1016/j.bmcl.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 31.Biswal BK, Wang M, Cherney MM, Chan L, Yannopoulos CG, Bilimoria D, Bedard J, James MN. Non-nucleoside inhibitors binding to hepatitis C virus NS5B polymerase reveal a novel mechanism of inhibition. J. Mol. Biol. 2006;361:33–45. doi: 10.1016/j.jmb.2006.05.074. [DOI] [PubMed] [Google Scholar]
- 32.Ontoria JM, Rydberg EH, Di Marco S, Tomei L, Attenni B, Malancona S, Martin Hernando JI, Gennari N, Koch U, Narjes F, Rowley M, Summa V, Carroll SS, Olsen DB, De Francesco R, Altamura S, Migliaccio G, Carfi A. Identification and biological evaluation of a series of 1H-benzo[de]isoquinoline-1,3(2H)-diones as hepatitis C virus NS5B polymerase inhibitors. J. Med. Chem. 2009;52:5217–5227. doi: 10.1021/jm900517t. [DOI] [PubMed] [Google Scholar]
- 33.Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Chen Y, Kucukguzel SG. 4-Thiazolidinones: a novel class of hepatitis C virus NS5B polymerase inhibitors. Front. Biosci. 2008;13:3857–3868. doi: 10.2741/2974. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Bopda-Waffo A, Basu A, Krishnan R, Silberstein E, Taylor DR, Talele TT, Arora P, Kaushik-Basu N. Characterization of aurintricarboxylic acid as a potent hepatitis C virus replicase inhibitor. Antivir. Chem. Chemother. 2009;20:19–36. doi: 10.3851/IMP1286. [DOI] [PubMed] [Google Scholar]
- 35.Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Costa PR, da Silva AJ, Sarafianos SG, Noel F. Identification and characterization of coumestans as novel HCV NS5B polymerase inhibitors. Nucleic Acids Res. 2008;36:1482–1496. doi: 10.1093/nar/gkm1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J. Comput. Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 37.Ring CS, Sun E, McKerrow JH, Lee GK, Rosenthal PJ, Kuntz ID, Cohen FE. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc. Natl. Acad. Sci. USA. 1993;90:3583–3587. doi: 10.1073/pnas.90.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoichet BK, Stroud RM, Santi DV, Kuntz ID, Perry KM. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993;259:1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- 39.Cheng CC, Shipps GW, Jr, Yang Z, Kawahata N, Lesburg CA, Duca JS, Bandouveres J, Bracken JD, Jiang CK, Agrawal S, Ferrari E, Huang HC. Inhibitors of hepatitis C virus polymerase: synthesis and characterization of novel 2-oxy-6-fluoro-N-((S)-1-hydroxy-3-phenylpropan-2-yl)-benzamides. Bioorg. Med. Chem. Lett. 2010;20:2119–2124. doi: 10.1016/j.bmcl.2010.02.054. [DOI] [PubMed] [Google Scholar]
- 40.Betzi S, Eydoux C, Bussetta C, Blemont M, Leyssen P, Debarnot C, Ben-Rahou M, Haiech J, Hibert M, Gueritte F, Grierson DS, Romette JL, Guillemot JC, Neyts J, Alvarez K, Morelli X, Dutartre H, Canard B. Identification of allosteric inhibitors blocking the hepatitis C virus polymerase NS5B in the RNA synthesis initiation step. Antiviral Res. 2009;84:48–59. doi: 10.1016/j.antiviral.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Soriano V, Peters MG, Zeuzem S. New therapies for hepatitis C virus infection. Clin Infect. Dis. 2009;48:313–320. doi: 10.1086/595848. [DOI] [PubMed] [Google Scholar]
- 42.Lee JC, Tseng CK, Chen KJ, Huang KJ, Lin CK, Lin YT. A cell-based reporter assay for inhibitor screening of hepatitis C virus RNA-dependent RNA polymerase. Anal. Biochem. 2010;403:52–62. doi: 10.1016/j.ab.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Prykhod'ko AO, Yakovenko OYa, Golub AG, Bdzhola VG, Yarmoluk SM. Evaluation of 4H-4-chromenone derivatives as inhibitors of protein kinase CK2. Biopolym. Cell. 2005;21:287–292. [Google Scholar]
- 44.Yakovenko O, Oliferenko AA, Golub AG, Bdzhola VG, Yarmoluk SM. The new method of distribution integrals evaluations for high throughput virtual screening. Ukrainica Bioorganica Acta. 2007;1:52–62. [Google Scholar]
- 45.Yakovenko O, Oliferenko AA, Bdzhola VG, Palyulin VA, Zefirov NS. Kirchhoff atomic charges fitted to multipole moments: implementation for a virtual screening system. J. Comput. Chem. 2008;29:1332–1343. doi: 10.1002/jcc.20892. [DOI] [PubMed] [Google Scholar]
- 46.Lindahl E, Hess B, Spoel Dvd. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 47.Ferrari E, He Z, Palermo RE, Huang HC. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J. Biol. Chem. 2008;283:33893–33901. doi: 10.1074/jbc.M803094200. [DOI] [PMC free article] [PubMed] [Google Scholar]