SUMMARY
Despite the crucial impact of leptin signaling on metabolism and body weight, little is known about the structure of the liganded leptin receptor (LEP-R) complex. Here we applied single-particle electron microscopy (EM) to characterize the architecture of the extracellular region of LEP-R alone and in complex with leptin. We show that unliganded LEP-R displays significant flexibility in a hinge region within the cytokine homology region 2 (CHR2) that is connected to rigid membrane-proximal FnIII domains. Leptin binds to CHR2 in order to restrict the flexible hinge and the disposition of the FnIII ‘legs’. Through a separate interaction, leptin engages the Ig-like domain of a second liganded LEP-R, resulting in the formation of a quaternary signaling complex. We propose that the membrane proximal domain rigidification in the context of a liganded cytokine receptor dimer is a key mechanism for the transactivation of Janus kinases (Jaks) bound at the intracellular receptor region.
Keywords: Leptin Receptor, Cytokine Receptor, Electron Microscopy, Structure
Leptin, a class I cytokine, is secreted from adipose tissue at levels that are proportional to body fat, and after crossing the blood-brain barrier it engages the leptin receptor (LEP-R) in order to modulate energy expenditure and food intake (Bates et al., 2003; Halaas et al., 1995; Morton et al., 2005). Based on controlling homeostasis and growth, leptin affects a range of diverse processes such as glucose level regulation, reproduction, bone formation and wound healing (Ahima et al., 1996; Lord et al., 1998; Peelman et al., 2006a).
Leptin adopts a four-helix bundle structure (Zhang et al., 1997), sharing structural homology to several helical cytokines of the hematopoietin family, such as interleukin-6 (IL-6), leukemia inhibitory factor (LIF), and ciliary neurotrophic factor (CNTF). Based on structural analyses and comparisons with homologous cytokines, leptin possesses three conserved epitopes (I-III) that can be potentially employed for receptor engagement and activation (Bravo and Heath, 2000; Iserentant et al., 2005). Previous biochemical studies suggested that epitope II constitutes the primary binding site of leptin to LEP-R (Peelman et al., 2004). However, the roles of the remaining epitopes in engaging LEP-R are unclear.
LEP-R belongs to the class I cytokine receptor family, which includes glycoprotein 130 (gp130), the LIF receptor (LIF-R), the CNTF receptor (CNTF-R), and others (Baumann et al., 1996; Tartaglia et al., 1995; Wang et al., 2009). These receptors do not possess intrinsic kinase activity, but rely on activating Janus kinases (Jaks) that are constitutively bound to the receptor intracellular domains (ICDs). The signature module of class I cytokine receptors is the extracellular cytokine homology region (CHR), which consists of two domains with a fibronectin type III (FnIII) fold containing the classical motif for cytokine binding (Wang et al., 2009). Along with oncostatin M receptor (OSM-R) and LIF-R, LEP-R is an unusual class I receptor since it contains two CHR modules that are both membrane-distal and separated by an immunoglobulin-like domain (IgD). Both CHR modules represent potential ligand binding sites, however, only CHR2 has been shown to be required for leptin binding (Fong et al., 1998; Iserentant et al., 2005; Peelman et al., 2004). Furthermore, unlike LIF-R and gp130, LEP-R possesses two, rather than three, FnIII membrane-proximal domains. Although the IgD (D3) and the two membrane-proximal FnIII domains are not prerequisites for high-affinity leptin binding, they have been shown to be essential for LEP-R activation (Zabeau et al., 2005; Zabeau et al., 2004).
Earlier studies have provided a wealth of information on the structural organization of cytokine receptor complexes (Boulanger et al., 2003a; Skiniotis et al., 2005; Skiniotis et al., 2008; Tamada et al., 2006). Recently, a crystal structure of human LEP-R CHR2 in complex with a Fab fragment from a leptin blocking monoclonal antibody provided insights into the mechanism of antagonism (Carpenter et al., 2012). However, owing to the existence of the three conserved epitopes on leptin, the structure of the signaling leptin/LEP-R complex has been a matter of debate, with the two main models proposing either a 2:2 or a 2:4 stoichiometry between leptin and LEP-R (Couturier and Jockers, 2003; Mistrik et al., 2004; Peelman et al., 2006b). Here we used single-particle electron microscopy (EM) to visualize the extracellular portion of LEP-R alone and in complex with the cytokine, thus elucidating the architecture of the signaling assembly and obtaining valuable insights into the mechanism of signal transduction.
RESULTS
Assembly of the Leptin/LEP-R complex
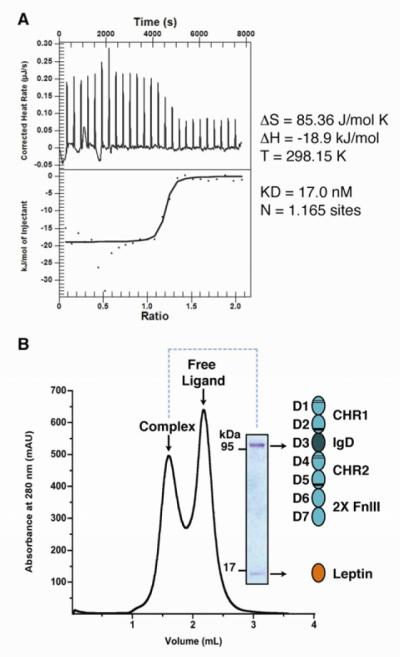
For the present study we used a baculovirus system to express and purify the extracellular region (domains D1-D7) of murine LEP-R and also a truncated construct (D1-D5) lacking the two membrane-proximal FnIII domains (Figure S1). To evaluate leptin/LEP-R complex formation we employed isothermal titration calorimetry (ITC) to measure the thermodynamics of the interaction. For both constructs, the ITC measurements suggested that leptin engages LEP-R by high affinity interactions, with a KD value of ~17 nM (Figures 1A, S1). Importantly, ITC indicates a ratio of ~1:1 between receptor and ligand, providing the first indication for stoichiometric complex formation. We thus incubated purified LEP-R[D1-D7] and LEP-R[D1-D5] with recombinant murine leptin and used size exclusion chromatography to isolate liganded receptor complexes for EM analysis (Figures 1B, S1).
Figure 1. Purification and thermodynamic analysis of the assembly of Leptin/LEP-R complexes.
A) Isothermal titration calorimetry for the assembly of Leptin and LEP-R. The inset table lists the thermodynamic parameters for the binding isotherm. B) Size exclusion chromatography profile and SDS-PAGE analysis of purified liganded LEP-R[D1-D7].
Rigid Membrane-Proximal Domains Connected to a Flexible CHR2
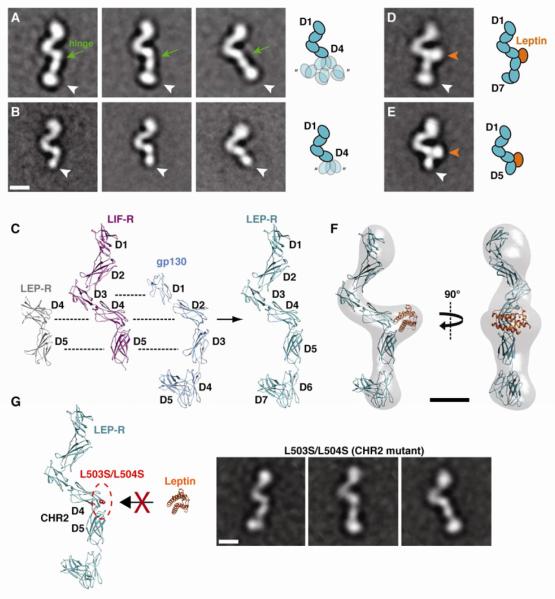
In a first step we examined unliganded LEP-R preparations by negative stain EM (Figure S2). Class averages of the D1-D7 construct revealed a single preferred orientation of a distinct rod-like structure of approximately 21 nm in length, which displays a characteristic ‘V’ shape closer to one end (Figure 2A). Comparison of EM averages from the D1-D7 construct and the shorter D1-D5 construct (~16.5 nm) reveals the orientation of the LEP-R termini, with the ‘V’ shape being closer to the N-terminus (Figure 2B). A higher density lobe at the C-terminus of the D1-D7 construct suggests that the two membrane-proximal FnIII domains likely assume a sharp bend, similar to what has been observed in the crystal structure of the gp130 ectodomain (Xu et al., 2010). Beyond the membrane-proximal domains, the observed structure is highly reminiscent to the extended “flying V” architecture observed in the crystal structures of murine and human LIF-R (Huyton et al., 2007; Skiniotis et al., 2008) (Figure 2C). In this configuration, the D1-D2 and D4-D5 CHR modules adopt the canonical bent elbow shape commonly seen in other cytokine receptors, while the IgD (D3) is centrally positioned at the base of the ‘V’. Given the highly similar extracellular domain architecture of LIF-R[D1-D5] and gp130[D1-D6], and also the similarities with the recently solved structure of the CHR2 region of LEP-R (Carpenter et al., 2012), we aligned and merged their available crystal structures to produce a homology model for the entire extracellular region of LEP-R (Figure 2C). This model is in striking agreement with the projection averages of LEP-R, indicating the common domain organization of the tall cytokine receptor family.
Figure 2. Conformational dynamics of LEP-R in unliganded and liganded states.
A) Representative 2D class averages of unliganded LEP-R[D1-D7] reveal significant flexibility in a hinge between D4 and a rigid D5-D7 module (white arrowheads). B) 2D class averages of unliganded LEP-R[D1-D5] confirm the domain assignments and the variable disposition of D5 (white arrowheads) in regards to D4. C) Comparison of the crystal structures from LEP-R, gp130 and LIF-R extracellular domains and homology model for LEP-R[D1-D7]. D) Representative 2D class average of the binary leptin/LEP-R[D1-D7] complex. The cytokine binds to CHR2 resulting in the stabilization of the rigid D5-D7 module in a single conformation. E) Representative 2D class average the binary leptin/LEP-R[D1-D5] complex. The orange and white arrowheads point to the leptin density and the LEP-R C-terminus, respectively. F) 3D reconstruction of the binary leptin/LEP-R[D1-D7] complex with docked leptin/LEP-R homology model. G) The double mutation L503S/L504S on D4 of Lep-R abolishes leptin binding via epitope II. EM class averages of this mutant after incubation with leptin reveal only monomeric receptor chains with no ligand bound at CHR2 (compare to A and D). All scale bars correspond to 5 nm.
Interestingly, D5 of CHR2 and the membrane-proximal FnIII domains (D6-D7) are well defined and assume the same angle, suggesting that they behave as a rigid body (Figure 2A). However, the orientation of the D5-D7 module in respect to the central ‘V’ is highly variable and assumes a continuum of angles with a range of ~40° in the plane of the carbon support of the EM grid. This observation suggests that the linker connecting the two domains composing CHR2 (D4-D5) is highly flexible and allows for variability in the relative configuration of the connected domains. Indeed, examination of class averages of the truncated construct lacking the FnIII ‘legs’ (D6-D7) clearly reveals that D5 assumes variable positions around D4 with the same angular range observed in the full-length extracellular construct (Figure 2B).
Leptin Engages CHR2 to Stabilize the Membrane-Proximal Domains
In a next step we used single-particle EM analysis to examine leptin/LEP-R complexes (for a detailed description see materials and methods and Figures S3-S5). A fraction of the particles from liganded LEP-R[D1-D7] and LEP-R[D1-D5] reveal a single preferred orientation of monomeric chains with an additional distinct globular density towards the middle of the receptor, at the junction between domains D4 and D5 (Figure 2D,E; approximately 15% and 25% of particles, respectively). The extra density can only be attributed to a leptin molecule bound to CHR2. This type of interaction has been previously observed in the signaling complexes of IL-6 with gp130 (Boulanger et al., 2003b; Skiniotis et al., 2005) and GCSF with GCSF-R (Tamada et al., 2006), where the cytokines use epitope II to interact with the elbow of the CHR module. To support this interpretation, we calculated a 3D reconstruction of the liganded monomeric LEP-R chain and compared it to our homology model including a site II interaction of leptin (Figures 2F, S3). Docking of the model in the 3D map shows a very good fit, reinforcing our analysis of the leptin/LEP-R interaction. Furthermore, we produced and analyzed a LEP-R[D1-D7] construct bearing the mutations L503S/L504S at the CHR2, originally described by Tavernier and colleagues (Iserentant et al., 2005). As expected, incubation of this mutant with leptin did not result in binary complex formation, and EM analysis showed that there was no additional globular density attached to the region that we attributed to CHR2 (Figures 2G, S4). Interestingly, none of the averages from monomeric receptor chains display leptin density at the level of the IgD (D3), located at the base of the flying ‘V’ architecture (Figure 2D-E). These observations suggest that the CHR2 of LEP-R represents the primary site for leptin binding via a high-affinity interaction, as also supported by our binding thermodynamics data (Figures 1A, S1). Importantly, the liganded monomeric LEP-R chains from both extracellular constructs do not display any variability in the relative disposition of the D4 and D5 domains of CHR2. Class averages from this population reveal a distinct single conformation of the D5-(D6-D7) module with a fixed angle in relation to the membrane-distal domains (Figure S3). Thus, leptin binding on CHR2 constrains D5 and the rigidly attached membrane-proximal FnIII domains (D6-D7) in a fixed orientation towards the plane of the membrane.
Liganded LEP-R Assembles into a 2:2 Quaternary Complex
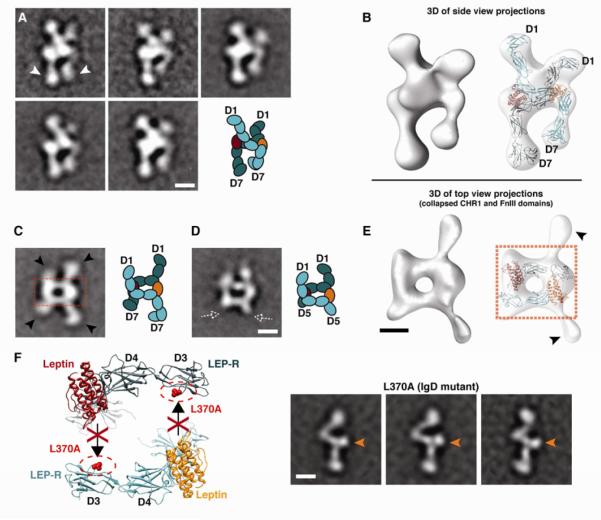
The majority of leptin/LEP-R complexes display variable views of dimeric LEP-R chains. A prominent set of class averages reveals a characteristic side view of the complex, where the two LEP-R chains appear to be crossing at the level of the CHR2 module, while both the CHR1 and the membrane-proximal modules remain unengaged (Figure 3A). This interpretation was confirmed by 3D reconstructions and molecular docking, showing that the LEP-R chains cross over to opposite sides at the level of CHR2 and IgD (Figure 3B). In most class averages the two LEP-R chains appear asymmetric, presumably due to the projection angle and distortion of the receptor chains on the carbon support of the EM grid, as was also the case with our study of the gp130/IL6/IL6-R complex (Skiniotis et al., 2005). In support of this notion, averages of relatively few particles in our population show a perfectly symmetric dimeric complex formation (Figure S3). Nevertheless, in all projection averages where the two LEP-R chains are clearly distinguished the membrane-proximal domains appear to be constrained and point towards each other (Figure 3A). This conformation reflects the same fixed geometry observed in the liganded monomeric LEP-R chains (Figure 2D-F). In the context of the signaling LEP-R dimer, leptin-induced stabilization of CHR2 appears to result in fixing the two membrane-proximal domains towards each other and in close proximity at their C-terminal tips.
Figure 3. Architecture of the quaternary leptin/LEP-R signaling complex.
A) 2D class averages of side-view projections reveal the cross-over configuration of two LEP-R extracellular chains. The two chains connect at the CHR2 level, while the membrane-proximal domains (white arrowheads) point towards each other and arrive in close proximity at their C-terminal tips. B) 3D reconstruction of the leptin/LEP-R side-view projections shown in (A). Flexible docking of two liganded LEP-R chains into the 3D density map shows the complex configuration (right). C) Representative 2D class average of a top view of the quaternary leptin/LEP-R[D1-D7] complex. In this type of projections the N-terminal CHR1 and C-terminal FnIII domains are collapsed on the carbon support (black arrowheads). The boxed area shows the rectangular formation that is reminiscent of the anti-parallel gp130/IL-6 or GCSF/GCSF-R interaction. D) Representative 2D class average of a top view of the quaternary leptin/LEP-R[D1-D5] complex, as in (C). The dashed white arrowheads point to missing densities from the omitted C-terminal domains of the truncated construct, as compared to (C). E) 3D reconstruction from top-view leptin/LEP-R[D1-D7] projections shown in (C), and docked gp130/IL-6 crystal structure into the central rectangular density of the 3D map (right). F) Mutation L370A on the IgD (D3) of LEP-R abolishes leptin binding via epitope III. EM class averages of this mutant after incubation with leptin reveal only monomeric receptor chains with ligand bound at CHR2 (orange arrows). All scale bars correspond to 5 nm.
An abundant series of class averages reveals a top view of the complex (Figure 3C), reminiscent to the top views we characterized in our earlier study of the gp130/IL6/IL6-R complex (Skiniotis et al., 2005). Here we also recognize a central rectangular formation with emanating projections from membrane-proximal FnIII and N-terminal CHR1 domains that collapse in a distinct fashion on the carbon support (pointed by arrows in Figure 3C). The rectangular cap architecture represents the top view of the tetrameric arrangement between leptin and LEP-R that follows the identical topological blueprint of the gp130/IL6 (Skiniotis et al., 2005) or GCSF/GCSF-R interaction (Tamada et al., 2006). In this arrangement, while epitope II of leptin interacts with the CHR2 (D4-D5) of LEP-R, epitope III engages the IgD (D3) of the second, opposing receptor. This set of interactions, which was also observed in class averages of complexes formed by the truncated LEP-R chains (Figure 3D), results in the formation of a closed tetrameric cap with antiparallel subunits of leptin and LEP-R. This organization is further supported by the good fit of the gp130/IL-6 structure (Boulanger et al., 2003b) in the 3D EM reconstruction of the collapsed top view of the leptin/LEP-R complex (Figure 3E), and also by the good match between our 2D projections and reprojections of a quaternary 3D model in this arrangement (Figure S5). To fully confirm this interpretation, we produced and analyzed a LEP-R[D1-D7] construct bearing the IgD mutation L370A, which has been previously shown to abolish leptin signaling (Peelman et al., 2006b). EM analysis of this IgD mutant LEP-R reveals that while it forms the binary complex with leptin through CHR2, it is unable to form the quaternary complex that is based on the interaction of the IgD with epitope III of leptin (Figures 3F, S4). Thus, the quaternary signaling leptin/LEP-R complex forms with a 2:2 stoichiometry by following the same organizing principles as gp130/IL6 and GCSF/GCSF-R complexes (Figure 4).
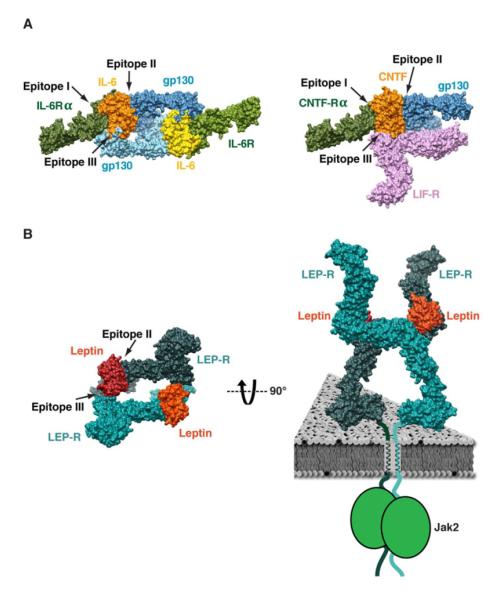
Figure 4. Signaling architecture of tall cytokine receptors.
Receptor organization and ligand epitope usage in the signaling complexes of gp130/IL-6/IL-6Rα (A; left), gp130/LIF-R/CNTF/CNTF-Rα (A; right), and leptin/LEP-R (B; left). Leptin employs only epitopes II and III to engage the CHR2 and IgD of LEP-R, respectively. Leptin-induced stabilization of each CHR2 in the quaternary complex results in the constrained and close disposition of the membrane-proximal domains, which likely favors intracellular Jak2 trans-phosphorylation (B; right).
DISCUSSION
Here we employed single-particle EM to characterize the architecture of the extracellular region of LEP-R alone and in complex with leptin. The unliganded receptor appears highly similar to the crystal structures of extracellular regions from gp130 (Xu et al., 2010), LIF-R (Huyton et al., 2007; Skiniotis et al., 2008), and GCSF-R (Tamada et al., 2006) (Figure 2). This result is perhaps not unexpected, considering the high sequence similarity and common domain organization of tall cytokine receptors. Nevertheless, this finding reinforces the notion that the overall architecture of these receptors is relatively constrained, although the extracellular chains are composed of an array of linker-connected FnIII and Ig-like domains. A puzzling issue however is that crystal structures of gp130 and LIF-R show the same receptor conformation in the presence and absence of the ligand, with a CHR2 configuration that is similar to the one observed in the recently determined crystal structure of LEP-R CHR2 in complex with a Fab fragment (Carpenter et al., 2012). Here we show that LEP-R displays significant flexibility in the hinge region connecting the D4 and D5 domains composing CHR2, which presumably allows the membrane-proximal domains to assume variable configurations in the absence of ligand. In contrast, leptin binding on the CHR2 of LEP-R rigidifies the position of D5, thereby stabilizing the membrane-proximal FnIII domains (D6-D7) in a single conformation (Figure 2D-F). The obtained LEP-R homology model, based on gp130 and LIF-R unliganded crystal structures, fits well in the 3D reconstruction of monomeric LEP-R in complex with leptin (Figure 2C,F). This observation suggests that crystallization conditions may have induced the unliganded receptors to assume the same conformation as in the presence of ligand.
The 2D projection analysis reveals that all liganded monomeric LEP-R chains display stable leptin binding on CHR2. This finding indicates that CHR2 is the primary site for leptin binding with a high-affinity interaction, as suggested by our ITC experiments (Figure 1A, S1) and earlier biochemical studies (Fong et al., 1998; Iserentant et al., 2005). Based on the homologous interactions observed for gp130 (Boulanger et al., 2003b) and GCSF-R (Tamada et al., 2006), the LEP-R CHR2 interaction must be maintained through epitope II of leptin, as also supported by mutagenesis studies (Iserentant et al., 2005) (Figure 4). 3D reconstructions and modeling suggest that conserved epitope III of leptin is used for engaging the IgD (D3) of the second receptor chain that is juxtaposed in an antiparallel fashion. In the case of gp130 homodimers and heterodimers (e.g., gp130/LIF-R), epitope I is used to engage the CHR of a non-signaling α-receptor (e.g., IL6-Rα or CNTF-Rα) that is required for signaling complex formation (Boulanger et al., 2003b; Skiniotis et al., 2008) (Figure 4A). However, the leptin/LEP-R signaling complex does not include a non-signaling α-receptor. This omission has led to the proposal that four LEP-R chains participate in the signaling complex through the additional engagement of epitope I. This is clearly not the case, as we show here that the signaling complex between leptin and LEP-R forms with a 2:2 stoichiometry, by engaging only leptin epitopes II and III (Figure 4B). Curiously, the N-terminal CHR1 of LEP-R does not appear to participate in any interactions, which has also been the case with the CHR1 of LIF-R in the gp130/LIF-R/CNTF/CNTF-Rα complex (Skiniotis et al., 2008). It will thus be interesting to examine whether CHR1 regions have a different type of functionality independent of signaling.
It is also worth noting that our results explain the absence of observed leptin density interacting with the IgD (D3) in monomeric LEP-R chains. Similarly to the gp130/IL6 system (Boulanger et al., 2003b), the epitope III interaction has undetectably low affinity for LEP-R IgD alone, and is only stabilized by the avidity afforded by the “two-point attachment” between preformed and antiparallel leptin/LEP-R dimeric complexes (Figure 4B). In this context, the mode of complex formation is likely cooperative, with leptin binding first to the CHR2 of one LEP-R with a 1:1 stoichiometry, followed by two liganded LEP-Rs engaging at the membrane distal regions through IgD-epitope III interactions.
Earlier work on gp130 and gp130/LIF-R signaling complexes has shown that the membrane-proximal FnIII domains of two juxtaposed receptors bend towards each other to reach the same position at the membrane level (Skiniotis et al., 2005; Skiniotis et al., 2008). Within this family of receptors, LEP-R is the only member possessing two, rather than three, FnIII domains connecting the distal cytokine binding regions to the cell membrane. Perhaps this “handicap” is utilized to differentiate LEP-R in the types of intracellular signaling it exerts. Nevertheless, we show here that the FnIII ‘legs’ in the leptin/LEP-R quaternary signaling complex also point towards each other and come in close proximity at their C-terminal tips. For the tall class of cytokine receptor liganded complexes the present study shows that this configuration of the FnIII ‘legs’ is induced and stabilized by the binding of the cytokine on the flexible CHR. In the absence of ligand, the membrane proximal FnIII domains and the preceding C-terminal domain of CHR appear to behave like rigid rods that assume variable relative orientations in regards to the membrane plane (Figure 2A-B). Ligand binding on the CHR fixes the membrane-proximal domains in a single configuration that facilitates precise juxtaposition at the membrane level (Figures 2D-E, 3A-B, 4).
Our EM studies with full-length gp130 and LIF-R constructs have suggested that the dimeric arrangement of the extracellular membrane-proximal domains was significantly stabilized by the presence of the receptor trans-membrane regions. As has been shown for the erythropoietin receptor, this is likely facilitated by dimerization properties of the receptor single-pass α-helices spanning the membrane (Constantinescu et al., 2001b). It is thus reasonable to assume that LEP-R and most cytokine receptors are preformed non-signaling dimers at the cell membrane. Elegant mutagenesis experiments by Constantinescu et al. (2001) on the erythropoietin receptor have revealed that the exact disposition and pitch of the TM helices is crucial for intracellular signaling (Constantinescu et al., 2001a). Given the leptin-induced stabilization and precise disposition of the membrane-proximal LEP-R regions observed here, we postulate that ligand binding on the membrane-distal regions of receptor dimers is rigidly transmitted towards the receptor TM helices (Figure 4B). This likely represents a common mechanism for cytokine and hormone receptors to stabilize an intracellular conformation that favors Jak trans-phosphorylation.
EXPERIMENTAL PROCEDURES
For a detailed description see Supplemental Experimental Procedures.
Protein Expression & Purification
LEP-R constructs were expressed in Sf9 cells and purified using nickel affinity purification followed by size exclusion chromatography.
Isothermal Titration Calorimetry
Titrations were performed on a NanoITC™ – Low Volume calorimeter (TA Instruments) at 25 °C
Electron Microscopy and Image Processing
Negative stained samples were imaged with a Tecnai T12 electron microscope operated at 120 kV using low-dose procedures. Reference-free alignment and classifications of 2D projections were carried out using SPIDER. (Frank et al., 1996). 3D reconstructions were calculated initially by the random conical tilt method (Radermacher et al., 1987) and further refined using FREALIGN (Grigorieff, 2007).
Molecular Modeling and Docking
Leptin/LEP-R models were built by aligning the homologous domains from crystal structures of gp130/IL-6 (Boulanger et al., 2003b), LIF-R (Huyton et al., 2007; Skiniotis et al., 2008), and leptin (Zhang et al., 1997). All docking operations in the EM densities were performed manually with visual inspection of the best fit.
Supplementary Material
HIGHLIGHTS.
The extracellular architecture of LEP-R is strikingly similar to gp130 and LIF-R
Unliganded LEP-R is highly flexible in a hinge region within CHR2
Leptin employs epitope I to bind to the CHR2 of LEP-R and rigidify its conformation
Two binary Leptin/LEP-R complexes engage to form a quaternary signaling complex
Acknowledgments
We thank M.G. Myers for the leptin receptor clone and for insightful discussions, C. Brown for LIC vectors, Verna Frasca at GE Healthcare and Krishnapriya Chinnaswamy for the ITC experiments. This work was supported by NIDDK R01 DK090165 (to G.S.); the University of Michigan Biological Sciences Scholars Program (G.S.); and the National Institute of General Medical Sciences (NIGMS) Pharmacological Sciences Training Program Grant, GM07767 (to L.V.M.). G.S. is a Pew Scholar of Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahima RS, Prabakaran D, Mantzoros C, Qu D, Lowell B, Maratos-Flier E, Flier JS. Role of leptin in the neuroendocrine response to fasting. Nature. 1996;382:250–252. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AW, Wang Y, Banks AS, Lavery HJ, Haq AK, Maratos-Flier E, et al. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003;421:856–859. doi: 10.1038/nature01388. [DOI] [PubMed] [Google Scholar]
- Baumann H, Morella KK, White DW, Dembski M, Bailon PS, Kim H, Lai CF, Tartaglia LA. The full-length leptin receptor has signaling capabilities of interleukin 6-type cytokine receptors. Proc Natl Acad Sci U S A. 1996;93:8374–8378. doi: 10.1073/pnas.93.16.8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell. 2003a;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003b;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- Bravo J, Heath JK. Receptor recognition by gp130 cytokines. EMBO J. 2000;19:2399–2411. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B, Hemsworth GR, Wu Z, Maamra M, Strasburger CJ, Ross RJ, Artymiuk PJ. Structure of the human obesity receptor leptin-binding domain reveals the mechanism of leptin antagonism by a monoclonal antibody. Structure. 2012;20:487–497. doi: 10.1016/j.str.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Constantinescu SN, Huang LJ, Nam H, Lodish HF. The erythropoietin receptor cytosolic juxtamembrane domain contains an essential, precisely oriented, hydrophobic motif. Mol Cell. 2001a;7:377–385. doi: 10.1016/s1097-2765(01)00185-x. [DOI] [PubMed] [Google Scholar]
- Constantinescu SN, Keren T, Socolovsky M, Nam H, Henis YI, Lodish HF. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc Natl Acad Sci U S A. 2001b;98:4379–4384. doi: 10.1073/pnas.081069198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier C, Jockers R. Activation of the leptin receptor by a ligand-induced conformational change of constitutive receptor dimers. J Biol Chem. 2003;278:26604–26611. doi: 10.1074/jbc.M302002200. [DOI] [PubMed] [Google Scholar]
- Fong TM, Huang RR, Tota MR, Mao C, Smith T, Varnerin J, Karpitskiy VV, Krause JE, Van der Ploeg LH. Localization of leptin binding domain in the leptin receptor. Molecular pharmacology. 1998;53:234–240. doi: 10.1124/mol.53.2.234. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of structural biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. Journal of structural biology. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, Lallone RL, Burley SK, Friedman JM. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- Huyton T, Zhang JG, Luo CS, Lou MZ, Hilton DJ, Nicola NA, Garrett TP. An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc Natl Acad Sci U S A. 2007;104:12737–12742. doi: 10.1073/pnas.0705577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserentant H, Peelman F, Defeau D, Vandekerckhove J, Zabeau L, Tavernier J. Mapping of the interface between leptin and the leptin receptor CRH2 domain. J Cell Sci. 2005;118:2519–2527. doi: 10.1242/jcs.02386. [DOI] [PubMed] [Google Scholar]
- Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- Mistrik P, Moreau F, Allen JM. BiaCore analysis of leptin-leptin receptor interaction: evidence for 1:1 stoichiometry. Anal Biochem. 2004;327:271–277. doi: 10.1016/j.ab.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Morton GJ, Blevins JE, Williams DL, Niswender KD, Gelling RW, Rhodes CJ, Baskin DG, Schwartz MW. Leptin action in the forebrain regulates the hindbrain response to satiety signals. The Journal of clinical investigation. 2005;115:703–710. doi: 10.1172/JCI200522081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelman F, Couturier C, Dam J, Zabeau L, Tavernier J, Jockers R. Techniques: new pharmacological perspectives for the leptin receptor. Trends in pharmacological sciences. 2006a;27:218–225. doi: 10.1016/j.tips.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Peelman F, Iserentant H, De Smet AS, Vandekerckhove J, Zabeau L, Tavernier J. Mapping of binding site III in the leptin receptor and modeling of a hexameric leptin.leptin receptor complex. J Biol Chem. 2006b;281:15496–15504. doi: 10.1074/jbc.M512622200. [DOI] [PubMed] [Google Scholar]
- Peelman F, Van Beneden K, Zabeau L, Iserentant H, Ulrichts P, Defeau D, Verhee A, Catteeuw D, Elewaut D, Tavernier J. Mapping of the leptin binding sites and design of a leptin antagonist. J Biol Chem. 2004;279:41038–41046. doi: 10.1074/jbc.M404962200. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Boulanger MJ, Garcia KC, Walz T. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat Struct Mol Biol. 2005;12:545–551. doi: 10.1038/nsmb941. [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol Cell. 2008;31:737–748. doi: 10.1016/j.molcel.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, Kuroki R. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. Proc Natl Acad Sci U S A. 2006;103:3135–3140. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kershaw NJ, Luo CS, Soo P, Pocock MJ, Czabotar PE, Hilton DJ, Nicola NA, Garrett TP, Zhang JG. Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J Biol Chem. 2010;285:21214–21218. doi: 10.1074/jbc.C110.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeau L, Defeau D, Iserentant H, Vandekerckhove J, Peelman F, Tavernier J. Leptin receptor activation depends on critical cysteine residues in its fibronectin type III subdomains. J Biol Chem. 2005;280:22632–22640. doi: 10.1074/jbc.M413308200. [DOI] [PubMed] [Google Scholar]
- Zabeau L, Defeau D, Van der Heyden J, Iserentant H, Vandekerckhove J, Tavernier J. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Molecular endocrinology (Baltimore, Md. 2004;18:150–161. doi: 10.1210/me.2003-0078. [DOI] [PubMed] [Google Scholar]
- Zhang F, Basinski MB, Beals JM, Briggs SL, Churgay LM, Clawson DK, DiMarchi RD, Furman TC, Hale JE, Hsiung HM, et al. Crystal structure of the obese protein leptin-E100. Nature. 1997;387:206–209. doi: 10.1038/387206a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.