Abstract
An innovative approach to enhance the selectivity of matrix metalloproteinase (MMP) inhibitors comprises targeting these inhibitors to catalytically required substrate binding sites (exosites) that are located outside the catalytic cleft. In MMP-2, positioning of collagen substrate molecules occurs via a unique fibronectin-like domain (CBD) that contains three distinct modular collagen binding sites. To characterize the contributions of these exosites to gelatinolysis by MMP-2, seven MMP-2 variants were generated with single, or concurrent double and triple alanine substitutions in the three fibronectin type II modules of the CBD. Circular Dichroism spectroscopy verified that recombinant MMP-2 wild-type (WT) and variants had the same fold. Moreover, the MMP-2 WT and variants had the same activity on a short FRET peptide substrate that is hydrolyzed independently of CBD binding. Among single-point variants, substitution in the module 3 binding site had greatest impact on the affinity of MMP-2 for gelatin. Simultaneous substitutions in two or three CBD modules further reduced gelatin binding. The rates of gelatinolysis of MMP-2 variants were reduced by 20–40% following single-point substitutions, by 60–75% after double-point modifications, and by >90% for triple-point variants. Intriguingly, the three CBD modules contributed differentially to cleavage of dissociated α-1(I) and α-2(I) collagen chains. Importantly, kinetic analyses (kcat/Km) revealed that catalysis of a triple- helical FRET peptide substrate by MMP-2 relied primarily on the module 3 binding site. Thus, we have identified three collagen binding site residues that are essential for gelatinolysis and constitute promising targets for selective inhibition of MMP-2.
Keywords: Matrix metalloproteinase-2, MMP-2, proteolysis, exosite, gelatin, gelatinolysis
1. Introduction
The family of matrix metalloproteinases (MMPs) consists of 25 endopeptidases which are structurally and functionally related (Steffensen et al., 2001; Sternlicht and Werb, 2001). The MMP family consists of members with complimentary specificities, i.e., collagenases and gelatinases, which taken together are capable of cleaving collagen in different conformations. MMPs also have activities against a number of non-collagen macromolecules of the extracellular matrix, and exert important regulatory functions by cleaving growth factors and chemokines (Butler and Overall, 2009). Whereas the proteolytic activities of MMPs contribute to normal tissue regulation and remodeling, significantly elevated expression and activities of MMPs have been strongly associated with excessive tissue degradation in several pathological conditions. These pathologies include chronic inflammation such as arthritis and periodontal disease, as well as tumor expansion and metastasis in cancer (Sternlicht and Werb, 2001; Egeblad and Werb, 2002).
Efforts to develop MMP inhibitors for treatment of these diseases produced compounds, such as the hydroxamic acid type inhibitors, that efficiently inhibited MMPs. In spite of inhibitory activities on tumors, low selectivity between MMPs resulted in extensive side effects among the patients and necessitated discontinuation of the studies (Brown, 1998; Coussens et al., 2002). This low selectivity originated from the fact that inhibitors targeting the enzyme active sites face the challenge of very similar chemistry and configuration of these sites across the MMPs (Overall and Lopez-Otin, 2002).
To overcome this inhibitor design dilemma, it is pertinent to consider a developing body of information on substrate binding sites, which are located outside the active site cleft and are required for positioning the substrate molecules during hydrolysis (exosites) (Overall and Lopez-Otin, 2002; Xu et al., 2007; Lauer-Fields et al., 2008; Lauer-Fields et al., 2009). MMP-2 and MMP-9 primarily utilize a unique collagen binding domain (CBD) consisting of three in-tandem fibronectin-like type II modules inserted into the catalytic domain for positioning collagen (Collier et al., 1992; Murphy et al., 1994; Steffensen et al., 1995) and elastin (Shipley et al., 1996) substrate molecules. Domain deletion of the CBD in MMP-2 attenuated cleavage of gelatin (denatured collagen) by ~90% (Murphy et al., 1994) and of elastin (Shipley et al., 1996). Among those MMPs which most efficiently cleave the interstitial triple-helical collagens, MMP-1, MMP-8, MMP-13, and MMP-14, the hemopexin-like (PEX) C-terminal domain has been shown to contribute mostly as an exosite (Murphy et al., 1992). Moreover, we found recently that the PEX of MMP-2 is critical for cleavage of fibronectin (Steffensen et al., 2011). Whereas PEX is not critical for gelatin cleavage, additional studies point to contributions of both CBD and PEX during MMP-2 cleavage of native type I collagen (Patterson et al., 2001). Collectively, these results provide a convincing rationale for investigation of exosites as highly promising targets of selective MMP inhibitors.
Extending those observations, our laboratory has conducted a series of experiments to identify precise binding site residues on the CBD from MMP-2. By screening a one-bead one-peptide combinatorial random library (Lam et al., 1996) with the CBD as bait, we identified a CBD-binding peptide with 71% sequence identity to residues 715-721 of the human α1(I) collagen chain (Xu et al., 2007). A 13 amino acid synthetic peptide from this region termed P713 specifically blocked the CBD-mediated binding of MMP-2 to gelatin, and also inhibited the gelatinolytic activities of MMP-2 (Xu et al., 2007). A triple-helical version of P713 exhibited similar properties (Lauer-Fields et al., 2008). Subsequent NMR studies of CBD in complex with P713 enabled us to identify the specific collagen binding site residues in all three modules of the CBD (Xu et al., 2009). The gelatin binding property of the tri-modular CBD was significantly reduced by alanine substitutions of single residues in each of the three putative modular binding sites and was not fully compensated by potential redundancies exerted by the other two modules. Concurrent substitutions of key residues in all three modular binding sites eliminated gelatin binding (Xu et al., 2009).
On the basis of these key observations on gelatin binding from isolated CBD, we have investigated the functional implications of these putative exosites in full length MMP-2. The present experiments demonstrate that highly localized alanine substitutions of single amino acids are sufficient to significantly inhibit the catalytic activities of MMP-2. Moreover, by characterizing the critical and differential contributions of individual exosites and residues to MMP-2 functions, we have verified that these sites are promising targets for development of selective inhibitors for MMP-2.
2. Results
2.1 Recombinant MMP-2 WT and variants with alanine substitutions in the collagen binding domain are structurally homologous and catalytically functional
To investigate the contributions of the collagen binding sites in individual CBD modules to the gelatin binding properties and gelatinolytic activities of MMP-2, we generated a series of seven MMP-2 variants containing single, double and triple point alanine substitutions (Fig. 1). The variant forms of MMP-2 expressed well in Escherichia coli. The refolded and purified proteins were homogenous, monomeric, and displayed masses by SDS/PAGE that equaled that of WT MMP-2 (Fig. 2A).
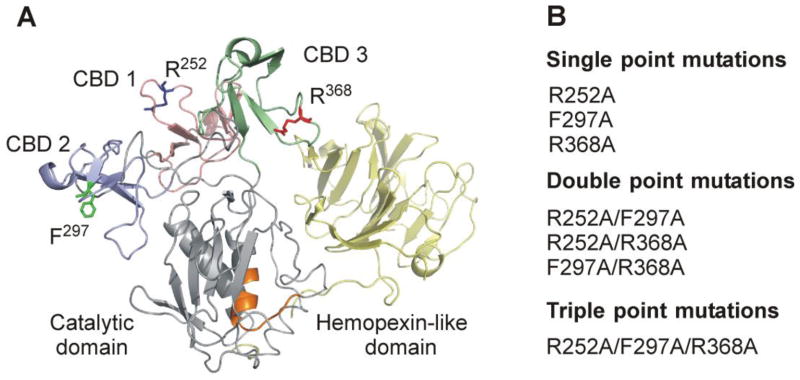
Fig. 1.
Structural overview of MMP-2 highlighting the locations of modified collagen binding site residues. (A) Crystal structure of MMP-2 (PDB 1CK7) (Morgunova et al., 1999) with modified collagen binding site residues (R252, F297, R368) presented in stick form (PyMOL). The three fibronectin type II modules of the collagen binding domain are shown in pink, blue, and green, respectively, and the enzyme active site is depicted in orange. (B) Alanine substitutions were introduced into one or concurrently into two or three of the CBD modules in full-length MMP-2 by a PCR-based site-directed mutagenesis strategy.
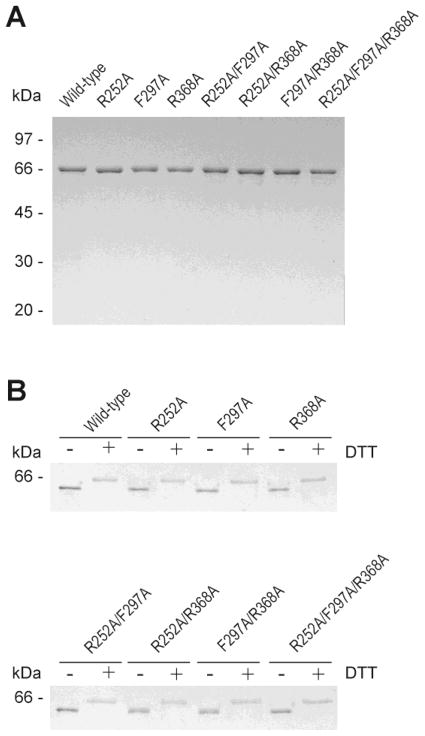
Fig. 2.
SDS/PAGE analysis of wild-type and variants of MMP-2. (A) The purity of recombinant proteins was confirmed by 10% SDS/PAGE under reducing conditions. Bands were visualized by Coomassie Blue staining. (B) In order to verify the proteins’ structural integrity, migration of wild-type and MMP-2 variants was compared by SDS/PAGE using 10% gels in the presence (+) or absence (−) of 100 mM dithiothrietol (DTT) followed by silver staining.
Cysteines in MMP-2 form seven disulfide bonds, six of which are located in the three CBD modules. Proper disulfide bond formation is critical for gelatin binding and cleavage by MMP-2 as reflected by our former observation that reduction and alkylation of cysteines eliminated gelatin binding properties of CBD (Steffensen et al., 1995) and MMP-2 activities on gelatin (Xu et al., 2004). All of the recombinant MMP-2 variants migrated as WT MMP-2 under reducing and non-reducing conditions indicating that the variant proteins had correct cysteine bond formation (Fig. 2B). Moreover, identical activities of WT and variants of MMP-2 in enzyme activity assays with the NFF-1 substrate, which does not utilize the CBD for catalysis, confirmed that all proteins were functionally folded (Table 2).
Table 2.
Hydrolysis of fluorogenic substrates by wild-type MMP-2 and variants.
| MMP-2 variants | NFF-1 | DQ-gelatin | ||
|---|---|---|---|---|
|
| ||||
| Rate (RFU · s−1) | Relative activity (%) | Rate (RFU · s−1) | Relative activity (%) | |
| Wild-type | 0.303 ± 0.024 | 100 | 0.306 ± 0.011 | 100 |
| R252A | 0.302 ± 0.020 | 100 | 0.269 ± 0.023 | 88 |
| F297A | 0.269 ± 0.005 | 89 | 0.213 ± 0.014 | 70 |
| R368A | 0.290 ± 0.024 | 96 | 0.178 ± 0.002 | 58 |
| R252A/F297A | 0.291 ± 0.006 | 96 | 0.140 ± 0.007 | 46 |
| R252A/R368A | 0.294 ± 0.035 | 97 | 0.105 ± 0.013 | 34 |
| F297A/R368A | 0.316 ± 0.002 | 104 | 0.100 ± 0.010 | 33 |
| R252A/F297A/R368A | 0.275 ± 0.002 | 91 | 0.029 ± 0.007 | 9 |
Substitution of amino acids has the potential to induce changes in the protein structure that could confound interpretation of the subsequent functional assays. Far-UV Circular Dichroism (CD) spectra from analyses of MMP-2 WT and variants verified that all variants of MMP-2 containing the alanine substitutions had a similar folded structure as WT (Fig. 3).
Fig. 3.
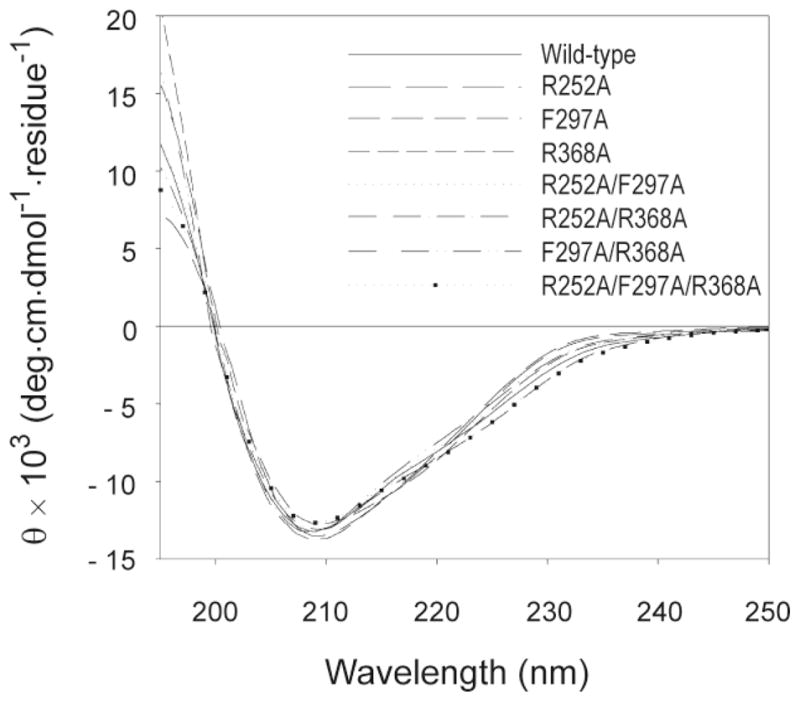
CD spectral analysis of wild-type and variant MMP-2 enzymes. CD studies were performed on a Jasco J-810 spectropolarimeter using a 1 mm rectangular cell. Protein samples were dissolved at 0.25 mg/ml in 50 mM Tris-HCl (pH 7.4). Wavelength scans were performed at 20 °C from 195 to 250 nm at scan rate of 50 nm/min. For each sample three scans were averaged and corrected for background absorbance.
2.2 All CBD modules contribute to the binding of MMP-2 to denatured type I gelatin
The interactions of WT and variant MMP-2 enzymes with gelatin were analyzed in microwell plate-based protein-protein binding assays. Fig. 4 presents gelatin binding of MMP-2 WT and variants as a function of concentration. Consistent with our previous observations (Xu et al., 2005), WT MMP-2 bound gelatin in concentration-dependent and saturable manner with an apparent Kd of 1.03 nM (Table 1). In comparison to WT, variants of MMP-2 with single point alanine substitutions in each of the CBD modular binding sites (R252A, F297A or R368A) had 2.5 – 3.5 fold decreased binding affinities to gelatin reflected by apparent Kds of 2.58, 2.80 and 3.56 nM, respectively. Variants with concurrent substitutions in two CBD modules (R252A/F297A, R252A/R368A or F297A/R368A) showed 5.7 – 36.5 fold reductions in gelatin binding compared with WT MMP-2. It is noteworthy that combined binding site substitutions in CBD modules 2 + 3 greatly reduced the gelatin affinity relative WT by 36.5 fold (Kd 37.6 nM). Finally, simultaneous alanine substitutions in the collagen binding sites of all three CBD modules (R252A/F297A/R368A) virtually eliminated the binding of MMP-2 to gelatin (Fig. 4, Table 1). Thus, as observed previously for isolated CBD domain (Xu et al., 2009), the collagen binding sites of all three CBD modules contribute gelatin binding in full-length MMP-2.
Fig. 4.
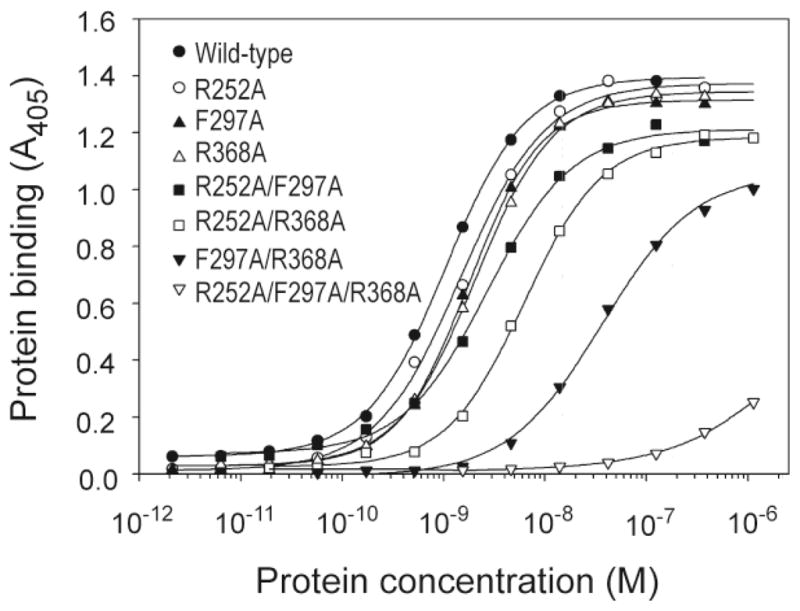
Binding of wild-type and MMP-2 variants to denatured type I collagen. The binding of wild-type as well as single, double and triple point variants of MMP-2 to denatured type I collagen was analyzed by plate-binding assays. Denatured type I collagen (0.5 μg/well) was immobilized in 96-microwell plates and allowed to react with varying concentrations of MMP-2. Bound protein was quantified at 405 nm after reaction with alkaline phosphatase-conjugated mouse monoclonal anti-His6-tag antibody and pNPP as substrate. Apparent Kd values are presented in Table 1.
Table 1.
Gelatin binding of wild-type MMP-2 and variants.
| MMP-2 variant |
Kd
|
|
|---|---|---|
| Apparent (M × 10−9) | Relative (versus wild-type) | |
| Wild-type | 1.03 ± 0.07 | 1.0 |
| R252A | 2.58 ± 0.99 | 2.5 |
| F297A | 2.80 ± 0.95 | 2.7 |
| R368A | 3.56 ± 1.35 | 3.5 |
| R252A/F297A | 5.84 ± 2.77 | 5.7 |
| R252A/R368A | 7.49 ± 1.76 | 7.3 |
| F297A/R368A | 37.6 ± 2.0 | 36.5 |
| R252A/F297A/R368A | ND | ND |
ND, not determined
2.3 CBD modules contribute independent and cooperative effects to MMP-2 cleavage of type 1 gelatin
Prior studies by our laboratory and others have demonstrated that the collagen binding domain (CBD) of MMP-2 is important for gelatinolysis (Murphy et al., 1994; Xu et al., 2004) and cleavage of elastin (Shipley et al., 1996). Yet, although the collagen binding sites have been localized on the isolated CBD domain (Xu et al., 2009), it remains to be understood how modifications of discrete sites defined by single amino acids impact the activity of MMP-2. To address these questions, we analyzed MMP-2 activities on fluorescent labeled and biotinylated gelatin, and on FRET peptide substrates for more detailed kinetic analyses. The overall goal was to characterize modular contributions to catalysis of gelatin and collagen and to define promising targets for MMP-2 inhibitors.
The proteolytic activities of MMP-2 WT and variants were first analyzed by gelatin zymography and then in solution assays with fluorescent labeled gelatin substrate (DQ-gelatin). Concurrent modification of all three binding sites yielded ~90% reduction in gelatin cleavage by both assays indicating that virtually all substrate positioning properties required for gelatinolysis may be attributed to these collagen binding sites (Fig. 5, Table 2). We attribute the ~10% residual activity to cleavage of short gelatin fragments, which do not require the CBD for hydrolysis (Xu et al., 2007).
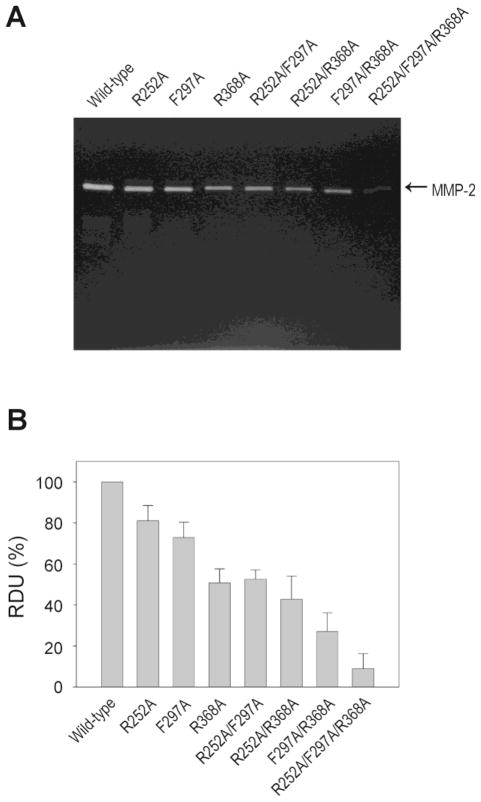
Fig. 5.
Degradation of denatured type I collagen by wild-type and MMP-2 variants. (A)Effects of alanine substitutions on the enzymatic activity of MMP-2 were analyzed by zymography using 10% minislab gels co-polymerized with gelatin. Gels were incubated 4 h at 37 °C with collagenase assay buffer and stained with Coomassie Blue. (B) Gels were scanned and band intensities were quantified using Kodak 1D imaging software. Data are expressed in per cent of activity of wild-type MMP-2. Bars represent means with S.D. from three separate experiments.
The strongest effects of substitution of single residues were measured following the R368A modification in module 3 and the simultaneous substitutions of F297A and R368A in modules 2 and 3, respectively. These variants had decreases in rates of cleavage of DQ-gelatin of 42% and 67%, respectively, and corresponding changes in cleavage when quantified by zymography (Fig. 5, Table 2). The other variants with single and double point substitutions also had reduced gelatinolytic activities that were consistent with the relative reduction in gelatin binding properties (Table 1).
Our analyses included control reactions with the short (11 amino acid) single chain substrate NFF-1 (Nagase et al., 1994). Our observation that none of the substitutions led to significant reductions in hydrolysis of NFF-1 (<10%) confirmed our former observations that CBD is not required for activity on short peptides (Xu et al., 2007), and verified that the catalytic site activities were fully retained in the MMP-2 variants (Table 2).
2.4 Collagen binding sites in different CBD modules selectively control the rates of cleavage of α1(I) and α2(I) collagen chains by MMP-2
Heat-denaturation dissociates the α1(I) and α2(I) chains in type I collagen (Danielsen, 1982); unwinding of the triple helix is also thought to occur following cleavage by collagenases at or near physiological temperature (Sakai and Gross, 1967). Prior data indicate the MMP-2 has different catalytic activities on the α1(I) and α2(I) chains (Gioia et al., 2007). Therefore, we analyzed the effects of substituting collagen binding site residues in the CBD on hydrolysis of these collagen α-chains. Biotinylated human type I collagen was heat-denatured and incubated with MMP-2 WT or variants over an enzyme:substrate (E:S) range of 1:200 – 10:1 (Fig. 6A), and the hydrolysis of the two α-chains was quantified. Our analysis showed that the amount of the enzyme required for degradation of 50% of the α-chains was 3 – 4 times higher following single substitutions, 7 – 14 times higher after double point modifications, and more then 60 times higher for triple substitution compared to wild-type MMP-2.
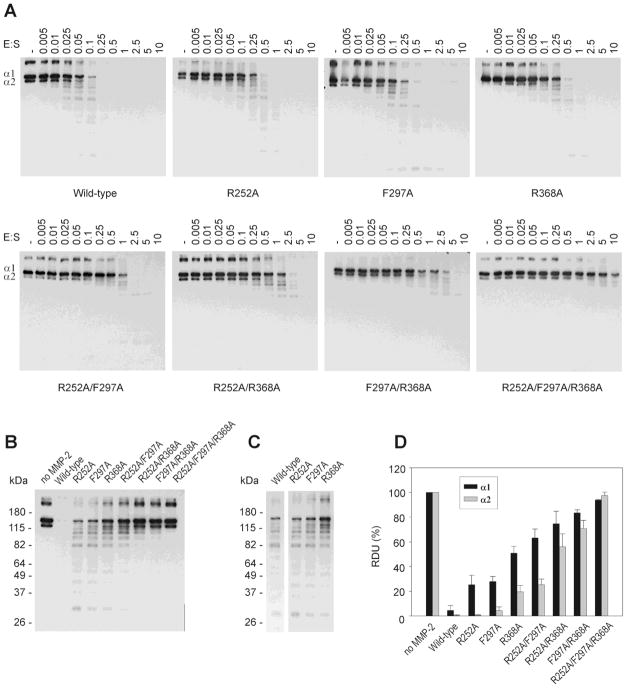
Fig. 6.
Effects of single, double and triple point collagen binding site modifications in the CBD of MMP-2 on the cleavage of α1 and α2 chains in denatured type I collagen. (A) Biotinylated denatured type I collagen (0.025 pmol) was incubated without enzyme (control) or with a concentration range of wild-type and variant MMP-2 (0 – 0.25 pmol) for 20 min at 28°C. The reactions were stopped by adding 2 × sample buffer and separated by SDS/PAGE on 8% gels. The collagen bands were detected by Western blotting using HRP-conjugated streptavidin and ECL as substrate. (B) Equal amounts of biotinylated denatured type I collagen (0.025 pmol) were incubated with wild-type or MMP-2 variants at a constant 1:10 enzyme:substrate ratio as described above. (C) Fragmentation pattern from cleavage of denatured type I collagen by MMP-2 WT and variants with single-point alanine substitutions in each of the three CBD modular collagen binding sites. (D) Quantification of the intensities of the α1 and α2 chains of type I collagen (Panel B) from radiographic films using Kodak 1D imaging software. Presented are means and S.D. expressed relative to untreated control collagen (n = 3).
Quantification of α-chain degradation at a constant E:S ratio of 1:10 for all MMP-2 WT and variants demonstrated that substitution of residues in the type I collagen binding sites in general impacted the hydrolysis of the α1(I) chain more than the α2(I) chain (Fig. 6B, D). Single binding site substitutions profoundly inhibited cleavage of the α1 chain by 20 – 50%; the reductions in cleavage were 25%, 27%, and 55% for R252A, F297A, and R368A, respectively. The corresponding effects on cleavage of the α2 chain ranged from 0–20%. Concurrent point substitutions at two binding sites yielded additional information on the effects of and cooperation between individual binding sites. The R252A/F297A modification had very different effects on the cleavage of the two α-chains, resulting in ~70% reduced cleavage of the α1 chain but only 25% reduction for the α2 chain. For the substitutions R252A/R368A and F297A/R368A, the inhibition of cleavage were significant (60–90%), but the α-chain selectivity was smaller in magnitude.
Alignment of blots representing similar extents of gelatin degradation by WT and single point substitutions in each of the three CBD modules showed that the pattern of fragmentation generated by the MMP-2 variants closely resembled that of the WT enzyme indicating that the three CBD modules did not affect cleavage site specificity (Fig. 6C).
Taken together, the large effects on catalysis resulting from the R368A substitution indicate that the collagen binding site and residue R368 in CBD module 3 are particularly important for cleavage of type I gelatin. Moreover, our data indicate that substrate positioning via the CBD collagen binding sites is critical for cleavage of the collagen α-chains and is of relative greater importance for the α1 than the α2 chain of type I collagen.
2.5 Cleavage of collagen-like triple helical peptide by MMP-2 is primarily dependent on substrate interactions with the CBD module 3 collagen binding site
Results from prior experiments analyzing catalysis of native collagen by MMP-2 suggest that the reaction is slow at best and requires very selective conditions with high E:S ratios for cleavage to occur (Patterson et al., 2001; Tam et al., 2004). Moreover, it has been proposed that MMP-2 or MMP-9 bind but do not cleave native type I collagen in fibrillar form (Collier et al., 2011). In light of these experimental uncertainties, we elected to address the potential effects of collagen binding site mutations on substrates with a helical conformation by studying cleavage of a triple-helical peptide rather than native type I collagen. The synthetic triple-helical FRET peptide substrate, fTHP-15, which replicates the types I - III collagen cleavage site and adjacent sequence, provides an important tool for exploration of the mechanism related to cleavage of triple-helical collagen molecules and enables in-depth kinetic analyses by its Mca flurophore and Dnp quencher pair. The cleavage of fTHP-15 has been shown previously to be CBD-dependent in MMP-9 (Lauer-Fields et al., 2008).
We determined the kcat, Km, kcat/Km parameters from the hydrolysis of fTHP-15 by the MMP WT and variants in enzyme kinetic assays (Table 3). The R368A substitution in CBD module 3 induced a strong (~75%) reduction in the kcat/Km (20,042 M−1 · s−1) from WT (79,490 M−1 · s−1). This single-point change exceeded that of double-point binding substitutions in CBD modules 1+2 (53,411 M−1 · s−1) and approached those of modules 1+3 (18,432 M−1 · s−1) and modules 2+3 (18,680 M−1 · s−1). These observations showed that most of the catalytic effects could be attributed to the R368A substitution. Further analyses revealed that the kcat/Km reductions could not be explained by the kcat values, which were reduced by ~10–25% and 25–33% for single and double-point substitutions, respectively. Rather, the reduced kcat/Km values observed from the R368A substitution, either by itself or in double-point variants, were primarily the result of ~3-fold increased Km values.
Table 3.
Kinetic parameters for hydrolysis of triple-helical FRET peptide substrate (fTHP-15) by wild-type MMP-2 and variants.
| MMP-2 variants | fTHP-15 | ||
|---|---|---|---|
|
| |||
| kcat/Km (M−1 · s−1) | kcat (s−1) | Km (μM) | |
| Wild-type | 79,490 ± 4,462 | 0.305 ± 0.009 | 3.85 ± 0.36 |
| R252A | 65,624 ± 3,787 | 0.270 ± 0.006 | 4.13 ± 0.33 |
| F297A | 58,297 ± 5,686 | 0.240 ± 0.001 | 4.14 ± 0.42 |
| R368A | 20,042 ± 2,574 | 0.238 ± 0.001 | 11.9 ± 0.15 |
| R252A/F297A | 53,411 ± 2,460 | 0.226 ± 0.004 | 4.23 ± 0.12 |
| R252A/R368A | 18,432 ± 441 | 0.205 ± 0.058 | 11.1 ± 0.15 |
| F297A/R368A | 18,680 ± 568 | 0.209 ± 0.013 | 11.2 ± 0.37 |
| R252A/F297A/R368A | 15,076 ± 735 | 0.197 ± 0.028 | 13.1 ± 0.25 |
These analyses suggest that MMP-2 cleavage of a synthetic substrate consisting of collagen-like α-chains that associate into a triple helical conformation also depend on interactions with R368. Further studies are needed to characterize the precise contributions of R368 to MMP-2 binding and cleavage of native type I collagen. It is interesting to note that, for efficient MMP-1 hydrolysis of fTHP-15 and type I collagen, a key Arg residue (R291) has been identified in the MMP-1 PEX domain (Lauer-Fields et al., 2009).
3. Discussion
There is mounting evidence that specific substrate binding sites (exosites) located outside the catalytic site clefts are essential for positioning substrate molecules for cleavage by matrix metalloproteinases (Overall et al., 2000; Xu et al., 2007; Lauer-Fields et al., 2008; Robichaud et al., 2011; Bertini et al., 2012). Therefore, compounds that block exosite-mediated substrate interactions have gained interest as novel MMP inhibitors with enhanced selectivity (Overall and Lopez-Otin, 2002).
The binding interactions of gelatin and elastin to the fibronectin-like collagen binding domain (CBD) of MMP-2 are closely associated with their cleavage by this enzyme (Steffensen et al., 1995; Shipley et al., 1996; Steffensen et al., 2002; Xu et al., 2005). Our laboratory and other investigators have used NMR analyses of CBD in complex with collagen-like peptides to localize critical collagen binding residues in MMP-2 (Tordai and Patthy, 1999; Briknarova et al., 2001; Gehrmann et al., 2002; Xu et al., 2009). The function of the identified residues was initially verified by reduced gelatin binding for alanine-substituted variants (Tordai and Patthy, 1999). Likewise, residues in the CBD of MMP-9 that were selected for study by others based on charge, hydrophobicity, and position predicted from the NMR structure of fibronectin-like modules in the bovine seminal protein PDC 109 (Pickford et al., 1997) were found to be critical for gelatin binding (Collier et al., 1992).
Extending those initial results, the present studies investigated the relative contributions of the three individual CBD modules as well as key residues in each of the three collagen binding sites to the catalytic activities of MMP-2. Specifically, we targeted amino acids in CBD that bound a peptide from the CBD-binding region in the α1 chain sequence 715–721 of human type I collagen (Xu et al., 2007; Xu et al., 2009).
While alanine substitutions generally are not thought to cause structural distortions, other investigators (Tordai and Patthy, 1999) found that modifications of structurally critical CBD residues (Y302A, Y323A, D326A and Y329A) induced extensive CD spectral changes that pointed to altered folding of the E. coli expressed recombinant. In comparison, neither our earlier highly targeted substitutions in the collagen binding sites of isolated CBD that were detected by NMR analyses of CBD in complex with a CBD binding collagen peptide from the α1(I) collagen chain (Xu et al., 2007; Xu et al., 2009) nor the present modifications of the same residues in full-length MMP-2 introduced structural perturbations as measured by 1D-NMR or CD spectroscopy, respectively. Therefore, our results are consistent with the CD analysis by Torday and Patthy (1999) that demonstrated significant functional, but virtually no structural effects for alanine substitutions of residues located in the hydrophobic binding site of CBD module 2.
Our experiments showed that wild-type full-length MMP-2 has higher affinity to gelatin than isolated CBD. This may result from stabilizing interactions of the CBD with other functional domains including the catalytic and PEX domains of the enzyme. However, the relative changes in affinity for gelatin (Kd app) for variants of MMP-2 in the present experiments paralleled those measured previously for variants of CBD (Xu et al., 2009); WT > R252A > F297A > R368A > R252A/F297A > R252A/R368A > F297A/R368A > R252A/F297A/R368A. We also found greater contributions to collagen binding of CBD module 3 (R368A) compared to the other two modules. These results confirmed that CBD is the major collagen binding domain of full-length MMP-2 and that CBD module 3 is particularly important for the interactions.
Earlier investigations of the CBD from MMP-9 found that recombinant module 2 had greater gelatin binding properties than modules 1 and 3 and, accordingly, that alanine substitutions of putative binding site residues in module 2 had greatest functional impact (Collier et al., 1992). The additional observation of stronger binding of isolated module 2 compared to combined CBD modules 1+2+3 may have been a function of the properties of the recombinant proteins as indicated by the authors. Nonetheless, a comparison of their data with our results points to potential functional differences between the CBDs from MMP-2 and -9 that could impact substrate selectivity and catalytic activities of the two enzymes.
The reduction in gelatin binding among the MMP-2 variants translated to decreasing cleavage of gelatin. When measured by the rate of cleavage of the fluorescent DQ-gelatin substrate, the MMP-2 variants ranked R252A > F297A > R368A > R252A/F297A > R252A/R368A > F297A/R368A > R252A/F297A/R368A (Table 2); these results were confirmed by analysis using gelatin zymography (Fig. 5). Importantly, the triple-point modifications in CBD reduced the gelatinolytic activities of MMP-2 by >90% as also detected by others following deletion of the CBD domain from MMP-2 (Murphy et al., 1994). We ascribe the ~9% residual gelatinolytic activity to cleavage of short collagen α-chain peptide fragments in the gelatin preparation, which are hydrolyzed in a CBD independent manner (Xu et al., 2004). Single point substitutions in individual CBD modules partially reduced the gelatinolytic activity of MMP-2. Double-point modifications had additive effects that were most pronounced when they included the R368A modification in CBD module 3. Thus, distinct collagen binding site residues in the CBD of MMP-2 are essential for the gelatinolytic activities. It is noteworthy from these studies of limited and highly localized amino acid changes that CBD binding indeed affects cleavage of gelatin by MMP-2 implying that the identified CBD binding sites may be sensitive targets for specific MMP-2 inhibitors.
The CBD has been linked to the enzymatic activity of MMP-2 by two proposed mechanisms. First, CBD binds and positions substrate molecules relative to the active site for cleavage site by MMP-2 (Overall, 2001; Overall, 2002; Xu et al., 2004). This function has been confirmed in experiments with single chain and denatured collagen substrates. Secondly, it has been proposed that CBD has triple helicase activities by which the collagen triple-helix is relaxed or unwound and cryptic cleavage sites on individual α-chains become available for cleavage by MMP-2 (Tam et al., 2004). This corresponds to the helicase activities exerted by functional regions including the PEX domain in other MMPs (Knauper et al., 1997; Pelman et al., 2005; Minond et al., 2006; Arnold et al., 2011; Bertini et al., 2012). Consistent with the proposed role of CBD in positioning longer but not short substrates (Xu et al., 2007), we found that cleavage of the short 11-amino acid fluorescent substrate NFF-1 (Nagase et al., 1994) by MMP-2 was not reduced following the collagen binding site modifications. Even the triple point variant of MMP-2 effectively cleaved NFF-1 (91% of WT).
Recent data have provided an enhanced understanding of α-chain cleavage in native collagen by several MMPs (Gioia et al., 2007; Bertini et al., 2012). Since MMP-2 proteolysis was proposed to involve CBD-mediated helicase activity, we investigated the contributions of the three CBD modules to MMP-2 activities on the α(I) and α2(I) chains in the context of denatured type I collagen. We found that CBD was required for cleavage of both chains by MMP-2 as shown by the absence of cleavage in the triple point variant of MMP-2. We observed that eliminating collagen binding to single CBD modular binding sites had substantial effect on α1 chain cleavage, but had little effect on α2 chain cleavage suggests that positioning of the α2 chain is less specific to the modular binding sites and can utilize the binding site redundancies on the CBD. This scenario may explain in turn the observation by Gioia et al. (Gioia et al., 2007) that the α2 chain was preferentially cleaved by MMP-2 and that the Km was 60 fold lower for the α1 than the α2 chain. In our experiments, both chains were most impacted by the elimination of the collagen binding site in module 3 (R368A>F297A>R252A). On this basis, we propose that there is modular selectivity in α1 and α2 chain binding, which impacts their hydrolysis by MMP-2.
As indicated in the Results section, MMP-2 cleavage of native type I collagen in monomeric or fibrillar forms is controversial (Aimes and Quigley, 1995; Patterson et al., 2001; Tam et al., 2004; Collier et al., 2011) or at least a very weak catalytic reaction. Therefore, to enhance our understanding of the effects of the contributions of the collagen binding sites to the catalysis of triple helical collagen molecules, we elected to analyze MMP-2 cleavage of a well-characterized collagen-like FRET substrate peptide, fTHP-15. In this substrate, three collagen chains approximately 40 amino acids in length fold to form a structure consistent with a collagen triple helix (Lauer-Fields et al., 2008; Robichaud et al., 2011). The fTHP contains the consensus interstitial collagenase cleavage sites from types I-III collagens, slightly altered for increased solubility. Efficient cleavage of fTHP-15 by MMP-9 requires the CBD (Lauer-Fields et al., 2008). In our experiments, cleavage of fTHP-15 by MMP-2 was significantly modified for CBD binding site variants demonstrating that this fTHP, as for MMP-9, also requires the CBD in MMP-2 for cleavage. All single and double point MMP-2 variants with the R368A substitution in module 3 had significantly reduced rate of fTHP-15 hydrolysis comparable to the triple point variant, which had only 19% of the wild-type catalytic activity on fTHP-15. Kinetic analyses revealed that the kcat/Km effects resulted from up to three-fold increases in the Km (Table 3). The kcat values were reduced, but by less than 30% compared to wild-type MMP-2. Single binding site substitutions of R252A (module 1) and F297A (module 2) had little effect on the Km for the interaction with MMP-2 even when they were combined in the double point variant, R252A/ F297A.
In summary, the functional impact of the collagen binding site in the third CBD module and the key residue R368 on gelatin binding and cleavage by MMP-2 were profound in the present studies. This is consistent with the results from prior NMR structural analyses that showed strong chemical shifts of R368 when recombinant proteins containing CBD modules 2 and 3 (Briknarova et al., 2001) or all three modules (Xu et al., 2009) were studied in complex with collagen-derived peptides. Our approach of using alanine substitutions enabled us to characterize contributions of collagen binding sites on the three CBD modules of MMP-2, to assign functions to specific binding site residues, and to confirm the involvement of specific residues in the proteolytic activities of MMP-2. This detailed information gained from the present series of experiments sets the stage for a drug discovery strategy that aims to identify compounds that block those specific residues and binding sites in CBD as a means for selective inhibition of MMP-2.
4. Experimental procedures
4.1 Site-specific alanine substitution in MMP-2
Seven MMP-2 variants were generated with single, double and triple point alanine substitutions of collagen binding site residues in the fibronectin-like type II modules of MMP-2 (Fig. 1). Three single point substitutions in each of the CBD modules (R252A, F297A, R368A) were introduced by overlap-extension PCR using the same pairs of primers as detailed previously (Xu et al., 2009) and, as template, plasmid pRSETA-MMP-2Δpro (Xu et al., 2005). This plasmid encodes constitutively active MMP-2 without the prodomain (MMP-2Δpro; wild-type). This strategy was pursued to avoid confounding effects in gelatin binding experiments caused by prodomain interactions with the CBD in proMMP-2 (Morgunova et al., 1999). Second point mutations encoding concurrent substitutions in two CBD modules (R252A/F297A, R252A/R368A and F297A/R368A) were introduced by the same approach with the modified R252A and F297A plasmids as templates. For the construct with triple point mutations, which encode concurrent changes in the collagen binding sites of all three CBD modules (R252A/F297A/R368A), the F297A/R368A plasmid served as template. All seven modified expression constructs were verified by double strand DNA sequencing.
4.2 Recombinant protein expression and purification
All recombinant MMP-2 variants were expressed in Escherichia coli BL21(DE3) at 37 °C using Super Broth medium (3.2% peptone, 2% yeast extract, and 0.5% NaCl) supplemented with 100 μg/ml ampicillin. When the OD600 reached 0.8, protein expression was induced by adding 0.4 mM isopropyl β-D-thiogalactoside (IPTG) followed by additional incubation for 5 h. After lysis of cells by sonication, protein-containing inclusion bodies were isolated by centrifugation and solubilized with 8 M urea in 50 mM Tris/HCl, 150 mM NaCl, pH 7.4. All recombinant proteins contained a His6 fusion tag and were purified by affinity chromatography on Ni2+-Sepharose (GE Healthcare, Piscataway, NJ). Following thorough washes, MMP-2 containing fractions were eluted with 150 mM imidazole in chromatography buffer (50 mM Tris/HCl, 150 mM NaCl, pH 7.4). The purified MMP-2 variants were refolded by drop-wise addition into ice-cold refolding buffer (50 mM Tris/HCl, pH 7.4, 2.5 mM GSH, 0.5 mM GSSG) to a final dilution ratio of 1:3 (v/v) and then dialyzed against refolding buffer overnight at 4 °C. After refolding, samples were subjected to a second step of purification by Ni2+-affinity chromatography. The MMP-2 variants were eluted with 80 mM imidazole in chromatography buffer. The purity and monomeric versus polymeric forms of MMP-2 were assessed following separation by 10 % SDS/PAGE under reducing (0.1 M dithiothreitol, DTT) and non-reducing conditions and visualization by Coomassie Brilliant Blue R-250 or silver staining. Fractions containing monomeric MMP-2 were combined, dialyzed against 50 mM Tris/HCl, 150 mM NaCl, pH 7.4, quantified by the BCA assay (Pierce, Rockford, IL), and stored at −80 °C in small assay size aliquots. Enzymes were exchanged into collagenase assay buffer (CAB, 50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 5 mM CaCl2, 0.05% Brij 35) immediately prior to each experiment.
4.3 Circular Dichroism Spectroscopy
Recombinant MMP-2 WT and variants were analyzed by circular dichroism (CD) to verify that the variants comprised a similar folded structure as WT protein, free of major structural perturbations. Analyses were performed on a Jasco J-810 spectropolarimeter (Jasco, Eston, MD) with a rectangular 0.1 cm path length quartz cell. Samples of WT and MMP-2 variants were exchanged into 50 mM Tris/HCl, pH 7.4 by overnight dialysis and adjusted to a protein concentration of 250 μg/ml. Data were collected at 4 °C from λ = 250 to 195 nm at a rate of 50 nm/min. For each sample, three scans were averaged and background absorbance subtracted. The observed CD signal was converted to mean molar ellipticity per residue (θ, deg cm2 dmol−1 residue−1) using the following equation: θ = θd /10(clnr), where θd is observed ellipticity in degrees, c is protein concentration in mol/L, l is optical path length in cm, and nr is number of amino acid residues in the protein (590 for MMP-2 WT and variants). The spectra were generated from the acquired CD data by SigmaPlot (SPSS, Chicago, IL).
4.4 Enzyme substrate binding assays
Interactions between MMP-2 variants and denatured type I collagen (gelatin) were analyzed in microplate protein binding assays as detailed previously (Steffensen et al., 1995) with the following minor modifications. Briefly, 96-well plates were coated overnight at 4 °C with 0.5 μg/well human type I collagen (Biodesign, Saco, ME) that had been heat-denatured at 56 °C for 30 min (gelatin). After blocking non-specific binding sites with 2.5% BSA in PBS (10 mM Phosphate Buffer, 137 mM NaCl, 2.7 mM KCl, pH 7.4), the coated gelatin was reacted with a concentration range (2 ×10−12 – 3 × 10−6 M) of recombinant MMP-2 WT and variants. Of note, our recombinant MMP-2 enzyme displays minimal and non-significant gelatinolytic activities in this binding assay buffer. Bound MMP-2 was detected with an alkaline phosphatase-conjugated mouse monoclonal anti-His6-tag antibody (Abcam, Cambridge, MA) and quantified at 405 nm using 1 mg/ml sodium p-nitrophenyl phosphate (pNPP) as substrate (Sigma–Aldrich, St. Louis, MO) and an Opsys MR plate reader (Dynex, Chantilly, VA). All experiments were performed in duplicate and repeated at least twice. Apparent Kd values were calculated with SigmaPlot (SPSS) from plots of protein binding vs. protein concentrations by 4-parameter nonlinear curve fitting using the equation y = (a−d)/[1+(x/c)b]+d (a, asymptotic maximum; b, slope parameter; c, value at inflexion point; d, asymptotic minimum).
4.5 Zymography
Gelatinolytic activities of MMP-2 WT and variants were assayed by zymography as detailed previously (Steffensen et al., 1998). MMP-2 samples (10 ng/lane) were separated under non-reducing conditions on 10% polyacrylamide gels that were co-polymerized with 0.1% gelatin (Sigma-Aldrich, St. Louis, MO). Gels were then washed thoroughly with 5% Triton X-100, and equilibrated and incubated overnight in CAB. Gels were scanned after staining with Coomassie Blue R-250. Intensities of dye-free bands representing hydrolysis of gelatin by MMP-2 were quantified using Kodak 1D imaging software (Kodak, Rochester, NY). Conditions were adjusted to ensure that band intensities were in the linear range of the assay.
4.6 Cleavage of biotinylated gelatin
For biotinylation, 0.25 ml aliquots of 5 mg/ml human type I collagen (Biodesign, Saco, ME) were dialyzed against PBS (pH 8.2) and then incubated with 300 μg of Sulfo-NHS-LC-Biotin (Pierce, Rockford, IL) for 20 min at 25 °C followed by 2 h at 4 °C. Free biotin was removed by dialysis against CAB, and biotinylation of the protein was verified by reaction with alkaline phosphatase-conjugated streptavidin (Pierce, Rockford, IL) in solid phase assays using pNPP (Sigma-Aldrich, St. Louis, MO) as substrate.
In preparation for enzyme activity assays, biotin-labeled type I collagen was heat denatured. To confirm that the native type I collagen was fully denatured, aliquots of native and heat-denatured type I collagen were incubated with 0.1 or 0.01 μl/ml trypsin (type XII porcine pancreas, Sigma-Aldrich, St. Louis, MO) (enzyme:substrate ratio ~ 1:2 and 1:20) for 19 h at 25 °C. Analysis by 6% SDS/PAGE revealed that gelatin was fully degraded by the enzyme reflecting disruption of the triple-helical structure whereas native protein was resistant to trypsin cleavage (not shown).
In catalytic reactions, heat-denatured biotin-labeled gelatin (0.025 pmol) was incubated with a concentration range of MMP-2 WT and variants (0.0001 – 0.25 pmol) for 20 min at 28 °C. Reactions were stopped by adding 2 × SDS/PAGE sample buffer. The samples were heated for 10 min at 70 °C and separated by 8% SDS/PAGE. Intact and degraded collagen α-chains were detected by Western blotting using HRP-conjugated streptavidin (1:10,000; Jackson Immunoresearch Laboratories, West Grove, PA) and ECL substrate (PerkinElmer, Wellesley, MA). Blots were scanned and band intensities were quantified using the Kodak 1D imaging software.
4.7 Enzyme activity assays with fluorogenic substrates
The specific activities of MMP-2 WT and variants were assayed in reactions containing the following fluorescent substrates: Fluorescein-labeled type I gelatin (DQ-gelatin, Molecular Probes, Eugene, OR), single chain FRET peptide substrate NFF-1 (Mca-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Lys-(Dnp)-Gly-NH2)(Nagase et al., 1994)), and triple-helical FRET peptide substrate fTHP-15 ((Gly-Pro-Hyp)5-Gly-Pro-Lys(Mca)-Gly-Pro-Gln-Gly-Leu-Arg-Gly-Gln-Lys(Dnp)-Gly-Val-Arg-(Gly-Pro-Hyp)5-NH2 (Lauer-Fields et al., 2001; Minond et al., 2006; Lauer-Fields et al., 2009). These FRET substrates contain the fluorophore Mca (7-metoxycoumarin-4-yl)acetyl and the quencher Dnp (2,4-dinitrophenyl). Typical enzyme activity assays were performed in 100 μl reactions containing 10 nM MMP-2 and 1 μg DQ-gelatin, 5 μM NFF-1 or 5 μM fTHP-15 in CAB. The fluorescent signal, which correlates positively with the amount of substrate cleavage, was quantified for 30 min at 25 °C on a SpectraMAX Gemini XS plate reader (Molecular Devices, Sunnyvale, CA) with λex at 485 nm and λem at 515 nm for DQ-gelatin, and λex at 325 nm and λem at 393 nm for NFF-1 and fTHP-15. Rates of hydrolysis were calculated in the linear range of the assays from plots of fluorescence versus time. Kinetic analysis was performed with by the same approach using substrate concentrations ranging from 0.4 to 12.5 μM for fTHP-15. Reaction velocities versus substrate concentrations were fitted to the Michaelis-Menten equation and kinetic constants were calculated using SigmaPlot (Enzyme kinetic edition; SPSS, Chicago, IL).
Highlights.
We identified collagen binding site residues that govern catalysis by MMP-2
Three collagen binding sites contributed differently to gelatinolysis by MMP-2
Cleavage of α1(I) and α2(I) collagen chains involve specific binding sites in MMP-2
Collagen binding sites are promising targets for selective MMP-2 inhibitors
Acknowledgments
We thank Dr. Stephen C. Hardies and Mandy Ronaldo, Department of Biochemistry, UTHSCSA, for DNA sequencing. This work was supported by the National Institutes of Health grants DE017139 (B.S.), DE16312 (B.S.), DE018135 (X.X.), HL07446 (T32 training grant, T.K.R.), CA098799 (G.B.F.), and the Multiple Sclerosis National Research Institute (G.B.F.).
Abbreviations
- MMP-2
matrix metalloproteinase-2
- CBD
collagen-binding domain
- PEX
hemopexin-like domain
- MMP-2Δpro
MMP-2 with deleted prodomain
- THP
triple-helical peptide
- FRET
fluorescence resonance energy transfer
- Dnp
2,4-dinitrophenyl
- Mca
(7-methoxycoumarin-4-yl)acetyl
- NFF-1
Mca-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Lys(Dnp)-Gly-NH2
- fTHP-15
(Gly-Pro-Hyp)5-Gly-Pro-Lys(Mca)-Gly-Pro-Gln-Gly-Leu-Arg-Gly-Gln-Lys(Dnp)-Gly-Val-Arg-(Gly-Pro-Hyp)5-NH2
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aimes RT, Quigley JP. Matrix metalloproteinase-2 is an interstitial collagenase. Inhibitor-free enzyme catalyzes the cleavage of collagen fibrils and soluble native type I collagen generating the specific 3/4- and 1/4-length fragments. J Biol Chem. 1995;270:5872–5876. doi: 10.1074/jbc.270.11.5872. [DOI] [PubMed] [Google Scholar]
- Arnold LH, Butt LE, Prior SH, Read CM, Fields GB, Pickford AR. The interface between catalytic and hemopexin domains in matrix metalloproteinase-1 (MMP-1) conceals a collagen binding exosite. J Biol Chem. 2011;286:45073–45082. doi: 10.1074/jbc.M111.285213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Fragai M, Luchinat C, Melikian M, Toccafondi M, Lauer JL, Fields GB. Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis. J Am Chem Soc. 2012;134:2100–2110. doi: 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briknarova K, Gehrmann M, Banyai L, Tordai H, Patthy L, Llinas M. Gelatin-binding region of human matrix metalloproteinase-2: solution structure, dynamics, and function of the COL-23 two-domain construct. J Biol Chem. 2001;276:27613–27621. doi: 10.1074/jbc.M101105200. [DOI] [PubMed] [Google Scholar]
- Brown PD. Matrix metalloproteinase inhibitors. Angiogenesis. 1998;1:142–154. doi: 10.1023/A:1018373520193. [DOI] [PubMed] [Google Scholar]
- Butler GS, Overall CM. Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry. 2009;48:10830–10845. doi: 10.1021/bi901656f. [DOI] [PubMed] [Google Scholar]
- Collier IE, Krasnov PA, Strongin AY, Birkedal-Hansen H, Goldberg GI. Alanine scanning mutagenesis and functional analysis of the fibronectin-like collagen-binding domain from human 92-kDA type IV. J Biol Chem. 1992;267:6776–6781. [PubMed] [Google Scholar]
- Collier IE, Legant W, Marmer B, Lubman O, Saffarian S, Wakatsuki T, Elson E, Goldberg GI. Diffusion of MMPs on the surface of collagen fibrils: the mobile cell surface-collagen substratum interface. PLoS ONE. 2011;6:e24029. doi: 10.1371/journal.pone.0024029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Danielsen CC. Difference in thermal stability of type-I and type-II collagen from rat skin. Biochem J. 1982;203:323–326. doi: 10.1042/bj2030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Gehrmann M, Briknarova K, Banyai L, Patthy L, Llinas M. The col-1 module of human matrix metalloproteinase-2 (MMP-2): structural/functional relatedness between gelatin-binding fibronectin type II modules and lysine-binding kringle domains. Biol Chem. 2002;383:137–148. doi: 10.1515/BC.2002.014. [DOI] [PubMed] [Google Scholar]
- Gioia M, Monaco S, Fasciglione GF, Coletti A, Modesti A, Marini S, Coletta M. Characterization of the mechanisms by which gelatinase A, neutrophil collagenase, and membrane-type metalloproteinase MMP-14 recognize collagen I and enzymatically process the two alpha-chains. Journal of Molecular Biology. 2007;368:1101–1113. doi: 10.1016/j.jmb.2007.02.076. [DOI] [PubMed] [Google Scholar]
- Knauper V, Cowell S, Smith B, Lopez-Otin C, O’Shea M, Morris H, Zardi L, Murphy G. The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J Biol Chem. 1997;272:7608–7616. doi: 10.1074/jbc.272.12.7608. [DOI] [PubMed] [Google Scholar]
- Lam KS, Lake D, Salmon SE, Smith J, Chen ML, Wade S, Abdul-Latif F, Knapp RJ, Leblova Z, Ferguson RD, Krchnak V, Sepetov NF, Lebl M. A One-Bead One-Peptide Combinatorial Library Method for B-Cell Epitope Mapping. Methods. 1996;9:482–493. doi: 10.1006/meth.1996.0056. [DOI] [PubMed] [Google Scholar]
- Lauer-Fields JL, Broder T, Sritharan T, Chung L, Nagase H, Fields GB. Kinetic analysis of matrix metalloproteinase activity using fluorogenic triple-helical substrates. Biochemistry. 2001;40:5795–5803. doi: 10.1021/bi0101190. [DOI] [PubMed] [Google Scholar]
- Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB. Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J Biol Chem. 2009;284:24017–24024. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer-Fields JL, Whitehead JK, Li S, Hammer RP, Brew K, Fields GB. Selective modulation of matrix metalloproteinase 9 (MMP-9) functions via exosite inhibition. J Biol Chem. 2008;283:20087–20095. doi: 10.1074/jbc.M801438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Visse R, Nagase H, Fields GB. The roles of substrate thermal stability and P2 and P1′ subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J Biol Chem. 2006;281:38302–38313. doi: 10.1074/jbc.M606004200. [DOI] [PubMed] [Google Scholar]
- Morgunova E, Tuuttila A, Bergmann U, Isupov M, Lindquist Y, Schneider G, Tryggvason K. Structure of human pro-matrix metalloproteinase-2: Activation mechanism revealed. Science. 1999;284:1667–1670. doi: 10.1126/science.284.5420.1667. [DOI] [PubMed] [Google Scholar]
- Murphy G, Allan JA, Willenbrock F, Cockett MI, O’Commell JP, Docherty AJP. The role of the C-terminal domain in collagenase and stromelysin specificity. J Biol Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- Murphy G, Nguyen Q, Cockett MI, Atkinson SJ, Allan JA, Knight CG, Willenbrock F, Docherty AJP. Assessment of the role of the fibronectin-like domain of gelatinase A by analysis of a deletion mutant. J Biol Chem. 1994;269:6632–6636. [PubMed] [Google Scholar]
- Nagase H, Fields CG, Fields GB. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin-1 (matrix metalloproteinase-3) J Biol Chem. 1994;269:20952–20957. [PubMed] [Google Scholar]
- Overall CM. Matrix metalloprotinase substrate binding domains, modules, and exosites. Overview and experimental strategies. In: Clark IM, editor. Matrix metalloproteinase protocols. Vol. 1. Humana Press; Totowa, N.J: 2001. pp. 79–120. Methods in molecular biology. [PubMed] [Google Scholar]
- Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- Overall CM, Wallon UM, Steffensen B, De Clerk Y, Tschesche H, Abbey RS. Substrate and TIMP interactions with human gelatinase A recombinant COOH-terminal hemopexin-like and fibronectin type II-like domains: Both the N- and C-domains of TIMP-2 bind the C-domain of gelatinase A. In: Edwards D, et al., editors. Inhibitors of Metalloproteinases in Development and Disease. Gordon & Breach; Amsterdam, Holland: 2000. pp. 57–69. [Google Scholar]
- Patterson ML, Atkinson SJ, Knauper V, Murphy G. Specific collagenolysis by gelatinase A, MMP-2, is determined by the hemopexin domain and not the fibronectin-like domain. FEBS Lett. 2001;503:158–162. doi: 10.1016/s0014-5793(01)02723-5. [DOI] [PubMed] [Google Scholar]
- Pelman GR, Morrison CJ, Overall CM. Pivotal molecular determinants of peptidic and collagen triple helicase activities reside in the S3′ subsite of matrix metalloproteinase 8 (MMP-8): the role of hydrogen bonding potential of ASN188 and TYR189 and the connecting cis bond. J Biol Chem. 2005;280:2370–2377. doi: 10.1074/jbc.M409603200. [DOI] [PubMed] [Google Scholar]
- Pickford AR, Potts JR, Bright JR, Phan I, Campbell ID. Solution structure of a type 2 module from fibronectin: Implications for the structure and function of the gelatin-binding domain. Structure. 1997;5:359–370. doi: 10.1016/s0969-2126(97)00193-7. [DOI] [PubMed] [Google Scholar]
- Robichaud TK, Steffensen B, Fields GB. Exosite interactions impact matrix metalloproteinase collagen specificities. J Biol Chem. 2011;286:37535–37542. doi: 10.1074/jbc.M111.273391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Gross J. Some properties of the products of reaction of tadpole collagenase with collagen. Biochemistry. 1967;6:518–528. doi: 10.1021/bi00854a021. [DOI] [PubMed] [Google Scholar]
- Shipley JM, Doyle GAR, Fliszar CJ, Ye QZ, Johnson LL, Shapiro SD, Welgus HG, Senior RM. The structural basis for the elastolytic activity of the 92-kDa and 72-kDa gelatinases. Role of the fibronectin type II-like repeats. J Biol Chem. 1996;271:4335–4341. doi: 10.1074/jbc.271.8.4335. [DOI] [PubMed] [Google Scholar]
- Steffensen B, Bigg HF, Overall CM. The involvement of the fibronectin type II-like modules of human gelatinase A in cell surface localization and activation. J Biol Chem. 1998;273:20622–20628. doi: 10.1074/jbc.273.32.20622. [DOI] [PubMed] [Google Scholar]
- Steffensen B, Chen Z, Pal S, Mikhailova M, Su J, Wang Y, Xu X. Fragmentation of fibronectin by inherent autolytic and matrix metalloproteinase activities. Matrix Biol. 2011;30:34–42. doi: 10.1016/j.matbio.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen B, Hakkinen L, Larjava H. Proteolytic events of wound healing - coordinated interactions between matrix metalloproteinases (MMPs), integrins, and extracellular matrix molecules. Crit Rev Oral Biol Med. 2001;12:373–398. doi: 10.1177/10454411010120050201. [DOI] [PubMed] [Google Scholar]
- Steffensen B, Wallon UM, Overall CM. Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J Biol Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- Steffensen B, Xu X, Martin P, Zardeneta G. Human fibronectin and MMP-2 collagen binding domains compete for collagen binding sites and modify cellular activation of MMP-2. Matrix Biol. 2002;21:399–414. doi: 10.1016/s0945-053x(02)00032-x. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J Biol Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- Tordai H, Patthy L. The gelatin-binding site of the second type-II domain of gelatinase A/MMP-2. Eur J Biochem. 1999;259:513–518. doi: 10.1046/j.1432-1327.1999.00070.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen Z, Wang Y, Bonewald L, Steffensen B. Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem J. 2007;406:147–155. doi: 10.1042/BJ20070591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen Z, Wang Y, Yamada Y, Steffensen B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and -2 (MMP-9 and -2) Biochem J. 2005;392:127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Mikhailova M, Ilangovan U, Chen Z, Yu A, Pal S, Hinck AP, Steffensen B. Nuclear magnetic resonance mapping and functional confirmation of the collagen binding sites of matrix metalloproteinase-2. Biochemistry. 2009;48:5822–5831. doi: 10.1021/bi900513h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang Y, Lauer-Fields JL, Fields GB, Steffensen B. Contributions of the MMP-2 collagen binding domain to gelatin cleavage. Substrate binding via the collagen binding domain is required for MMP-2 degradation of gelatin but not short peptides. Matrix Biol. 2004;23:171–181. doi: 10.1016/j.matbio.2004.05.002. [DOI] [PubMed] [Google Scholar]