Abstract
We previously developed an antibody-avidin fusion protein (ch128.1Av) that targets the human transferrin receptor 1 (TfR1) and exhibits direct cytotoxicity against malignant B cells in an iron-dependent manner. ch128.1Av is also a delivery system and its conjugation with biotinylated saporin (b-SO6), a plant ribosome-inactivating toxin, results in a dramatic iron-independent cytotoxicity, both in malignant cells that are sensitive or resistant to ch128.1Av alone, in which the toxin effectively inhibits protein synthesis and triggers caspase activation. We have now found that the ch128.1Av/b-SO6 complex induces a transcriptional response consistent with oxidative stress and DNA damage, a response that is not observed with ch128.1Av alone. Furthermore, we show that the antioxidant N-acetylcysteine partially blocks saporin-induced apoptosis suggesting that oxidative stress contributes to DNA damage and ultimately saporin-induced cell death. Interestingly, the toxin was detected in nuclear extracts by immunoblotting, suggesting the possibility that saporin might induce direct DNA damage. However, confocal microscopy did not show a clear and consistent pattern of intranuclear localization. Finally, using the long-term culture-initiating cell assay we found that ch128.1Av/b-SO6 is not toxic to normal human hematopoietic stem cells suggesting that this critical cell population would be preserved in therapeutic interventions using this immunotoxin.
Keywords: saporin, immunotoxin, transferrin receptor, oxidative stress, ribosome-inactivating protein
Introduction
Saporin is a ribosome-inactivating proein (RIP) isolated from the plant Saponaria officinalis that strongly blocks protein synthesis (Lombardi et al. 2010). It is a Type I RIP in that it consists of a single catalytic polypeptide chain and lacks a cell-binding chain. It has similar catalytic activity to that of ricin, a Type II RIP that consists of both the catalytic and cell-binding domains (de Virgilio et al. 2010). RIPs are N-glycosidases that depurinate specific adenine residues of the 23S/25S/28S ribosomal subunits leading to the irreversible block in protein synthesis. Saporin has also been reported to have DNase-like activity (Gasperi-Campani et al. 2005; Roncuzzi and Gasperi-Campani 1996), although this is controversial (Lombardi et al. 2010). It has also been reported that the glycosidase activity of saporin is not required for its cytotoxicity (Cimini et al. 2011; Sikriwal et al. 2008). There are several isoforms of saporin that have been identified and named based on the tissue of origin and chromatographic peak in ion-exchange chromatography (Lombardi et al. 2010). Saporin-6 (SO6), one of the most active forms of the toxin, is produced in the seeds of the plant and represents the major peak (peak 6) in chromatography analysis of seed extracts (Lombardi et al. 2010). This peak contains up to 4 different isoforms of the toxin that has either an aspartic or glutamic acid residue in position 48 and either a lysine or arginine residue at position 91. Due to its high cytotoxicity, high stability and resistance to denaturation (Santanche et al. 1997), and inability to readily enter cells, saporin is a promising therapeutic agent for delivery into cancer cells.
An antibody-avidin fusion protein (ch128.1Av) was previously produced as a delivery system for a broad range of biotinylated therapeutic agents, such as SO6, into cancer cells (Daniels et al. 2007; Ng et al. 2002; Ng et al. 2006). This fusion protein contains avidin genetically fused to the CH3 domains of the human IgG3 heavy chains. The antibody is specific for the human transferrin receptor 1 (TfR1, also known as CD71) and does not compete with the endogenous ligand transferrin (Tf) for receptor binding (Ng et al. 2006; Rodriguez et al. 2007). The TfR1 is a Type II transmembrane homodimeric protein involved in iron uptake and regulation of cell growth (Daniels et al. 2006b). It is widely expressed at low levels on many cell types, but shows increased expression on rapidly dividing cells including malignant cells due to their increased need for iron (Daniels et al. 2006b). Because of its central role in cancer pathology, its accessibility on the cell surface, and its ability to internalize through receptor-mediated endocytosis, the TfR1 has been used for the targeted delivery of numerous different therapeutic agents into cancer cells (Daniels et al. 2012; Daniels et al. 2006a). The TfR1 can be targeted in two ways, either through the use of conjugates containing Tf, or through the use of antibodies like ch128.1Av. In addition to its delivery potential, ch128.1Av is cytotoxic to certain human malignant B cells, including multiple myeloma (MM) and non-Hodgkin’s lymphoma (NHL) cells (Ng et al. 2002; Ng et al. 2006; Ortiz-Sanchez et al. 2009), an activity that is higher compared to that of its parental antibody (ch128.1) without avidin (Daniels et al. 2011; Ng et al. 2006). This activity is due to an alteration in the TfR cycling pathway, increased TfR degradation, and the induction of lethal iron starvation in sensitive cells (Daniels et al. 2007; Ng et al. 2006; Rodriguez et al. 2011). However, both ch128.1Av and its parental antibody demonstrated in vivo anti-cancer activity in two xenograft mouse models of disseminated human MM (Daniels et al. 2011). Taken together, ch128.1Av is a versatile approach for the treatment of B-cell malignancies in that it can be directly cytotoxic through the disruption of iron metabolism or it can be used as a universal delivery system for many therapeutic agents.
Previously we have shown that ch128.1Av delivers the active b-SO6 toxin into human malignant B cells resulting in protein synthesis inhibition, caspase activation (especially caspases 2 and 3), and the induction of apoptosis in both cells that are sensitive to the fusion protein alone and those that are resistant (Daniels et al. 2007). The cytotoxicity of b-SO6 conjugated to ch128.1Av in cells that are sensitive to the direct effects of ch128.1Av occurs much faster than that of the ch128.1Av alone. Additionally, the cytotoxicity of the conjugate could not be blocked by the addition of excess iron (Daniels et al. 2007), indicating that in contrast to ch128.1Av alone, iron starvation does not play a role in this cell death. These data suggest that the death induced by the conjugate is exclusively mediated by the toxin and not the direct cytotoxic effects of the fusion protein. A previous report on the gene expression analysis of ch128.1Av alone showed a transcriptional response consistent with iron deprivation mediated in part by p53 (Rodriguez et al. 2011). We now show that the ch128.1Av/b-SO6 immunotoxin induces a different transcriptional response, which is consistent with the induction of oxidative stress and DNA damage. The induction of lethal oxidative stress was confirmed through the analysis of cell death in the presence of an antioxidant. In addition, we have conducted studies that suggest nuclear localization of the toxin. Finally, we found that ch128.1Av/b-SO6 does not show toxicity to normal human hematopoietic stem cells or non-committed (early) progenitor cells.
Materials and Methods
Cell Lines
IM-9 (a human EBV-transformed B-lymphoblastoid cell line) and U266 (a human MM cell line) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Both malignant B-cell lines were grown in RPMI 1640 medium (Life Technologies, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum (Atlanta Biologicals Inc., Lawrenceville, GA) and grown in 5% CO2 and 37°C.
Recombinant antibody-avidin fusion protein production and immunotoxin formation
The antibody-avidin fusion protein ch128.1Av (formerly known as anti-human TfR IgG3-Av) has been previously described (Ng et al. 2002; Ng et al. 2006). It consists of a mouse/human chimeric IgG3 antibody genetically fused to avidin via its CH3 domains. The IgG3 contains the variable regions of the murine antibody 128.1. A similar non-targeting isotype control fusion protein specific for the hapten dansyl (DNS): 5-dimethylamino naphthalene-1-sulfonyl chloride (IgG3-Av) has been previously reported (Ng et al. 2006). Both fusion proteins, expressed in murine myeloma cells, were purified from cell culture supernatants using affinity chromatography. Proteins were dialyzed into buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.8) and protein concentrations were determined by the bicinchoninic acid (BCA) Protein Assay (Thermo Fisher Scientfic, Walnut, CA). Mono-biotinylated saporin (b-SO6, mw ~30 kDa) was purchased from Advanced Targeting Systems (San Diego, CA) as a custom conjugate of one biotin per toxin molecule. ch128.1Av or IgG3-Av was conjugated to b-SO6 in a 1:1 molar ratio on ice for 30 minutes before the addition to cell culture medium as previously described (Daniels et al. 2007).
Microarray hybridization and data quality control
IM-9 and U266 cells were incubated for 1, 3, 9, or 24 hours with 10 nM ch128.1Av alone or conjugated to b-SO6. Control samples consisted of cells incubated with an equal volume of buffer alone for the same time points. Total mRNA was collected from all samples using the RNeasy Kit (Qiagen, Valencia, CA). RNA was quantified and the integrity evaluated using a Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). RNA was hybridized onto HumanRef-8 v2 Expression BeadChips (Illumina, Inc., San Diego, CA) and global gene expression profiles for these samples were collected using the BeadArray software package (Illumina). Quality control, preprocessing, data normalization, and statistical analysis of differential expression was performed as described previously (Rodriguez et al. 2011). All changes were deemed significant (p < 0.05) based on a regularized Bayesian test. Data for ch128.1Av-treated cells has already been reported (Rodriguez et al. 2011).
Proliferation and apoptosis assays
IM-9 and U266 cells were incubated for 48 hours with 10 nM ch128.1Av alone or conjugated to b-SO6. Control cells were incubated with an equal volume of buffer alone. Inhibition of cell proliferation was monitored using the [3H]-thymidine incorporation assay as previously described (Daniels et al. 2007). Apoptosis was assessed using Annexin V staining and flow cytometry as described previously (Daniels et al. 2007). For antioxidant studies, IM-9 and U266 cells were treated with 10 or 1 nM ch128.1Av, b-SO6, or the ch128.1Av/b-SO6 complex in the presence or absence of 2 mM of the antioxidant N-acetylcysteine (NAC) for 48 hours. As a commonly used protein synthesis inhibitor, cells were treated with cycloheximide (CHX, ThermoFisher Scientific, Walnut, CA). Different concentrations of CHX were used for the two cell lines since U226 can be resistant to apoptosis due to the high level of Bcl-xL expression (Catlett-Falcone et al. 1999). U266 cells were treated with either 100 or 10 μg/mL CHX and IM-9 cells were treated with 1.0 or 0.1 μg/mL CHX (in the presence or absence of 2 mM NAC). Apoptosis was then assessed by flow cytometry. Ten thousand events were recorded for each sample on a Becton–Dickinson FACScan Analytic Flow Cytometer (BD Biosciences, San Jose, CA) in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility. Data were analyzed using the FCS Express V3 software (De Novo Software, Los Angeles, CA).
Validation by real time, quantitative PCR (QPCR)
New RNA samples were collected using the RNeasy Kit (Qiagen) from IM-9 and U266 cells treated with either 10 nM ch128.1Av, b-SO6, or the ch128.1Av/b-SO6 complex for 24 hours. U266 and IM-9 cells were also treated with 100 μg/mL or 1.0 μg/mL CHX, respectively, for 24 hours as a control protein synthesis inhibitor. cDNA was prepared using 2 μg of RNA and the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s protocol. For all QPCR reactions, cDNA samples were diluted 1:100 since this dilution was pre-determined to be overall the best for target and housekeeping genes in untreated cells. The Universal Probe Library Real Time PCR System (Roche Applied Science, Indianapolis, IN) was used for validation of gene expression. This system uses sequence-specific primers that recognize the gene of interest as well as fluorescently labeled probes that attach to the piece of DNA that is being amplified. The online Assay Design Center (Roche Applied Science) that uses the gene accession numbers was used to identify the universal probe and primer sequences for each reaction. This information is given in Table 1. Primers were synthesized by Integrated DNA Technologies (Coralville, IA). The GAPDH reference gene kit labeled with Yellow555 (Roche Applied Science) was used as a housekeeping gene. All other probes were labeled with FAM. Real time PCR reactions (total volume of 20 μL) were prepared using the LightCycler 480 Probes Master Mix (Roche Applied Science) as instructed by the manufacturer and run on the LightCycler 480 in the GenoSeq UCLA Genotyping and Sequencing core facility. The LightCycler 1.5 Software (Roche Applied Science) was used to analyze all data. This software uses the ΔΔCT method to calculate the fold change in gene expression compared to the reference gene in treated versus control cells treated with buffer alone.
Table 1.
Gene, probe, and primer information for real time PCR analysis.
| Gene | Accession # | UPL Probe # | 5′ Primer | 3′ Primer |
|---|---|---|---|---|
|
| ||||
| TXNIP | NM_006472.3 | 85 | cttctggaagaccagccaac | gaagctcaaagccgaacttg |
| CDC14B | AF023158.1 | 66 | gggtgccattgcagtacatt | agatcctgacccacgcaat |
| RGS1 | NM_002922.3 | 84 | tgaaatcttccaagtccaagg | tccaaagacattttgaccagttt |
| GADD45β | AF087853.1 | 10 | cattgtctcctggtcacgaa | taggggacccactggttgt |
| HIST2H4 | AF525682.1 | 25 | gagtgagagggacctgagca | cctctcgacatttcgtcattt |
| BHLHB2 | AB043885.1 | 66 | tggattcccctgagttaaggt | tcaggaaaatcctttgacagataa |
| TSC22D3 | BC072446.1 | 10 | tggtggccatagacaacaag | tctcggatctgctccttcag |
| KLF6 | BC000311.2 | 85 | gatgagttaaccaggcacttcc | agaggtgcctcttcatgtgc |
| NFKBIE | ENST00000275015.4 | 79 | gctctgttgcctggcttt | agccagatggagtgctgtct |
| FYTTD1 | BC039734.1 | 25 | gcaatgaaaacctcgacaaaa | attgctgggcaccacttt |
| THUMP2 | BC013299.2 | 86 | gacttgactttcagagtatcttgtcg | aattccaattacttttcctacctcct |
UPL: universal probe library (Roche Applied Sciences)
Confocal microscopy
ch128.1Av (10 nM) conjugated to b-SO6 was labeled with Zenon® (excitation 594 nm; Life Technologies) and the labeled complex was incubated with IM-9 or U266 cells for 1 or 16 hours. Cells were collected, fixed with 3.7% paraformaldehyde, permeabilized with 0.2% Triton-X and incubated with a goat anti-saporin antibody labeled with Alexa Fluor® 488 (Advanced Targeting Systems) for at least 2 hours followed by the anti-goat IgG labeled with Alexa Fluor® 488 (Life Technologies) for 1 hour. After washing, cells were mounted on slides in Vectashield® mounting medium with 1.5 μg/mL DAPI (Vector Labs, Burlingame, CA). Images were obtained using a confocal microscope (Leica, Wetzlar, Germany) equipped with an oil-immersion NA 1.4 60X objective. Images were analyzed using ImageJ software (NIH). Confocal laser scanning microscopy was performed in the California NanoSystems Institute Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA.
Western Blot analysis of nuclear extracts
Both IM-9 and U266 cells were treated in 60 mm dishes with 10 nM ch128.1Av/b-SO6 for either 1 or 16 hours. Nuclear and cytoplasmic extracts were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) as described by the manufacturer. Control cells were incubated with an equal volume of buffer alone. Protein concentrations were determined using the BCA protein assay (Thermo Fisher Scientfic). Proteins (20 μg for cytoplasmic fractions and 2 μg for nuclear extracts) were then separated on 4–12% Bis-Tris NuPAGE gels (Life Technologeis) in MOPS buffer. Proteins were transferred to Whatman Protran nitrocellulose membranes (Thermo Fisher Scientific) and probed with a goat anti-saporin antibody labeled with horseradish peroxidase (HRP; Advanced Targeting Systems). A rabbit anti-human GAPDH antibody (Cell Signaling Technology, Boston, MA) was used as a control for cytoplasmic protein and was detected using a donkey anti-rabbit IgG-HRP (GE Healthcare Life Sciences, Piscataway, NJ). The murine antibody targeting the human TATA-box binding protein (TBP; Life Technologies) was used as a nuclear protein control as previously described (Dansithong et al. 2008) and was detected using a sheep anti-mouse IgG-HRP antibody (GE Healthcare Life Sciences). For all Western Blots the ChemiGlow West Chemiluminescent Substrate (ProteinSimple, San Jose, CA) was used as described by the manufacturer and blots were developed on a Kodak X-OMAT 2000A (Rochester, NY).
Long-term culture-initiating cell assay (LTC-IC)
Bone marrow mononuclear cells (BMMC, StemCell Technologies, Vancouver, Canada) were treated with 10 nM ch128.1Av, b-SO6, ch128.1Av/b-SO6 or IgG3-Av/b-SO6 for 1 hour in Iscove’s Modfied Dulbecco’s Medium with 2% FBS. Cells were then washed 3 times and the assay was carried out as recommended in the LTC-IC Procedure Manual (StemCell Technologies) and as described previously (Daniels et al. 2011). In brief, treated cells were seeded on a M2-10B4 murine fibroblast (ATCC) feeder layer in human long-term medium (StemCell Technologies). Cells were cultured for 5 weeks with half media changes weekly. Both nonadherent and adherent cells were harvested and seeded in quadruplicate in MethoCult GF+H4435 (“Complete PLUS” methylcellulose medium with recombinant cytokines and erythropoietin; StemCell Technologies). After an 18-day incubation total colony numbers were determined using an Olympus CK2 inverted microscope (Olympus America Inc., Center Valley, PA). The assay was conducted using BMMC from 3 different donors.
Results
Global gene expression analysis in cells treated with the ch128.1Av/b-SO6 conjugate
Two cell lines were chosen for the global gene expression analysis. IM-9 cells are highly sensitive to the effects of the fusion protein alone, while U266 cells are resistant (Daniels et al. 2007; Ng et al. 2006; Rodriguez et al. 2011). Both malignant B-cell lines have been previously shown to be sensitive to apoptosis-induced by the ch128.1Av/b-SO6 conjugate (Daniels et al. 2007). RNA was collected for analysis from both cells lines at various time points after treatment with ch128.1Av/b-SO6 (Figure 1A). Cells incubated with buffer alone were used as controls. Treated cells were also monitored simultaneously for the expected cytotoxic effects at 48 hours using proliferation and apoptosis assays (Figure 1B–C). As expected, ch128.1Av was only cytotoxic to IM-9 cells, while both cell lines were sensitive to the effects of the conjugate.
Figure 1. Time course protocol for mRNA isolation and verification of the cytotoxic effects of the fusion protein alone and conjugated to b-SO6.
Cells were treated with 10 nM ch128.1 alone or conjugated to b-SO6 for the indicated times in duplicate samples in a 48-well plate at 200,000 cells/well (A). At each time point RNA was collected for microarray analysis. Control wells were treated with buffer alone. Additional wells for each treatment were incubated in parallel for 48 hours to measure the anti-proliferative (B) and pro-apoptotic (C) effect of ch128.1Av and ch128.1Av/b-SO6 on IM-9 and U266 cells. The anti-proliferative effect was measured by [3H]-thymidine incorporation and values expressed as the % of buffer control. Error bars represent standard deviation. Apoptosis was determined by flow cytometry in cells labeled with Annexin-V/Propidium Iodide. Percentage of total cells is shown in the corner of each quadrant.
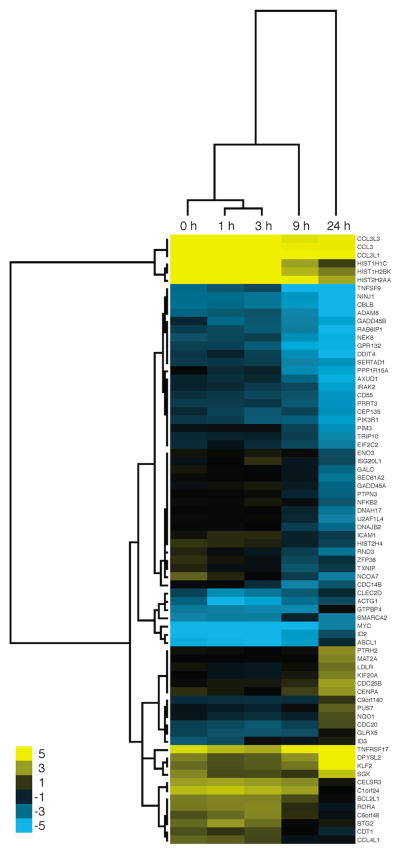
HumanRef-8 v2 Expression BeadChips were used to monitor gene expression changes. The data normalization process is described and shown in Supplementary Figure 1. The number of differentially expressed genes (LFC > 1, p < 0.05) in IM-9 cells was higher than those of U266. Most of the changes in IM-9 cells occurred at 9 and 24 hours and included both up and down regulation of genes (Figure 2A). Limited changes were detected in U266 cells treated with the conjugate and all changes were upregulation of the specified genes (Figure 2B). A focused view of the expressional changes (LFC > 1, p < 0.05) that occurred in IM-9 cells after 9 hours of treatment with the conjugate (compared to buffer alone) is shown in Figure 2C. Gene expression changes (LFC > 1, p < 0.05) in the two cells lines for the same treatment times are compared in Figure 3 and ontologies are shown in Figure 4. A Venn diagram (Supplementary Figure 2) shows the gene expression changes that were observed in IM-9 and U266 cells. A set of 11 genes were upregulated in both cell lines: TXNIP, CDC14B, HIST2H4, RGS1, THUMPD2, KLF6, BHLHB1, FYTTD1, GADD45B, TSC22D3, and NFKBIE. No genes were found to be downregulated in both cell lines. The expression levels of these 11 genes were further analyzed by QPCR to confirm the gene expression changes (Figure 5, Table 2). In the QPCR analysis, the changes observed in the 11 genes in IM-9 cells were of higher magnitude than those of U266 and were consistent with the microarray data. All 11 genes tested by QPCR showed upregulation upon treatment with the ch128.1Av/b-SO6 conjugate, but not with the fusion protein or the toxin alone. These genes are different from those that were previously shown to be differentially expressed in response to the fusion protein alone, which are consistent with a response to iron deprivation (Rodriguez et al. 2011). However, 10 of the genes were also upregulated in both cell lines upon treatment with the commonly used protein synthesis inhibitor CHX (Supplementary Table 1). HIST2H4 was the only gene not consistently upregulated by CHX in both cell lines. Thus, this transcriptional response appears to be in response to protein synthesis inhibition in general and not a specific response to the ch128.1Av/b-SO6 conjugate.
Figure 2. Gene expression analysis in cells treated with ch128.1Av complexed to b-SO6.
Changes in gene expression with a greater than 1.0 Log base 2-fold change (LFC) after 1, 3, 9, or 24 hours of ch128.1Av/b-SO6 treatment with respect to their time matched control (buffer alone) are shown for IM-9 (A) and U266 (B) cells clustered by time point and magnitude of change. Changes in IM-9 greater than 1.0 LFC at 9 hours (C) post treatment are also shown for comparison with changes in U266 at 24 hours. Clustering was conducted using the Cluster program and visualized using Java TreeView.
Figure 3. Comparison of gene expression changes in IM-9 and U266 at the various time points.
Direct comparison in gene expression changes between IM-9 and U266 cells treated with ch128.1Av/b-SO6 in which genes with a variance greater than 0.4 are clustered over the various time points and the magnitude of difference between the RNA levels in each cell line. In this case genes whose transcripts are present at a higher level in U266 than in IM-9 are shown in yellow. Clustering was conducted using the Cluster program and visualized using Java TreeView.
Figure 4. Gene clustering according to ontologies.
The same list of genes shown in Figure 3 are now shown clustered by their corresponding ontologies, with statistically enriched ontologies listed at right alongside their corresponding p-values. Clustering was conducted using the Cluster program and visualized using Java TreeView.
Figure 5. Validation of gene expression changes by real time PCR.
Freshly isolated RNA from U266 (A) and IM-9 (B) cells treated with either 10 nM ch128.1Av, b-SO6, or ch128.1Av/b-SO6 for 24 hours was converted to cDNA and gene expression determined using relative quantification analysis on a Roche LightCycler 480 System. Data is shown as the fold change in target gene expression compared to the housekeeping gene GAPDH. All samples were tested in triplicate and the standard deviations are shown. Data are representative of two independent experiments carried out with different RNA preparations.
Table 2.
Comparison of microarray and QPCR data for the 11 genes commonly upregulated in IM-9 and U266 cells.
| Gene | U266 Micoarray (Fold Change) | U266 QPCR (Fold Change) | IM-9 Microarray (Fold Change) | IM-9 QPCR (Fold Change) |
|---|---|---|---|---|
|
| ||||
| TXNIP | 3.0 | 8.2 | 8.4 | 22.4 |
| CDC14B | 2.9 | 2.2 | 15.2 | 2.9 |
| HIST2H4 | 2.3 | 2.0 | 9.4 | 5.7 |
| RGS1 | 4.1 | 14.5 | 6.8 | 18.8 |
| THUMPD2 | 2.1 | 5.3 | 3.6 | 6.2 |
| KLF6 | 2.3 | 5.6 | 4.6 | 8.6 |
| BHLHB1 | 2.0 | 2.9 | 2.9 | 1.9 |
| FYTTD1 | 2.1 | 2.0 | 3.5 | 2.5 |
| GADD45B | 2.1 | 13.1 | 13.5 | 25.5 |
| TSC22D3 | 2.9 | 12.8 | 2.9 | 8.1 |
| NFKBIE | 2.1 | 4.9 | 6.4 | 7.5 |
The effect of the antioxidant N-acetylcysteine on saporin-induced apoptosis
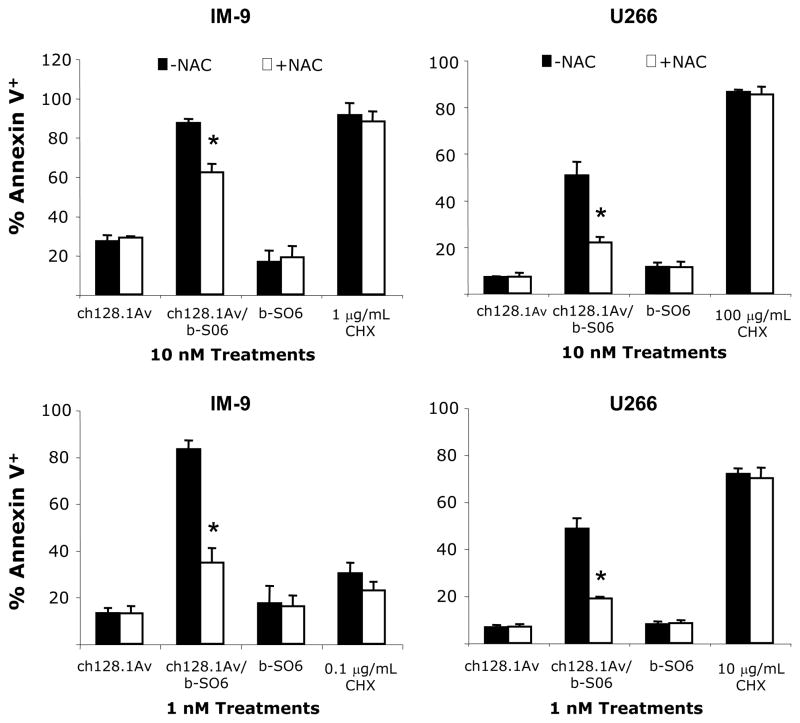
The gene expression profile of IM-9 and U266 cells treated with the ch128.1Av/b-SO6 conjugate is consistent with the induction of oxidative stress and DNA damage triggered by the toxin. In order to examine whether oxidative stress plays a role in the apoptosis mediated by the toxin, the antioxidant NAC was co-incubated with the conjugate or CHX for the full 48 hours and apoptosis assessed. Cells were treated with 1nM ch128.1Av/b-SO6 since this concentration consistently induces a high level of apoptosis in both cell lines. Additionally, we used a high concentration (10 nM) of the conjugate as to determine if the effects of NAC were dose-dependent. We also used a high and a low dose of CHX. As expected, ch128.1Av alone had no effect on U266 cells. In IM-9 cells ch128.1Av had an effect that was not blocked by NAC. However, in both cell lines ch128.1Av/b-SO6 strongly induced apoptosis that could be blocked by NAC in a dose-dependent manner (Figure 6). CHX induced apoptosis in both cell lines; however, NAC did not block this cell death.
Figure 6. Protection of ch128.1Av/b-SO6-induced cell death by the antioxidant NAC.
IM-9 (left panels) and U266 (right panels) cells were treated with 10 nM (top panels) or 1 nM (bottom panels) ch128.1Av, b-SO6, or ch128.1Av/b-SO6 for 48 hours with or without the addition of 2 mM NAC. CHX was used as a common protein synthesis inhibitor. The percentage of Annexin V positive cells was determined using flow cytometry. Data are the average of three independent experiments. Error bars indicate the standard deviation. * p < 0.01, Student’s t-test.
Analysis of saporin localization
To address the possibility that b-SO6 can cause direct DNA damage upon delivery by ch128.1Av into the cells, we performed localization studies by confocal microscopy. If saporin is found within the nucleus, it would be possible that the toxin can cause direct DNA damage. Therefore, we followed the internalization of ch128.1Av/b-SO6 in IM-9 (Supplementary Figure 3) and U266 (Supplementary Figure 4) cells over a 24-hour period. The ch128.1Av/b-SO6 complex was intracellularly localized after 30 minutes. The complex was seen in punctate structures in the cytoplasm and accumulated around the nucleus. The toxin appeared to colocalize with ch128.1Av; however, no strong intranuclear localization of saporin was detected by confocal microscopy at any time point tested. Nevertheless, Western Blot analysis showed that the toxin was detected in nuclear extracts in both IM-9 and U266 cells as early as 1 hour after treatment (Figure 7). After 16 hours post-exposure, there appeared to be higher levels of b-SO6 in the nuclear extracts.
Figure 7. Detection of b-SO6 in nuclear extracts of cells treated with ch128.Av/b-SO6 conjugate.
U266 (left panels) and IM-9 (right panels) were treated with 10 nM ch128.1Av/b-SO6 for either 1 hour or 16 hours followed by the preparation of nuclear and cytoplasmic extracts. Saporin within these extracts was detected by Western Blot analysis. Control cells (“C”) were incubated with an equal amount of buffer for 16 hours. GAPDH was used a control for cytoplasmic protein, while TBP was used as a control for nuclear protein.
In vitro toxicity to hematopoietic stem cells and early progenitors
Since the ch128.1Av/b-SO6 conjugate targets malignant hematopoietic cancers via interaction with TfR1, we evaluated the potential toxicity of the conjugate on normal hematopoietic stem and early progenitor cells using the long-term cell-initiating culture (LTC-IC) assay. The LT-IC assay enumerates the number of or pluripotent hematopoietic stem and noncommitted, early progenitor cells within a given sample. The number of colonies at the end of the assay correlates with the number of viable pluripotent progenitor cells after treatment. Exposure to ch128.1Av/b-SO6 did not result in a reduction of colony number (Table 3) when we tested BMMC from 3 different donors, suggesting that the conjugate is not toxic to this population of cells. ch128.1Av or b-SO6 alone, as well as the isotype control conjugate (IgG3-Av/b-SO6), also did not affect colony formation.
Table 3.
Effect of ch128.1Av on normal pluripotent hematopoietic progenitor cells from three separate donors as determined by the LTC-IC assay.
| Donor 1 | Donor 2 | Donor 3 | |
|---|---|---|---|
|
| |||
| Untreated | 7 ± 0.96 | 1 ± 0.82 | 13 ± 1.71 |
| Buffer | 13 ± 1.00 | 3 ± 1.83 | ND |
| ch128.1Av | 15 ± 2.20 | 3 ± 1.50 | 13 ± 2.16 |
| b-SO6 | 15 ± 2.63 | 9 ± 2.08 | 21 ± 1.15 |
| IgG3-Av/b-SO6 | 10 ± 1.83 | 8 ± 1.71 | 20 ± 2.71 |
| ch128.1Av/b-SO6 | 12 ± 1.50 | 10 ± 2.65 | 21 ± 2.5 |
Data represent the mean of quadruplicates ± the standard deviation. LTC-IC indicates long-term cell-initiating culture; ND, not determined. Data for the untreated, buffer, and ch128.1Av alone have been previously reported (Daniels et al. 2011). All samples were tested simultaneously in each donor.
Discussion
We have previously shown that the delivery of the single chain RIP b-SO6 using ch128.1Av effectively blocks protein synthesis, activates caspases 2 and 3, and to lesser extent caspases 8 and 9, and induces apoptosis in malignant B cells (Daniels et al. 2007). In order to further explore the mechanism of cell-death induced by this immunotoxin, we conducted a global gene expression analysis in two different human malignant B-cell lines, one that is sensitive to the cytotoxic effects of the fusion protein alone (IM-9) and one that is not (U266). Eleven genes that were differentially expressed in both cell lines were identified. Since these genes were similarly activated in both cell lines, they may represent a generalized response to the toxin in malignant B cells. The proteins encoded by 11 genes upregulated in both cell lines in response to the immuntoxin are involved in the cellular response to oxidative stress, DNA damage, or are involved with RNA processing. KLF6 is a zinc finger-containing transcription factor whose expression increases upon treatment with hydrogen peroxide (Cullingford et al. 2008; Ghaleb and Yang 2008). It also acts as a tumor suppressor that has been shown to interact with Cyclin D1 causing cell cycle arrest (Benzeno et al. 2004). TXNIP can also act as a tumor suppressor and plays a role in redox homeostasis through its interaction with the antioxidant thioredoxin (Zhou and Chng 2012). The expression of TXNIP is increased upon treatment with various anti-cancer agents and it has been shown to render cells more sensitive to oxidative stress (Yoshioka et al. 2006). CDC14B is a phosphatase known to regulate the cell cycle. Its expression was shown to increase upon treatment with the plant toxin curcumin, which induced DNA damage in lung carcinoma cells (Skommer et al. 2007). It has also been suggested that CDC14B may be required for DNA repair of double stranded breaks (Mocciaro et al. 2010; Wei et al. 2011). BHLHB2 has been previously shown to be upregulated in response to DNA damaging agents in a p53-independent manner (Thin et al. 2007). It is also a regulator of ionizing radiation-induced apoptosis and cell death caused by other stress stimuli including hypoxia and serum starvation (Yamada and Miyamoto 2005). GADD45B is induced by various stress stimuli including DNA damage, DNA alkylating agents, UVB radiation, and the multi-kinase inhibitor sorafenib (Ou et al. 2010; Thyss et al. 2005; Zumbrun et al. 2009). Additionally, GADD45B was induced by treatment with either the verotoxin or shiga toxin 2, toxins produced by Escherichia coli that inhibit protein synthesis (the shiga toxin 2 is an N-glycosyidase), in human colorectal carcinoma cells or brain microvascular endothelial cells, respectively (Bhattacharjee et al. 2005; Fujii et al. 2008). The gene encoding the core histone protein HIST2H4 was also upregulated in both IM-9 and U266 cells. HIST2H4 can regulate DNA repair and has been shown to be upregulated by photodynamic therapy, which induces an increase in cellular ROS levels (Cekaite et al. 2007). Reactive oxygen species (ROS) can interact with signaling molecules, other proteins, and nucleic acids causing a variety of changes in growth signaling and cellular damage that can lead to apoptosis or necrosis (Fogg et al. 2011; Mates et al. 2012; Ray et al. 2012). Specifically, ROS can cause DNA damage by inducing DNA strand breaks, mutations, deletions, gene amplification, and rearrangements. The upregulation of the above mentioned genes by b-SO6 delivered into cells by ch128.1Av suggests that the toxin may increase the oxidative stress level within the cell through the induction of ROS. Furthermore, these data, together with the fact that caspase-2 can function in response to DNA damage (Cullen and Martin 2009; Krumschnabel et al. 2009; Zhivotovsky and Orrenius 2005), is highly activated by ch128.1Av/b-SO6 (Daniels et al. 2007) suggest that the toxin induces DNA damage in treated cells.
Treatment with the ch128.1Av/b-SO6 conjugate also upregulated other genes involved in RNA binding and processing, as well as the inhibition of the NF-κB pathway. THUMPD2 encodes for a methyltransferase with a putative RNA-binding domain whose function is not understood (Aravind and Koonin 2001). FYTTD1 encodes a protein that is required for mRNA processing and export from the nucleus to the cytosol (Hautbergue et al. 2009). This upregulation of proteins involved in RNA binding and processing is understandable as a compensatory response, given the fact that b-SO6 strongly inhibits protein synthesis. Additionally, the NF-κB pathway is also affected by b-SO6 treatment. The NF-κB family of transcription factors play a role in immunity, inflammatory responses, and ultimately cell growth (Bubici et al. 2006). A unique interaction occurs between ROS and the NF-κB pathway. Depending on the circumstances, ROS can either induce or inhibit the NF-κB pathway (Bubici et al. 2006). The activity of these transcription factors is regulated by a family of inhibitors (IκBs), one of which is encoded by the gene NFKBIE (IκBε) (Li and Nabel 1997; Whiteside et al. 1997). This inhibitor blocks NF-κB activity by sequestering certain family members in the cytoplasm and keeping them from translocating to the nucleus. In our studies, b-SO6 also upregulated the protein encoded by the TSC22D3 gene. This protein is known to inhibit inflammation (Beaulieu et al. 2010) and NF-κB activity (Di Marco et al. 2007). Furthermore, TSC22D3 is upregulated by glucocorticoid treatment in MM cells (Grugan et al. 2008). Moreover, ch128.1Av/b-SO6 upregulates the RGS1 protein that is known to block signal transduction blocking the activity of GTPase activating proteins (Bansal et al. 2007). Recently, RGS1 levels increased in plant cells treated with ozone, which was shown to induce ROS levels (Booker et al. 2012). Taken together, our data suggest that b-SO6 induces a transcriptional response in malignant B cells consistent with the induction of oxidative stress/DNA damage that results in the blockage of signal transduction, cell cycle arrest, and ultimately apoptosis. However, 10 out of the 11 genes that were upregulated by the ch128.1Av/b-SO6 conjugate were also upregulated by the protein synthesis inhibitor CHX. This compound interferes with the translocation step in protein synthesis and thus, inhibits translational elongation. These data suggest that the observed transcriptional response is not specific to the effects of the toxin per se, but rather, is a response to a block in protein synthesis.
To our knowledge this is a pioneer study in terms of the gobal expression analysis in cancer cells treated with SO6 alone or bound to any delivery vehicle. However, a gene expression analysis has been conducted in human airway cells treated with the native ricin toxin for 24 hours (Wong et al. 2007). This analysis showed the upregulation of a variety of histones; however, HIST2H4 (upregulated by b-SO6 in our studies) was not one of them. Ricin did increase the expression of HIST2H2AA and TNFAIP3, which were observed to be upregulated by b-SO6 in our studies, but only in IM-9 cells. Furthermore, in murine kidney cells isolated from animals in which ricin was administered intravenously, gene expression changes included a decrease in ATF4 expression and an increase in ICAM-1 (Korcheva et al. 2005), which were also differentially expressed in our studies in IM-9 cells only. In general, the genes upregulated by ricin in these two studies did not overlap with the 11 genes that were seen to be upregulated in both IM-9 and U266 cells under our study conditions.
In order to confirm the role of ROS in the cell-death mediated by b-SO6, we co-incubated cells with the antioxidant NAC. Surprisingly, even though b-SO6 is a potent inhibitor of protein synthesis, the protective effect of NAC was very strong. This suggests that, in addition to the inhibition of protein synthesis, ROS play a major role in b-SO6-mediated cytotoxicity. In our study, NAC was unable to block the effects of CHX, even though the transcriptional response to CHX and the immunotoxin were similar. We observed that apoptosis induced by CHX occurred at a faster rate and thus, the effects of CHX may have been too strong for NAC to neutralize. Alternatively, there may be a difference in the type of ROS generated by the two treatments and NAC can only protect from those induced by b-SO6. Further studies are needed to understand this phenomenon. The fact that NAC can block apoptosis induced by our conjugate is consistent with the observation that NAC was able to block the DNA damage induced by abrin, a Type I RIP similar to b-SO6, in U937 human myeloleukemic cells (Bhaskar et al. 2008). Ricin-induced apoptosis is also mediated by the generation of ROS in the HeLa human cervical cancer cell line (Rao et al. 2005). However, ricin has also been shown to cause direct DNA damage that occurs prior to the induction of apoptosis (Brigotti et al. 2002). CHX was also used in that study and was shown to block protein synthesis but did not appear to induce DNA damage. This is consistent with our study showing that NAC did not block the cytotoxic effects of CHX. Taken together, these studies suggest that while RIPs are potent inhibitors of protein synthesis, they also induce oxidative stress that contributes their cytotoxic effects.
Although SO6 lacks a cell-binding domain, at high concentrations and under certain conditions, it can enter cells and cause cytotoxicity (Lombardi et al. 2010). It has been suggested that in some cases the LDL-related family of receptors may be involved in the uptake of the natural toxin in mammalian cells. It has also been suggested that the natural toxin is taken up mostly by receptor-independent endocytosis (Bolognesi et al. 2012). Since SO6 is known to act on the ribosomes to block protein synthesis, the toxin must ultimately reach the cytosol. However, its intracellular trafficking is still largely unknown (Lombardi et al. 2010). It has been suggested that SO6 can remove adenine from DNA through its N-glycosidase activity (Gasperi-Campani et al. 2005; Roncuzzi and Gasperi-Campani 1996), although this is controversial (Lombardi et al. 2010). However, if this is the case, SO6 must reach the nucleus. Although the catalytic activity of SO6 is similar to that of ricin, the intracellular trafficking of the two toxins clearly differs. Ricin (a Type II RIP that contains a cell-binding domain) undergoes retrograde transport through the endoplasmic reticulum where it exploits the ER-associated degradation pathway to reach the cytosol (Spooner et al. 2006; Vago et al. 2005). Previous studies have suggested that saporin (specific isoform not specified) may traffic through an endosomal compartment due to partial colocalization with disulphide isomerase, an endoplasmic reticulum marker, in GL15 human glioblastoma cells (Cimini et al. 2011) and LAMP2, a late endosome marker, in Vero monkey kidney cells (Vago et al. 2005). Saporin did not colocalize with the early endosome marker EEA1 or the Golgi marker Golgin 97 (Vago et al. 2005). Another study found SO6 localized intracellularly within 20 minutes (Bolognesi et al. 2012). At this time SO6 was detected in endocytic vesicles in the HeLa cytosol accumulating in a perinuclear vesicular structure via fluorescence microscopy detection. About 30% of SO6 colocalized with the endosomal compartment (ER marker BiP) while around 7% co-localized with the Golgi apparatus (Golgi marker GM130) as seen by confocal microscopy. After a 1-hour incubation, 4% of endocytosed SO6 was detected in the nucleus. Using transmission electron microscopy, gold-saporin molecules were seen migrating from the plasma membrane and clear vesicles and vacuoles into perinuclear late endosomes and lysosomes, with approximately 10% of cells showing saporin nuclear localization (Bolognesi et al. 2012). In addition, recombinant 125I-SO6 appears to migrate from the cytosol to the nuclear fraction over time in J774A.1 mouse macrophage cells as detected by immunoblotting (Bagga et al. 2003).
It stands to reason that conjugation of SO6 to a delivery vehicle would alter the natural trafficking of the toxin at least at the early stages. The delivery of saporin (specific isoform not specified) conjugated to polyamidoamine dendrimer (PAMAM) coupled with the photochemical internalization (PCI) technology, which breakdowns endosomal/lysosomal membranes by activating photosensitizers localized on the membranes, resulted in enhanced delivery of saporin into Ca9-22 gingival cancer cells (Lai et al. 2008). Furthermore, saporin was found in the nucleus under these conditions. In our colocalization studies, b-SO6 delivered by ch128.1Av accumulated in cytoplasmic vesicles and did not show a clear and consistent pattern of intranuclear localization. Since we used only 10 nM of the toxin in our studies, and such small percentage of saporin has been reported to enter the nucleus, it is possible that b-SO6 was not clearly observed in the nucleus due to the detection limits of confocal microscopy. However, we were able to detect b-SO6 in nuclear extracts of treated cells by Western Blot analysis, although we cannot determine if the toxin is associated with the nuclear membrane or if it is actually within the nucleus. Since the toxin may have reached the nucleus, it is possible that it can interact directly with DNA, although we do not have direct evidence for this at this time. Whether or not b-SO6 directly induces DNA damage, its high toxicity and inability to enter cells at low concentrations make it a meaningful therapeutic agent for delivery purposes.
Toxicity to normal cells is always a concern with any anti-cancer drug. In order to address this concern, we evaluated the toxicity of ch128.1Av/b-SO6 on hematopoietic stem and non-committed progenitor cells. Our data show that there was no observed toxicity on this population of cells. This is consistent with the fact that these non-committed progenitors express very low levels of the TfR1/CD71 (Gross et al. 1997; Knaan-Shanzer et al. 2008; Lansdorp and Dragowska 1992). We have previously shown that the conjugate is toxic to late (committed) progenitor cells of both the erythroid and myeloid lineages (CFU-e, BFU-e and CFU-GM) (Daniels et al. 2011) that are known to express the TfR1 (Daniels et al. 2006b). Our results are consistent with a previous in vitro study (that also used the LTC-IC assay), which demonstrated the depletion of committed progenitor cells while pluripotent progenitors were spared if human bone marrow cells were treated with an immuntoxin composed of SO6 chemically conjugated with the OKT9 antibody that targets the TfR1 (Benedetti et al. 1994). Together, these results suggests that while the immunotoxin may have toxicity against committed progenitor cells, these cells can be repopulated by the pluripotent progenitors that are not affected by treatment with the conjugate. These results also suggest that the ch128.1Av/b-SO6 conjugate may be a therapeutic option for the ex vivo purging of cancer cells during autologous stem cell transplantation, a treatment option for some patients affected with incurable B-cell malignancies such as MM, in which grafts can be contaminated with MM cells that may shorten progression-free survival (Bashey et al. 2008; Vogel et al. 2005).
In conclusion, in this study we further examined the mechanism of b-SO6-induced apoptosis when the toxin is delivered into malignant B cells using ch128.1Av. Our results here suggest that in addition to its RIP function, b-SO6 induces the generation of ROS, which may play a role in DNA damage and ultimately the induction of apoptosis in malignant B cells by the ch128.1Av/b-SO6 conjugate. Our studies confirm the dual activity of ch128.1Av, in that it can be a direct anti-cancer agent, as well as an effective delivery agent for the transport of active molecules into cancer cells. Thus, ch128.1Av alone or conjugated to therapeutic agents should be further explored as a potential therapy for B-cell malignancies.
Supplementary Material
A variance stabilizing transform was applied to the data to reduce the mean variance for each technical replicate, followed by a probe level quantile normalization within each array and between arrays (Lin et al. 2008). Shown is a graphical representation of normalized array data comparing two IM-9 (1674277005_A and 1674277005_B) and two U266 (1688628159_B and 1688628159_C) control array profiles. For each pairwise comparison, the log intensity ratio M (y axis) is plotted against the average log intensity A (x axis). Data normalization was conducted using the Lumi package from the Bioconductor Project (http://www.bioconductor.org). The top panel shows the data without normalization and the bottom panel after the analysis with the Lumi package.
The circle on the left lists all the genes that show a change in expression at 24 hours in IM-9 cells, while the circle on the right shows genes that change in U266 cells. The overlapping portion of the two circles shows a list of 11 genes for which the expression level changes in both cells lines.
Confocal microscopy images of cells incubated with Zenon® labeled ch128.1Av (red) alone or conjugated to b-SO6 for 1 or 16 hours. Saporin was detected using a goat anti-SO6 antibody labeled with Alexa Fluor® 488 (green). Nuclei are stained with DAPI (blue) within the mounting medium. The white bar indicates 25μm. Experiments were repeated at least twice.
Confocal microscopy images of cells incubated with Zenon® labeled ch128.1Av (red) alone or conjugated to b-SO6 for 1 or 16 hours. Saporin was detected using a goat anti-SO6 antibody labeled with Alexa Fluor® 488 (green). Nuclei are stained with DAPI (blue) within the mounting medium. The white bar indicates 25μm. Experiments were repeated at least twice.
Acknowledgments
This work was supported in part by NIH/NCI R01CA107023, R01CA57152, and K01CA138559. This work was also supported by the Howard Hughes Medical Institute Gilliam Fellowship, the Whitcome Fellowship of the Molecular Biology Institute at UCLA, the UCLA Amgen Scholars Program, and the Amgen Foundation. The UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility is supported by the NIH awards CA16042 and AI28697, the Jonsson Cancer Center, the UCLA AIDS Institute, and the UCLA School of Medicine. The UCLA CNSI Advanced Light Microscopy/Spectroscopy Shared Resource Facility is supported by NIH-NCRR shared resources grant (CJX1-44835-WS-29646) and NSF Major Research Instrumentation grant (CHE-0722519). Gustavo Helguera is member of the National Council for Scientific and Technological Research (CONICET), Argentina and is supported by PICT PRH 2008-00315, PIP 114-2011-01-00139, and UBACYT 200-2011-02-00027.
Abbreviations
- b-SO6
biotinylated saporin-6
- BCA
bicinchoninic acid
- BFU-e
burst forming unit-erythroid
- BHLHB2
basic-helix-loop-helix transcription factor B2
- BMMC
bone marrow mononuclear cells
- CDC14B
dual specificity protein tyrosine phosphatase family
- CFU-e
colony forming unit-erythroid
- CFU-GM
colony forming unit-granulocyte/macorphage
- CHX
cycloheximide
- ch128.1Av
mouse/human chimeric antibody avidin fusion protein targeting CD71
- DNS
dansyl hapten (5-dimethylamino naphthalene-1-sulfonyl chloride)
- FYTTD1
forty-two-three domain containing 1
- GADD45B
growth arrest DNA damage-inducible gene 45β
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HIST2H4
core histone 2H4
- HRP
horseradish peroxidase
- kDa
kilodlatons
- KLF6
Kruppel-like transcription factor 6
- LFC
log base 2-fold change
- LTC-IC
long-term culture-initiating cell
- MM
multiple myeloma
- mw
molecular weight
- NAC
N-acetylcysteine
- NFKBIE
NF-κB inhibitor epsilon (IκBε)
- NHL
non-Hodgkin’s lymhoma
- NIH
National Institutes of Health
- QPCR
quantitative polymerase chain reaction
- RGS1
regulator of G-protin signaling
- RIP
ribosomal-inactivating protein
- ROS
reactive oxygen species
- SO6
saporin-6
- TBP
TATA-box binding protein
- Tf
transferrin
- TfR1
transferrin receptor 1 (also known as CD71)
- THUMD2
THUMP domain containing 2
- TSC22D3
glucocorticoid-induced leucine zipper; TFGβ-stimulated clone 22 domain
- TXNIP
thioredoxin interacting protein
Footnotes
Conflicts of Interest
The authors have no conflicts to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravind L, Koonin EV. THUMP-a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- Bagga S, Seth D, Batra JK. The cytotoxic activity of ribosome-inactivating protein saporin-6 is attributed to its rRNA N-glycosidase and internucleosomal DNA fragmentation activities. J Biol Chem. 2003;278:4813–4820. doi: 10.1074/jbc.M207389200. [DOI] [PubMed] [Google Scholar]
- Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116:473–495. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashey A, Perez WS, Zhang MJ, Anderson KC, Ballen K, Berenson JR, To LB, Fonseca R, Freytes CO, Gale RP, Gibson J, Giralt SA, Kyle RA, Lazarus HM, Maharaj D, McCarthy PL, Milone GA, Nimer S, Pavlovsky S, Reece DE, Schiller G, Vesole DH, Hari P. Comparison of twin and autologous transplants for multiple myeloma. Biol Blood Marrow Transplant. 2008;14:1118–1124. doi: 10.1016/j.bbmt.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu E, Ngo D, Santos L, Yang YH, Smith M, Jorgensen C, Escriou V, Scherman D, Courties G, Apparailly F, Morand EF. Glucocorticoid-induced leucine zipper is an endogenous antiinflammatory mediator in arthritis. Arthritis Rheum. 2010;62:2651–2661. doi: 10.1002/art.27566. [DOI] [PubMed] [Google Scholar]
- Benedetti G, Bondesan P, Caracciolo D, Cherasco C, Ruggieri D, Gastaldi ME, Pileri A, Gianni AM, Tarella C. Selection and characterization of early hematopoietic progenitors using an anti-CD71/S06 immunotoxin. Exp Hematol. 1994;22:166–173. [PubMed] [Google Scholar]
- Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D, Friedman SL. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. [DOI] [PubMed] [Google Scholar]
- Bhaskar AS, Deb U, Kumar O, Lakshmana Rao PV. Abrin induced oxidative stress mediated DNA damage in human leukemic cells and its reversal by N-acetylcysteine. Toxicol In Vitro. 2008;22:1902–1908. doi: 10.1016/j.tiv.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee RN, Park KS, Uematsu S, Okada K, Hoshino K, Takeda K, Takeuchi O, Akira S, Iida T, Honda T. Escherichia coli verotoxin 1 mediates apoptosis in human HCT116 colon cancer cells by inducing overexpression of the GADD family of genes and S phase arrest. FEBS Lett. 2005;579:6604–6610. doi: 10.1016/j.febslet.2005.10.053. [DOI] [PubMed] [Google Scholar]
- Bolognesi A, Polito L, Scicchitano V, Orrico C, Pasquinelli G, Musiani S, Santi S, Riccio M, Bortolotti M, Battelli MG. Endocytosis and intracellular localisation of Type 1 ribosome-inactivating protein saporin-s6. J Biol Regul Homeost Agents. 2012;26:97–109. [PubMed] [Google Scholar]
- Booker F, Burkey K, Morgan P, Fiscus E, Jones A. Minimal influence of G-protein null mutations on ozone-induced changes in gene expression, foliar injury, gas exchange and peroxidase activity in Arabidopsis thaliana L. Plant Cell Environ. 2012;35:668–681. doi: 10.1111/j.1365-3040.2011.02443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigotti M, Alfieri R, Sestili P, Bonelli M, Petronini PG, Guidarelli A, Barbieri L, Stirpe F, Sperti S. Damage to nuclear DNA induced by Shiga toxin 1 and ricin in human endothelial cells. FASEB J. 2002;16:365–372. doi: 10.1096/fj.01-0521com. [DOI] [PubMed] [Google Scholar]
- Bubici C, Papa S, Dean K, Franzoso G. Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene. 2006;25:6731–6748. doi: 10.1038/sj.onc.1209936. [DOI] [PubMed] [Google Scholar]
- Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna JL, Nunez G, Dalton WS, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- Cekaite L, Peng Q, Reiner A, Shahzidi S, Tveito S, Furre IE, Hovig E. Mapping of oxidative stress responses of human tumor cells following photodynamic therapy using hexaminolevulinate. BMC Genomics. 2007;8:273. doi: 10.1186/1471-2164-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini A, Mei S, Benedetti E, Laurenti G, Koutris I, Cinque B, Cifone MG, Galzio R, Pitari G, Di Leandro L, Giansanti F, Lombardi A, Fabbrini MS, Ippoliti R. Distinct cellular responses induced by saporin and a transferrin-saporin conjugate in two different human glioblastoma cell lines. J Cell Physiol. 2011;227:939–951. doi: 10.1002/jcp.22805. [DOI] [PubMed] [Google Scholar]
- Cullen SP, Martin SJ. Caspase activation pathways: some recent progress. Cell Death Differ. 2009;16:935–938. doi: 10.1038/cdd.2009.59. [DOI] [PubMed] [Google Scholar]
- Cullingford TE, Butler MJ, Marshall AK, Tham el L, Sugden PH, Clerk A. Differential regulation of Kruppel-like factor family transcription factor expression in neonatal rat cardiac myocytes: effects of endothelin-1, oxidative stress and cytokines. Biochim Biophys Acta. 2008;1783:1229–1236. doi: 10.1016/j.bbamcr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels TR, Bernabeu E, Rodriguez JA, Patel S, Kozman M, Chiappetta DA, Holler E, Ljubimova JY, Helguera G, Penichet ML. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels TR, Delgado T, Helguera G, Penichet ML. The transferrin receptor part II: targeted delivery of therapeutic agents into cancer cells. Clin Immunol. 2006a;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Daniels TR, Delgado T, Rodriguez JA, Helguera G, Penichet ML. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin Immunol. 2006b;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Daniels TR, Ng PP, Delgado T, Lynch MR, Schiller G, Helguera G, Penichet ML. Conjugation of an anti transferrin receptor IgG3-avidin fusion protein with biotinylated saporin results in significant enhancement of its cytotoxicity against malignant hematopoietic cells. Mol Cancer Ther. 2007;6:2995–3008. doi: 10.1158/1535-7163.MCT-07-0330. [DOI] [PubMed] [Google Scholar]
- Daniels TR, Ortiz-Sanchez E, Luria-Perez R, Quintero R, Helguera G, Bonavida B, Martinez-Maza O, Penichet ML. An antibody-based multifaceted approach targeting the human transferrin receptor for the treatment of B-cell malignancies. J Immunother. 2011;34:500–508. doi: 10.1097/CJI.0b013e318222ffc8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansithong W, Wolf CM, Sarkar P, Paul S, Chiang A, Holt I, Morris GE, Branco D, Sherwood MC, Comai L, Berul CI, Reddy S. Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS One. 2008;3:e3968. doi: 10.1371/journal.pone.0003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Virgilio M, Lombardi A, Caliandro R, Fabbrini MS. Ribosome-inactivating proteins: from plant defense to tumor attack. Toxins (Basel) 2010;2:2699–2737. doi: 10.3390/toxins2112699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco B, Massetti M, Bruscoli S, Macchiarulo A, Di Virgilio R, Velardi E, Donato V, Migliorati G, Riccardi C. Glucocorticoid-induced leucine zipper (GILZ)/NF-kappaB interaction: role of GILZ homo-dimerization and C-terminal domain. Nucleic Acids Res. 2007;35:517–528. doi: 10.1093/nar/gkl1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg VC, Lanning NJ, Mackeigan JP. Mitochondria in cancer: at the crossroads of life and death. Chin J Cancer. 2011;30:526–539. doi: 10.5732/cjc.011.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii J, Wood K, Matsuda F, Carneiro-Filho BA, Schlegel KH, Yutsudo T, Binnington-Boyd B, Lingwood CA, Obata F, Kim KS, Yoshida S, Obrig T. Shiga toxin 2 causes apoptosis in human brain microvascular endothelial cells via C/EBP homologous protein. Infect Immun. 2008;76:3679–3689. doi: 10.1128/IAI.01581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperi-Campani A, Brognara I, Baiocchi D, Roncuzzi L. Mitochondrial DNA D-loop as a new target of Saporin 6 nuclease activity. Toxicon. 2005;45:475–480. doi: 10.1016/j.toxicon.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Yang VW. The Pathobiology of Kruppel-like Factors in Colorectal Cancer. Curr Colorectal Cancer Rep. 2008;4:59–64. doi: 10.1007/s11888-008-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Helm K, Gruntmeir JJ, Stillman WS, Pyatt DW, Irons RD. Characterization and phenotypic analysis of differentiating CD34+ human bone marrow cells in liquid culture. Eur J Haematol. 1997;59:318–326. doi: 10.1111/j.1600-0609.1997.tb01693.x. [DOI] [PubMed] [Google Scholar]
- Grugan KD, Ma C, Singhal S, Krett NL, Rosen ST. Dual regulation of glucocorticoid-induced leucine zipper (GILZ) by the glucocorticoid receptor and the PI3-kinase/AKT pathways in multiple myeloma. J Steroid Biochem Mol Biol. 2008;110:244–254. doi: 10.1016/j.jsbmb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Walsh MJ, Snijders AP, Chang CT, Jones R, Ponting CP, Dickman MJ, Wilson SA. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol. 2009;19:1918–1924. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaan-Shanzer S, van der Velde-van Dijke I, van de Watering MJ, de Leeuw PJ, Valerio D, van Bekkum DW, de Vries AA. Phenotypic and functional reversal within the early human hematopoietic compartment. Stem Cells. 2008;26:3210–3217. doi: 10.1634/stemcells.2007-0117. [DOI] [PubMed] [Google Scholar]
- Korcheva V, Wong J, Corless C, Iordanov M, Magun B. Administration of ricin induces a severe inflammatory response via nonredundant stimulation of ERK, JNK, and P38 MAPK and provides a mouse model of hemolytic uremic syndrome. Am J Pathol. 2005;166:323–339. doi: 10.1016/S0002-9440(10)62256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen’s view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai PS, Pai CL, Peng CL, Shieh MJ, Berg K, Lou PJ. Enhanced cytotoxicity of saporin by polyamidoamine dendrimer conjugation and photochemical internalization. J Biomed Mater Res A. 2008;87:147–155. doi: 10.1002/jbm.a.31760. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM, Dragowska W. Long-term erythropoiesis from constant numbers of CD34+ cells in serum-free cultures initiated with highly purified progenitor cells from human bone marrow. J Exp Med. 1992;175:1501–1509. doi: 10.1084/jem.175.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nabel GJ. A new member of the I kappaB protein family, I kappaB epsilon, inhibits RelA (p65)-mediated NF-kappaB transcription. Mol Cell Biol. 1997;17:6184–6190. doi: 10.1128/mcb.17.10.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A, Marshall RS, Savino C, Serena Fabrini M, Ceriotti A. Type I Ribosome-Inactivating Proteins from Saponaria officinialis. In: Lord JM, Hartley MR, editors. Toxic Plant Proteins. Springer-Verlag Berlin; Heidelbery, Germany: 2010. pp. 55–78. [Google Scholar]
- Mates JM, Segura JA, Alonso FJ, Marquez J. Oxidative stress in apoptosis and cancer: an update. Arch Toxicol. 2012 doi: 10.1007/s00204-012-0906-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, Gillespie D, Jallepalli P, Schiebel E. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol. 2010;189:631–639. doi: 10.1083/jcb.200910057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PP, Dela Cruz JS, Sorour DN, Stinebaugh JM, Shin SU, Shin DS, Morrison SL, Penichet ML. An anti-transferrin receptor-avidin fusion protein exhibits both strong proapoptotic activity and the ability to deliver various molecules into cancer cells. Proc Natl Acad Sci U S A. 2002;99:10706–10711. doi: 10.1073/pnas.162362999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PP, Helguera G, Daniels TR, Lomas SZ, Rodriguez JA, Schiller G, Bonavida B, Morrison SL, Penichet ML. Molecular events contributing to cell death in malignant human hematopoietic cells elicited by an IgG3-avidin fusion protein targeting the transferrin receptor. Blood. 2006;108:2745–2754. doi: 10.1182/blood-2006-04-020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Sanchez E, Daniels TR, Helguera G, Martinez-Maza O, Bonavida B, Penichet ML. Enhanced cytotoxicity of an anti-transferrin receptor IgG3-avidin fusion protein in combination with gambogic acid against human malignant hematopoietic cells: functional relevance of iron, the receptor, and reactive oxygen species. Leukemia. 2009;23:59–70. doi: 10.1038/leu.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou DL, Shen YC, Yu SL, Chen KF, Yeh PY, Fan HH, Feng WC, Wang CT, Lin LI, Hsu C, Cheng AL. Induction of DNA damage-inducible gene GADD45beta contributes to sorafenib-induced apoptosis in hepatocellular carcinoma cells. Cancer Res. 2010;70:9309–9318. doi: 10.1158/0008-5472.CAN-10-1033. [DOI] [PubMed] [Google Scholar]
- Rao PV, Jayaraj R, Bhaskar AS, Kumar O, Bhattacharya R, Saxena P, Dash PK, Vijayaraghavan R. Mechanism of ricin-induced apoptosis in human cervical cancer cells. Biochem Pharmacol. 2005;69:855–865. doi: 10.1016/j.bcp.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Helguera G, Daniels TR, Neacato II, Lopez-Valdes HE, Charles AC, Penichet ML. Binding specificity and internalization properties of an antibody-avidin fusion protein targeting the human transferrin receptor. J Control Release. 2007;124:35–42. doi: 10.1016/j.jconrel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Rodriguez JA, Luria-Perez R, Lopez-Valdes HE, Casero D, Daniels TR, Patel S, Avila D, Leuchter R, So S, Ortiz-Sanchez E, Bonavida B, Martinez-Maza O, Charles AC, Pellegrini M, Helguera G, Penichet ML. Lethal iron deprivation induced by non-neutralizing antibodies targeting transferrin receptor 1 in malignant B cells. Leuk Lymphoma. 2011;52:2169–2178. doi: 10.3109/10428194.2011.596964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncuzzi L, Gasperi-Campani A. DNA-nuclease activity of the single-chain ribosome-inactivating proteins dianthin 30, saporin 6 and gelonin. FEBS Lett. 1996;392:16–20. doi: 10.1016/0014-5793(96)00776-4. [DOI] [PubMed] [Google Scholar]
- Santanche S, Bellelli A, Brunori M. The unusual stability of saporin, a candidate for the synthesis of immunotoxins. Biochem Biophys Res Commun. 1997;234:129–132. doi: 10.1006/bbrc.1997.6597. [DOI] [PubMed] [Google Scholar]
- Sikriwal D, Ghosh P, Batra JK. Ribosome inactivating protein saporin induces apoptosis through mitochondrial cascade, independent of translation inhibition. Int J Biochem Cell Biol. 2008;40:2880–2888. doi: 10.1016/j.biocel.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Skommer J, Wlodkowic D, Pelkonen J. Gene-expression profiling during curcumin-induced apoptosis reveals downregulation of CXCR4. Exp Hematol. 2007;35:84–95. doi: 10.1016/j.exphem.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Spooner RA, Smith DC, Easton AJ, Roberts LM, Lord JM. Retrograde transport pathways utilised by viruses and protein toxins. Virol J. 2006;3:26–35. doi: 10.1186/1743-422X-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thin TH, Li L, Chung TK, Sun H, Taneja R. Stra13 is induced by genotoxic stress and regulates ionizing-radiation-induced apoptosis. EMBO Rep. 2007;8:401–407. doi: 10.1038/sj.embor.7400912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyss R, Virolle V, Imbert V, Peyron JF, Aberdam D, Virolle T. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 2005;24:128–137. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vago R, Marsden CJ, Lord JM, Ippoliti R, Flavell DJ, Flavell SU, Ceriotti A, Fabbrini MS. Saporin and ricin A chain follow different intracellular routes to enter the cytosol of intoxicated cells. FEBS J. 2005;272:4983–4995. doi: 10.1111/j.1742-4658.2005.04908.x. [DOI] [PubMed] [Google Scholar]
- Vogel W, Kopp HG, Kanz L, Einsele H. Myeloma cell contamination of peripheral blood stem-cell grafts can predict the outcome in multiple myeloma patients after high-dose chemotherapy and autologous stem-cell transplantation. J Cancer Res Clin Oncol. 2005;131:214–218. doi: 10.1007/s00432-004-0635-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Peddibhotla S, Lin H, Fang X, Li M, Rosen JM, Zhang P. Early-onset aging and defective DNA damage response in Cdc14b-deficient mice. Mol Cell Biol. 2011;31:1470–1477. doi: 10.1128/MCB.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside ST, Epinat JC, Rice NR, Israel A. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Korcheva V, Jacoby DB, Magun BE. Proinflammatory responses of human airway cells to ricin involve stress-activated protein kinases and NF-kappaB. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1385–1394. doi: 10.1152/ajplung.00207.2007. [DOI] [PubMed] [Google Scholar]
- Yamada K, Miyamoto K. Basic helix-loop-helix transcription factors, BHLHB2 and BHLHB3; their gene expressions are regulated by multiple extracellular stimuli. Front Biosci. 2005;10:3151–3171. doi: 10.2741/1772. [DOI] [PubMed] [Google Scholar]
- Yoshioka J, Schreiter ER, Lee RT. Role of thioredoxin in cell growth through interactions with signaling molecules. Antioxid Redox Signal. 2006;8:2143–2151. doi: 10.1089/ars.2006.8.2143. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. Caspase-2 function in response to DNA damage. Biochem Biophys Res Commun. 2005;331:859–867. doi: 10.1016/j.bbrc.2005.03.191. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chng WJ. Roles of thioredoxin binding protein (TXNIP) in oxidative stress, apoptosis and cancer. Mitochondrion. 2012 doi: 10.1016/j.mito.2012.06.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Zumbrun SD, Hoffman B, Liebermann DA. Distinct mechanisms are utilized to induce stress sensor gadd45b by different stress stimuli. J Cell Biochem. 2009;108:1220–1231. doi: 10.1002/jcb.22354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A variance stabilizing transform was applied to the data to reduce the mean variance for each technical replicate, followed by a probe level quantile normalization within each array and between arrays (Lin et al. 2008). Shown is a graphical representation of normalized array data comparing two IM-9 (1674277005_A and 1674277005_B) and two U266 (1688628159_B and 1688628159_C) control array profiles. For each pairwise comparison, the log intensity ratio M (y axis) is plotted against the average log intensity A (x axis). Data normalization was conducted using the Lumi package from the Bioconductor Project (http://www.bioconductor.org). The top panel shows the data without normalization and the bottom panel after the analysis with the Lumi package.
The circle on the left lists all the genes that show a change in expression at 24 hours in IM-9 cells, while the circle on the right shows genes that change in U266 cells. The overlapping portion of the two circles shows a list of 11 genes for which the expression level changes in both cells lines.
Confocal microscopy images of cells incubated with Zenon® labeled ch128.1Av (red) alone or conjugated to b-SO6 for 1 or 16 hours. Saporin was detected using a goat anti-SO6 antibody labeled with Alexa Fluor® 488 (green). Nuclei are stained with DAPI (blue) within the mounting medium. The white bar indicates 25μm. Experiments were repeated at least twice.
Confocal microscopy images of cells incubated with Zenon® labeled ch128.1Av (red) alone or conjugated to b-SO6 for 1 or 16 hours. Saporin was detected using a goat anti-SO6 antibody labeled with Alexa Fluor® 488 (green). Nuclei are stained with DAPI (blue) within the mounting medium. The white bar indicates 25μm. Experiments were repeated at least twice.