SUMMARY
Complex genome organizations participate in various nuclear processes including transcription, DNA replication and repair. However, the mechanisms that generate and regulate these functional genome structures remain largely unknown. Here, we describe how the Ku heterodimer complex, which functions in nonhomologous end joining, mediates clustering of long terminal repeat (LTR) retrotransposons at centromeres in fission yeast. We demonstrate that the CENP-B subunit, Abp1, functions as a recruiter of the Ku complex, which in turn loads the genome-organizing machinery condensin to retrotransposons. Intriguingly, histone H3 Lys56 (H3K56) acetylation, which functions in DNA replication and repair, interferes with Ku localization at retrotransposons without disrupting Abp1 localization and, as a consequence, dissociates condensin from retrotransposons. This dissociation releases condensin-mediated genomic associations during S phase and upon DNA damage. ATR (ATM and Rad3-related) kinase mediates DNA damage-response of condensin-mediated genome organization. Our study describes a function of H3K56 acetylation that neutralizes condensin-mediated genome organization.
INTRODUCTION
The eukaryotic genome exists in the cell nucleus as an elaborate, three-dimensional structure. It is becoming clear that 3D genome organization is connected to various nuclear processes such as transcription, DNA replication, and repair (Cook, 1999; Misteli, 2007). Despite the accumulating evidence supporting the importance of genome structures in nuclear processes, the mechanisms responsible for organizing the 3D genome structure remain elusive. In particular, the role of epigenetic regulation in the folding of higher-order chromatin structures has not been explored.
The fission yeast, Schizosaccharomyces pombe, serves as an excellent model organism for studying global genome organization. For instance, centromeres and telomeres cluster at the nuclear periphery (Funabiki et al., 1993). RNA polymerase III (Pol III)-transcribed genes such as tRNA and 5S rRNA genes, dispersed throughout chromosomal arm regions, localize to centromeres, and this centromeric association of Pol III-transcribed genes is mediated by the condensin complex (Iwasaki et al., 2010). More recently, the ELP (Enrichment of Ligation Products) genomic approach combining next-generation sequencing and the molecular biology procedure called chromosome conformation capture (3C)—also referred to as Hi-C—revealed the existence of chromosomal territories and significant associations among highly transcribed genes, co-regulated genes, and functionally related genes, respectively (Lieberman-Aiden et al., 2009; Tanizawa et al., 2010). Moreover, LTR retrotransposons and their derived solo-LTRs tend to associate with one another in the nucleus (Cam et al., 2008). The fission yeast genome has been colonized by Tf1 and Tf2 retrotransposons, referred to as Tf elements in this article. The sequenced genome contains 13 full-length Tf2s and more than 200 solo-LTRs distributed throughout the fission yeast genome (Bowen et al., 2003). Therefore, associations among dispersed Tf elements should have a great impact on global genome organization. Clustering of Tf elements involves CENP-B proteins, Abp1, Cbh1, and Cbh2, which are also required for the formation of centromeric heterochromatin in fission yeast (Cam et al., 2008; Nakagawa et al., 2002). However, the molecular mechanisms driving associations among Tf elements and the involvement of epigenetic regulations in this genome organization remain unexplored.
The Ku heterodimer complex, consisting of Ku70 and Ku80, functions in nonhomologous end joining (NHEJ) (Daley et al., 2005). Ku also plays roles in various other cellular processes, including telomere maintenance, transcription, and apoptosis (Downs and Jackson, 2004). In addition, it was shown that Ku mediates clustering and tethering of telomeres to the nuclear periphery in budding yeast (Laroche et al., 1998). Ku functions redundantly with silent information regulatory 4 (Sir4) in clustering and tethering of telomeres to the nuclear periphery (Hediger et al., 2002; Taddei et al., 2004). This organization involves the two nuclear membrane proteins Esc1 and Mps3, of which Mps3 is the SUN domain-containing protein (Bupp et al., 2007; Schober et al., 2009; Taddei et al., 2004). Ectopic binding of Ku to non-telomeric regions relocates its associated chromatin to the nuclear periphery, thereby demonstrating the potent activity of Ku in tethering of genomic loci to the nuclear periphery (Schober et al., 2009; Taddei et al., 2004).
In this study, we begin with investigating the roles of Ku in various genome organizations in fission yeast. Our analyses reveal the involvement of Ku in both telomere tethering to the nuclear periphery and clustering of Tf elements at centromeres. We show that clustering of Tf elements at centromeres involves Ku, condensin, and the CENP-B factor, Abp1. Intriguingly, histone H3K56 acetylation interferes with the binding of Ku and condensin to Tf elements, thereby releasing condensin-mediated genome organization during S phase and upon DNA damage. Upon DNA damage, ATR kinase mediates the destruction of Hst4 HDAC specific to H3K56, leading to DNA damage-response of condensin-mediated genome organization through H3K56 acetylation. Interestingly, clustering of Tf elements at centromeres is seemingly concerned with the efficient destruction of Hst4 upon DNA damage. In addition, our study suggests that Ku localization becomes diffuse upon DNA damage through H3K56 acetylation, and this diffusion of Ku probably facilitates the NHEJ process. Finally, we show that H3K56 acetylation also participates in telomere tethering to the nuclear periphery, indicating a global role for this specific histone modification in genome organization.
RESULTS
Distribution of Ku across the fission yeast genome
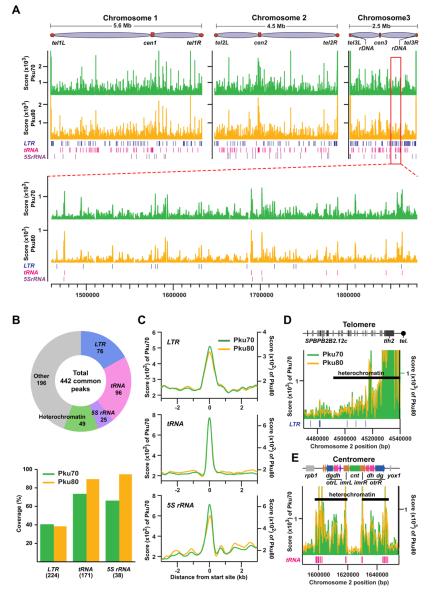
To understand the extent of Ku-mediated genome organizations, we first determined the distributions of Pku70 and Pku80 proteins throughout the fission yeast genome using the ChIP-seq approach (Figure 1A). Ku peaks were associated with genomic regions, including Pol III genes such as tRNA and 5S rRNA genes, LTRs of Tf retrotransposons, and heterochromatic loci, including telomeres and centromeres (Figures 1B-1E and S1A).
Figure 1. Genome-wide distribution of Ku binding.
(A) Chromosomal distributions of Pku70 (green) and Pku80 (yellow) were determined based on ChIP-seq analyses. ChIP was performed using strains carrying Pku70-Myc and Pku80-Myc proteins.
(B) Summary of PKu distributions across the fission yeast genome. Pie-chart shows the composition of Pku70/Pku80 common peaks (top). Coverages of LTR, tRNA, and 5S rRNA genes estimated by the presence of significant Pku binding peaks within 300 bp from the genes are shown in the bar graph (bottom). Each number in parenthesis indicates the total gene number.
(C) Average binding patterns of Pku proteins at LTR, tRNA, and 5S rRNA genes.
(D and E) Pku distributions at the right telomere (D) and centromere (E) of chromosome 2.
Immunofluorescence (IF) analysis indicated that both Pku70 and Pku80 proteins were not evenly diffuse in the nucleus, but rather were enriched at subnuclear domains often associated with the nuclear periphery (Figure S1B). Since Ku foci often locate at the nuclear periphery where heterochromatin is present, we investigated whether Ku is required for heterochromatin silencing at the heterochromatin loci such as centromeres, telomeres, and the mating-type region. Deletions of pku70 and pku80 had no effect on the silencing of the ura4+ marker genes inserted at the heterochromatic loci (Figure S1C). ChIP results further indicated that enrichment of Swi6, a homolog of heterochromatin protein 1 (HP1), and histone H3 Lys9 methylation at the heterochromatin loci were not affected in pku70Δ and pku80Δ cells (Figure S1D). Consistently, it has previously been shown that telomeric silencing is not affected by pku70Δ and pku80Δ in fission yeast, but is affected in budding yeast (Laroche et al., 1998; Manolis et al., 2001; Miyoshi et al., 2003). These data suggest that Ku might locate its associated genomic regions within the specific subnuclear domains, but is generally not required for heterochromatin silencing in fission yeast.
Involvement of Ku in telomere tethering to the nuclear periphery
Ku binds to telomeres and is often enriched at the nuclear periphery (Figures 1D and S1B; Miyoshi et al., 2003). It has been shown that telomeres are clustered and tethered to the nuclear periphery (Funabiki et al., 1993). We speculated that Ku might function in clustering and tethering of telomeres to the nuclear periphery in fission yeast. The FISH data revealed that telomere clustering was not affected by pku70Δ and pku80Δ, but telomere tethering to the nuclear periphery was significantly compromised by pkuΔ (P < 0.001, Mann-Whitney U test), suggesting that telomere clustering and tethering to the nuclear periphery are distinct processes (Figures S2A and S2B).
In budding yeast, telomere tethering involves Mps3, the integral, nuclear membrane protein bearing the Sad1-UNC-84 (SUN) domain (Bupp et al., 2007; Schober et al., 2009). The fission yeast homologue, Sad1, is known to interact with Bqt1 and mediates telomere tethering to the nuclear periphery in the horsetail nucleus during meiosis (Chikashige et al., 2006). Interestingly, telomere tethering was also significantly compromised by the sad1-1 mutation in vegetative cells (P < 0.001, Mann-Whitney U test), and double mutants carrying pkuΔ and sad1-1 mutations did not show a significant synergistic effect on telomere tethering (P > 0.05, Mann-Whitney U test; Figure S2B), suggesting that Ku and Sad1 function in the same pathway for telomere tethering to the nuclear periphery.
It has also been shown that Taz1, the fission yeast homologue of hTRF (human TTAGGG repeat factor), recruits Rap1 to telomeres, and Rap1 in turn associates with the inner membrane proteins Bqt3 and Bqt4, resulting in telomere tethering to the nuclear periphery in vegetative cells (Chikashige et al., 2009; Cooper et al., 1997; Kanoh and Ishikawa, 2001). Therefore, we examined whether Ku/Sad1 and Taz1/Rap1/Bqt3/Bqt4 pathways might cooperate to tether telomeres to the nuclear periphery. As previously shown, telomere tethering was significantly compromised in rap1Δ and bqt4Δ cells (Figure S2C; Chikashige et al., 2009). Moreover, telomere tethering to the nuclear periphery was exacerbated in double mutants carrying pkuΔ and either rap1Δ or bqt4Δ, compared to rap1Δ and bqt4Δ single mutants, although double mutants did not show the synergistic effect on telomere tethering compared to pkuΔ single mutants. These results suggest that the Ku/Sad1 and Taz1/Rap1/Bqt3/Bqt4 pathways mediate telomere tethering to the nuclear periphery, and that Ku seemingly participates in both pathways.
To explore the roles of Ku in the centromeric localization of Pol III genes, we performed a FISH analysis and visualized centromeres and the c417 Pol III gene locus, which consists of three tRNA and two 5S rRNA genes. We observed that the association frequencies between the Pol III gene locus and centromeres in pku70Δ and pku80Δ cells were similar to that in wild-type cells (Figure S2D). Moreover, deletions in pku70 and pku80 did not affect transcript levels of Pol III genes (Figure S2E). These data suggest that Ku is not involved in the centromeric localization of Pol III genes and their transcription.
Ku mediates clustering of Tf retrotransposons
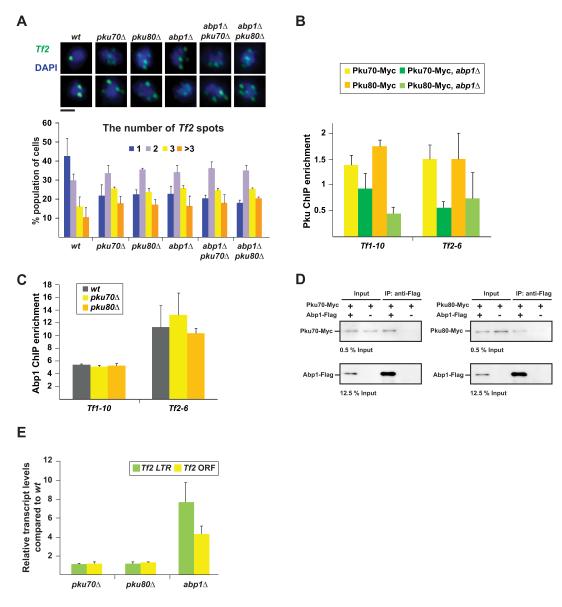
It was previously shown that Tf retrotransposons cluster into Tf bodies, referred to as Tf clustering in this article (Cam et al., 2008). Since Ku binds to Tf elements, we examined the role of Ku in Tf clustering. Interestingly, Tf clustering was impaired by pku70Δ and pku80Δ at a level similar to that observed in abp1Δ cells (Figure 2A). Double mutants carrying pkuΔ and abp1Δ did not show a cumulative effect on Tf clustering compared to single mutants. The ChIP analysis revealed that enrichment of Ku at Tf elements was decreased in abp1Δ cells, whereas Abp1 binding to Tf elements was not affected by pkuΔ (Figures 2B and 2C). Moreover, the co-immunoprecipitation (co-IP) assay showed that Ku associates with Abp1, and the significant association between Pku70 and Abp1 remained after DNase I treatment (Figures 2D, S2F, and S2G). These results support the hypothesis that Ku is recruited to Tf elements by Abp1 and mediates Tf clustering.
Figure 2. Ku mediates retrotransposon clustering.
(A) FISH analysis visualizing Tf2 elements (green). Typical microscopic images in the indicated strains are shown on top. Black bar indicates 1 μm. Number of Tf2 dots in the nucleus is summarized at the bottom.
(B) ChIP results showing Pku enrichment at Tf elements in wt and abp1Δ cells.
(C) ChIP results showing enrichment of Abp1-Pk at Tf elements in wt and pkuΔ cells.
(D) Co-immunoprecipitation results showing the interaction between Pku-Myc and Abp1-Flag.
(E) RT-PCR results indicating the transcript levels of Tf2 LTR and ORF regions in pkuΔ and abp1Δ cells. The transcript levels in mutants were normalized by those in wt cells. In panels A, B, C, and E, data are represented as mean +/− SD.
It has been shown that abp1Δ derepresses Tf elements, because Abp1 recruits Clr3 and Clr6 histone deacetylases (HDACs) to Tf elements for repression of Tf elements (Cam et al., 2008). In contrast, pkuΔ did not derepress Tf elements, although it did compromise Tf clustering, indicating that Tf clustering is independent of the repression of Tf elements (Figure 2E).
Clustering of Tf retrotransposons at centromeres
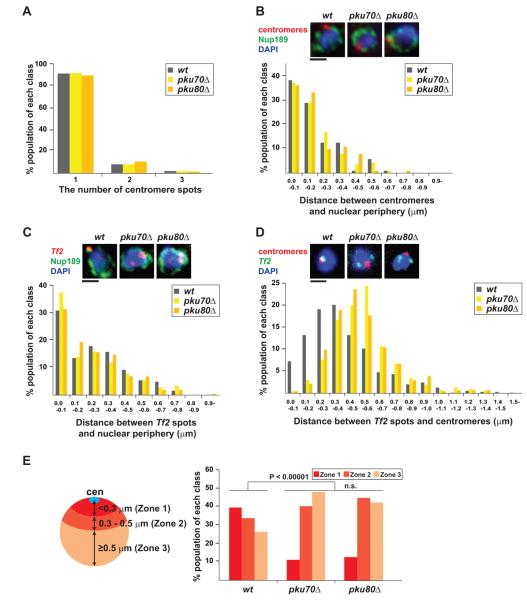
We next investigated where Tf elements cluster. Since Ku often localizes at the nuclear periphery, we speculated that Tf elements might co-localize with centromeres, also present at the nuclear periphery. Before addressing this possibility, we first examined whether Ku plays a role in clustering and tethering of centromeres to the nuclear periphery. We found that clustering and tethering of centromeres to the nuclear periphery were not affected in pku70Δ and pku80Δ cells, although Ku does localize at centromeres (Figures 1E, 3A, and 3B). We then investigated the positioning of Tf clustering in relation to the nuclear periphery and found that Tf clustering is frequently localized at the nuclear periphery (Figure 3C). However, tethering of Tf cluster to the nuclear periphery was not affected by pkuΔ, indicating that Ku is not involved in the association of Tf elements with the nuclear periphery. Remarkably, the association of Tf elements with centromeres was significantly compromised in pkuΔ cells (P < 0.00001, Mann-Whitney U test; Figures 3D and 3E). These data suggest that Ku mediates Tf clustering and the preferential association between Tf cluster and centromeres.
Figure 3. Clustering of retrotransposons at centromeres.
(A) Centromere clustering is not affected by pkuΔ. Centromeres were visualized by FISH in the indicated strains. Number of centromeric foci in the nucleus is summarized in a graph.
(B) Centromere anchoring to the nuclear periphery is not compromised in pkuΔ cells. Centromeres (red) and nuclear pore complex (NPC; green) were visualized by FISH and IF using anti-Nup189 antibody, respectively (top). Distance between centromeric signal and its nearby NPC focus was measured and summarized in a graph.
(C) FISH/IF analysis visualizing Tf2 elements (red) and the NPC subunit, Nup189 (green). Distance between Tf2 dot and its nearest NPC was measured.
(D) FISH analysis visualizing centromeres (red) and Tf2 elements (green). Distance between centromeric and Tf2 foci was measured and summarized in a graph.
(E) Distance from centromeres was divided into three zones based on the criteria depicted on the left. Distance between centromeres and Tf2 dot from FISH result shown in (D) was binned into one of the assigned zones. n.s. indicates P > 0.05. Black bar indicates 1 μm.
Moreover, Tf clustering at centromeres was not affected in sad1-1 cells (Figure S3). Even though Ku and Sad1 function in telomere tethering to the nuclear periphery, likely in the same pathway, they do not cooperate for Tf clustering at centromeres. Tf clustering at centromeres was also not affected by the rap1Δ and bqt4Δ mutations, which compromise telomere tethering to the nuclear periphery (Figures S2C and S3). These suggest that the two genome organizations involving telomeres and Tf elements are independent. It was previously shown that both Ku and the DNA ligase IV, Lig4, are required for NHEJ in fission yeast (Baumann and Cech, 2000; Manolis et al., 2001; Miyoshi et al., 2003). We observed that Tf clustering at centromeres was not affected by lig4Δ (Figure S3). Therefore, Ku, but not Lig4, is required for Tf clustering at centromeres, while both are involved in the NHEJ process.
Condensin participates in Tf clustering at centromeres
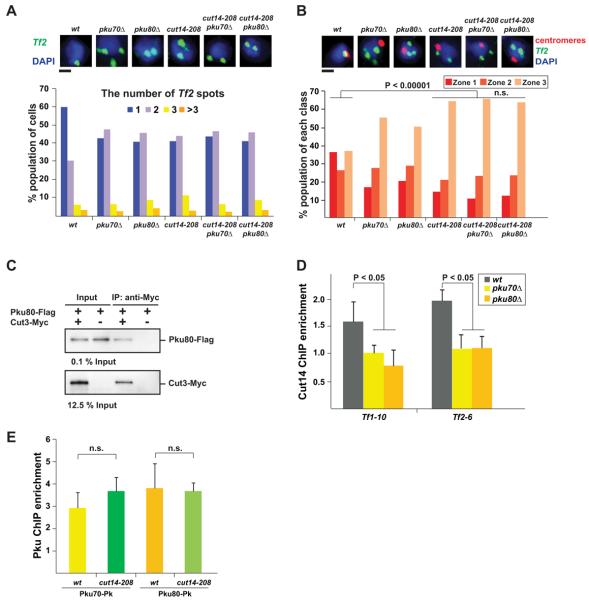
Next, we sought to identify the genome-organizing machinery responsible for Tf clustering at centromeres. In this regard, we have previously shown that the centromeric localization of Pol III genes is mediated by condensin (Iwasaki et al., 2010). We thus speculated that Tf clustering at centromeres might also be mediated by condensin. We found that Tf clustering and the association of Tf cluster with centromeres were significantly compromised in the cut14-208 condensin mutant (P < 0.00001, Mann-Whitney U test; Figures 4A and 4B), while they were not affected in rad21-K1 cohesin mutant (Figure S4), indicating that condensin, but not cohesin, participates in Tf clustering at centromeres. Double mutants carrying pkuΔ and cut14-208 did not show synergistic effect on Tf clustering at centromeres compared to cut14-208 single mutant. Moreover, the co-IP data showed that the condensin subunit Cut3 interacts with Pku80 (Figure 4C). The ChIP analysis revealed that enrichment of the Cut14 condensin subunit at the Tf elements was decreased in kuΔ cells, whereas Pku binding was not affected in cut14-208 condensin mutant (Figures 4D and 4E).
Figure 4. Tf clustering at centromeres is compromised in condensin mutant.
(A) The condensin mutation, cut14-208, impairs Tf clustering. FISH analysis was performed as described in Figure 2A. Black bar indicates 1 μm.
(B) Centromeric localization of Tf cluster is compromised in condensin mutant cells. The experiment was carried out as described in Figures 3D and 3E.
(C) Co-IP analysis investigating an interaction between Pku80-Flag and Cut3-Myc.
(D) ChIP result showing enrichment of the condensin subunit Cut14-Pk at Tf elements in wt and pkuΔ cells.
(E) ChIP result showing enrichment of Pku70-Pk and Pku80-Pk at Tf1-10 in wt and cut14-208 condensin mutant cells.
Data are represented as mean +/− SD.
n.s. indicates P > 0.05.
The ChIP-seq result indicated that Cut14 condensin peaks were associated with genomic regions, including LTRs of Tf retrotransposons, Pol III genes such as tRNA and 5S rRNA genes, and centromeres (Figures S5A-S5C). Importantly, binding peaks of Ku and condensin around LTRs were located at the same position and the co-localization frequency of Ku and condensin peaks was highly significant (Figures S5D-S5F). All together, these results suggest that Ku recruits condensin to Tf elements. It was previously shown that condensin mediates numerous interactions between DNA duplexes residing within a chromosome (Hirano, 2006). Therefore, our current hypothesis is that condensin, recruited by Ku, mediates Tf clustering at centromeres (See Discussion).
Genomic associations involving Tf retrotransposons
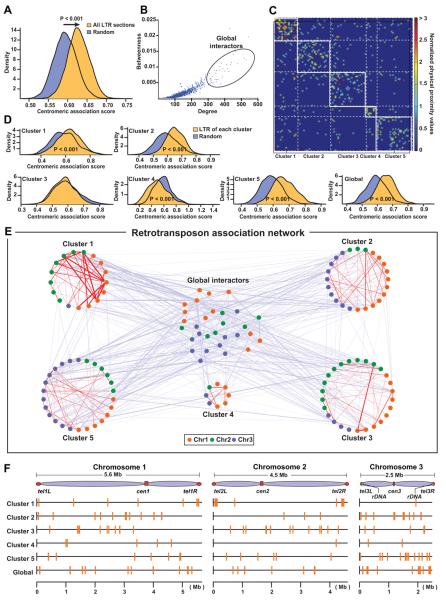
In order to better understand how centromeres and dispersed Tf elements contribute to the organization of the entire genome, we analyzed the ELP genomic data, which was generated by our approach similar to the Hi-C, combining next-generation sequencing and 3C (Tanizawa et al., 2010). Our data identified significant associations between genomic regions containing LTRs and centromeres (P < 0.001, Student’s t-test; Figure 5A). We next examined whether specific LTRs tend to cluster. Out of 143 LTR-containing genomic sections, we observed that some sections exhibited characteristics of global interactors, which tend to associate with many other sections (Figure 5B). In order to identify association clusters among Tf elements, we excluded global interactors, carried out a network clustering analysis (Newman and Girvan, 2004; Ruan and Zhang, 2008), and identified 5 clusters (Figure 5C). Importantly, the LTR-containing genomic sections classified in clusters 1, 2, and 5 significantly associate with centromeres, while the genomic sections from cluster 4 tend not to (Figure 5D). In addition, we visualized the entire association network involving the LTR-containing genomic sections (Figure 5E). Interestingly, we observed that genomic sections in clusters 1, 2, and 3 are mainly derived from two out of three chromosomes, indicating that clustering associations among Tf elements likely occur at junctions between chromosomal territories (Scherthan et al., 1994). For instance, cluster 2 contains 11 and 10 genomic sections from chromosomes 1 and 3, respectively, whereas only three sections come from chromosome 2. LTR-containing sections in respective clusters are dispersed throughout the genome except for cluster 1, which includes more sections derived from subtelomeric regions compared to other clusters (Figure 5F). Moreover, the ChIP-seq results revealed that average enrichments of Ku and condensin at the LTR-containing genomic sections in different clusters were similar, suggesting that frequent associations among specific Tf elements in the respective clusters are probably caused by other factors than the binding strength of Ku and condensin to LTRs, while Ku and condensin appear to be required for Tf clustering at centromeres (Figures S5G and S5H). Together, these genomic data reveal that Tf elements frequently cluster at centromeres and the composition of Tf elements participating in the respective clusters is probably concerned with chromosomal territories.
Figure 5. Genomic analyses on LTR clustering.
(A) LTR-containing genomic sections significantly associate with centromeres. Distributions for average centromeric association scores for both LTR-containing sections and random control were generated by repeated sampling. We carried out Student’s t-test and observed a significant difference between the two distributions.
(B) LTR-containing genomic sections with both high local and global connectivity measures, degree and betweenness, respectively, were designated as potential global interactors and excluded from subsequent clustering analyses.
(C) Heat map representation of the final clustering results. Total of five significant clusters were identified at a modularity level of 0.4. Normalized physical proximity values reflect association strength between genomic sections (Tanizawa et al. 2010).
(D) Significance of associations between centromeres and genomic sections in each cluster or global interactors were calculated using the same procedure exploited in (A).
(E) Graphical representation of the final clustering results. Each colored dot represents an LTR-containing genomic section. Each edge indicates significant association, and its thickness correlates with association strength.
(F) Genomic locations of LTR-containing genomic sections in the different clusters. Method details are described in Supplemental Experimental Procedures.
Histone H3K56 acetylation antagonizes Tf clustering at centromeres
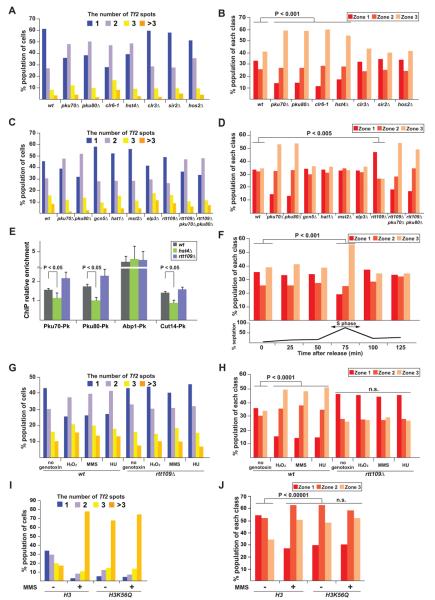
We next explored whether epigenetic regulations are involved in Tf clustering at centromeres. We investigated Tf clustering at centromeres in deletions of genes encoding HDACs and acetyltransferases (HATs). Interestingly, the hst4Δ and clr6-1 HDAC mutations, but not other HDAC mutations, significantly compromised Tf clustering and the association of Tf cluster with centromeres (P < 0.001, Mann-Whitney U test; Figures 6A and 6B). Moreover, only the rtt109Δ HAT mutations, but not other HAT mutations, significantly promoted the association of Tf cluster with centromeres (P < 0.005, Mann-Whitney U test), whereas none of the HAT mutations affected Tf clustering (Figures 6C and 6D). It has been shown that lysine 56 (K56) of histone H3 is specifically acetylated by Rtt109 and deacetylated by Hst4 in fission yeast, while Clr6 HDAC has broad substrate specificity (Haldar and Kamakaka, 2008; Xhemalce and Kouzarides, 2010; Xhemalce et al., 2007). Namely, the rtt109Δ results in the absence of H3K56 acetylation, while the hst4Δ causes H3K56 hyper-acetylation. Therefore, these results suggest that H3K56 acetylation interferes with Tf clustering at centromeres.
Figure 6. Regulation of Tf clustering at centromeres by H3K56 acetylation.
(A) The histone deacetylase mutations, clr6-1 and hst4Δ, impair Tf clustering. FISH analysis was performed as described in Figure 2A. The five HDAC mutants, clr6-1, hst4Δ, clr3Δ, sir2Δ, and hos2Δ, were subjected to the experiment.
(B) The clr6-1 and hst4Δ HDAC mutations affect centromeric localization of Tf elements. The analyses were carried out as described in Figures 3D and 3E using the indicated strains.
(C) The histone acetyltransferase mutations do not affect Tf clustering. The five HAT mutants, gcn5Δ, hat1Δ, mst2Δ, elp3Δ, and rtt109Δ, were subjected to the experiment.
(D) The HAT mutation, rtt109Δ, enhances centromeric localization of Tf elements.
(E) ChIP results showing enrichment of PKu-Pk, Abp1-Pk, and Cut14-Pk at Tf1-10 in wt, hst4Δ and rtt109Δ cells. Data are represented as mean +/− SD.
(F) Centromeric localization of Tf elements during the cell cycle. The cdc25-22 mutation was used for the cell-cycle synchronization. The septation index is shown at the bottom and the septation peak roughly coincides with S phase. FISH experiment was performed using cells in different stages of the cell cycle.
(G and H) Tf clustering at centromeres is compromised upon DNA damage. Wild-type and rtt109Δ cells were cultured in a nutrient-rich (YEA) medium containing 0.5 mM H2O2, 0.08% methyl methanesulfonate (MMS), and 200 mM hydroxyurea (HU). Tf clustering (G) and its centromeric localization (H) were investigated by FISH. (I and J) Histone H3K56Q mutation impairs Tf clustering at centromeres. Tf clustering (I) and its centromeric localization (J) were investigated by FISH. n.s. indicates P > 0.05.
We examined how H3K56 acetylation antagonizes Tf clustering at centromeres. Remarkably, binding of Ku was reduced and enhanced in hst4Δ and rtt109Δ cells, respectively, suggesting that H3K56 acetylation has an inhibitory effect on Ku binding to Tf elements (Figure 6E). Condensin enrichment was also affected in the mutants, probably because condensin is recruited to Tf elements through Ku (Figures 4C-4E). Interestingly, localization of Abp1, a recruiter of Ku to Tf elements, was not affected in the mutants. Therefore, recruitment of Ku to Tf elements entails not only Abp1, but also deacetylation of H3K56. These results suggest that H3K56 acetylation releases Tf clustering at centromeres through inhibition of Ku binding to Tf elements. Moreover, the co-IP result showed that Hst4 associates with Abp1, and the ChIP-chip analysis previously revealed that Hst4 is enriched at Tf elements (Figure S6A; Durand-Dubief et al., 2007). Therefore, it is likely that Hst4 is preferentially recruited to Tf elements through Abp1 and maintains H3K56 hypo-acetylation around Tf elements, which can promote localization of Ku and condensin at Tf elements, thereby leading to Tf clustering at centromeres.
We also examined whether other genome organizations might be regulated by H3K56 acetylation. We found that telomere tethering to the nuclear periphery, but not telomere clustering, was impaired in rtt109Δ cells (Figures S6B and S6C). Moreover, Ku binding to the telomere was not affected by rtt109Δ and hst4Δ, suggesting that H3K56 acetylation releases Ku from Tf elements, but not from telomeres (Figure S6D). These results suggest that H3K56 acetylation is required for telomere tethering to the nuclear periphery, and two genome organizations involving telomeres and Tf elements are regulated by the distinct mechanisms. It has been shown that telomere tethering to the nuclear periphery involves H3K56 acetylation in budding yeast, suggesting that the role of H3K56 acetylation in telomere tethering might be conserved among eukaryotes (Hiraga et al., 2008). We also observed that centromere clustering and tethering to the nuclear periphery, as well as centromeric localization of Pol III-transcribed genes, were not affected in hst4Δ and rtt109Δ cells (Figures S6E-S6G).
Modulation of Tf clustering during the cell cycle and upon DNA damage
It has been shown that H3K56 acetylation is induced by Rtt109 during S phase in fission yeast and functions in DNA replication (Li et al., 2008; Ransom et al., 2010; Xhemalce et al., 2007). If this histone modification disrupts Tf clustering at centromeres, Tf clustering should be disrupted during S phase. Indeed, the association of Tf cluster with centromeres was temporarily compromised during S phase (Figure 6F). This suggests that the acetylation status of H3K56 governs Tf clustering at centromeres in a cell cycle-specific manner.
Since H3K56 acetylation is also involved in DNA repair (Chen et al., 2008; Driscoll et al., 2007; Masumoto et al., 2005), Tf clustering at centromeres might be compromised upon DNA damage. We observed that treatments of fission yeast cells with genotoxic agents resulted in the disruption of Tf clustering at centromeres (Figures 6G and 6H). In contrast, the genotoxic treatments did not affect clustering of Tf elements at centromeres in rtt109Δ cells. We also investigated Tf clustering at centromeres in histone H3K56Q mutant. This mutation mimics acetylated lysine. Tf clustering at centromeres was compromised by the mutation regardless of MMS treatment (Figures 6I and 6J). These results collectively support that Tf clustering at centromeres is disassembled upon DNA damage through H3K56 acetylation.
ATR mediates disassembly of Tf clustering at centromeres
ATR (ATM and Rad3-related) kinase functions as one of the master regulators in DNA-damage response (Ciccia and Elledge, 2010; Cimprich and Cortez, 2008). We found that Tf clustering at centromeres was not affected by the MMS treatment in rad3Δ ATR mutant, indicating that DNA damage-response of the Tf element-mediated genome organization requires the ATR kinase (Figures 7A and 7B). The amount of Hst4 was decreased upon DNA damage in wild-type cells but not in rad3Δ cells, while Rtt109 protein level was not affected by the MMS treatment (Figures 7C and S7A). It has been shown that the Hst4 homologues in budding yeast are targeted by ubiquitin-mediated proteolysis upon DNA damage, and this protein degradation is triggered by the ATR kinase (Thaminy et al., 2007). Therefore, the fission yeast Hst4 is presumably destructed in a similar manner upon DNA damage. H3K56 acetylation induced by the Hst4 degradation upon DNA damage can lead to disassembly of Tf clustering at centromeres. This explains why DNA damage induces disassembly of Tf clustering at centromeres only in the presence of the ATR kinase. Interestingly, the Hst4 degradation upon the MMS treatment was compromised in pku70Δ, pku80Δ, and abp1Δ cells, in which Tf clustering at centromeres is diminished (Figure 7C). It is possible that Tf clustering at centromeres is implicated in the efficient destruction of Hst4 upon DNA damage.
Figure 7. Disruption of Tf clustering at centromeres through ATR.
(A) Tf clustering is not impaired upon DNA damage in rad3Δ cells. The MMS treatment was performed as described in Figure 6G. FISH analysis was performed as described in Figure 2A.
(B) Centromeric localization of Tf elements is not affected by the MMS treatment in rad3Δ cells. The analyses were carried out as described in Figures 3D and 3E.
(C) The MMS treatment reduces the amount of Hst4 in wt cells, but not in rad3Δ, pku70Δ, pku80Δ, and abp1Δ cells. Western blot analysis was performed to detect Hst4-Pk and tubulin using anti-Pk and TAT-1 antibodies, respectively.
(D) IF result showing Pku70-Pk localization (green) with and without MMS treatment in wt and rtt109Δ cells. Pku70-Pk staining area on a focal point, which reflects Pku70 occupancy in the nucleus, was measured and the derived data were classified into two groups, below 0.6 μm2 and more than 0.6 μm2. Typical images are shown on top.
(E) Nonhomologous end joining assay. Efficiencies of NHEJ in wt, pku70Δ, pku80Δ, rad3Δ, rtt109Δ, and hst4Δ cells were measured by a plasmid-based assay. Data are represented as mean +/− SD.
(F) A schematic model for retrotransposon-mediated genome organization and its regulatory mechanism through histone H3K56 acetylation. The CENP-B subunit Abp1 binds to Tf elements and recruits Ku, which in turn loads the genome-organizing machinery condensin onto chromatin. Condensin associating with Tf elements and centromeres mediates Tf clustering at centromeres. H3K56 acetylation by Rtt109 releases Ku and condensin from Tf elements without disrupting Abp1 binding, thereby disassembling this genome organization during S phase of the cell cycle and upon DNA damage. Once H3K56 is deacetylated by Hst4, stable interaction of the recruiter Abp1 with Tf elements would help efficiently reestablish condensin-mediated genome organization.
In panels B, D, and E, n.s. indicates P > 0.05.
Diffusion of Ku upon DNA damage
We observed that Ku localization was diffuse after DNA damage, but this diffusion was inhibited by rtt109Δ (Figure 7D). This result indicates that Ku mainly localizes within the subnuclear domains under normal conditions, but it relocates to distribute across the nucleoplasm upon DNA damage through H3K56 acetylation. The MMS treatment did not affect Pku80 protein levels in wt and rtt109Δ cells, while it slightly increased Pku70 (Figure S7B). This small increase in Pku70 was independent of the rtt109Δ mutation. Therefore, the expression level of Ku cannot account for their diffusion.
We hypothesized that the diffusion of Ku helps improve the efficiency of NHEJ in the repair of DNA double-strand breaks dispersed across the genome. We observed that the efficiency of NHEJ was significantly reduced and enhanced in rtt109Δ and hst4Δ cells, respectively (Figure 7E). This result suggests that H3K56 acetylation contributes to efficient NHEJ, probably through the Ku diffusion. In addition, NHEJ efficiency was reduced in rad3Δ cells, suggesting that the ATR kinase is required for efficient NHEJ (Figure 7E).
It has previously been shown that CENP-B promotes replication-fork progression through LTR (Zaratiegui et al., 2011). We observed that replication pausing was not affected by the pkuΔ, even though the plasmid containing LTR tends to associate with centromeres in wild-type cells and the association was significantly impaired in pkuΔ cells (Figures S7C and S7D). Moreover, the plasmid without LTR did not associate with centromeres, suggesting that LTR in the plasmid mediates its association with centromeres through Ku. These results suggest that Ku binding to LTRs or its resultant genome organization is not related to replication pausing at LTRs. In addition, frequencies of Tf1 transposition, as well as homologous recombination between Tf1 cDNA and genomic Tf elements, were not significantly affected by the pkuΔ in fission yeast (Table S1).
DISCUSSION
We have characterized the mechanisms that generate and regulate clustering of Tf retrotransposons at centromeres (Figure 7F). This study unveils a mechanism in which Abp1 recruits Ku to Tf elements. Moreover, Tf clustering at centromeres requires condensin, which is recruited to Tf elements through Ku. We have previously shown that Pol III-transcribed genes such as tRNA and 5S rRNA genes, dispersed throughout the genome, associate with centromeres, and this genome organization is also mediated by condensin in fission yeast (Iwasaki et al., 2010). Condensin mediates associations between two DNA duplexes through interactions with other condensin complexes (Hirano, 2006). Therefore, condensin likely functions in tethering dispersed genomic regions to centromeres, but factors recruiting condensin to genomic regions are dependent upon respective genetic elements. For instance, Pol III transcription machinery recruits condensin to Pol III genes in fission yeast (Iwasaki et al., 2010). On the other hand, Ku loads condensin to Tf elements. Condensin is also targeted to the kinetochore portions of centromeres through a mechanism involving kinetochore proteins (Nakazawa et al., 2008; Tada et al., 2011). Relatively more condensin molecules localize at centromeres than other genomic loci, which might explain why dispersed retrotransposons associated with condensin are frequently tethered to centromeres.
Our study reveals that H3K56 acetylation has an inhibitory effect on Tf clustering at centromeres. Tf clustering at centromeres is likely disassembled during the global deposition of acetylated histone H3 at lysine 56 during DNA replication, although it is also possible that H3K56 acetylation or acetylated histones are preferentially targeted to Tf elements. H3K56 acetylation interferes with Ku binding to chromatin, but does not affect Abp1 binding to Tf elements. Therefore, Abp1 provides sequence specificity for Ku binding, but the association of Ku with chromatin is restricted by H3K56 acetylation. H3K56 acetylation might interfere with the interaction between Ku and Abp1. Once Ku binding to chromatin is inhibited, condensin is also delocalized, causing disassembly of Tf clustering at centromeres. To re-assemble Tf clustering at centromeres after DNA replication and repair, stable binding of Abp1 likely facilitates the recruitment of Ku, condensin, and Hst4 to Tf elements. Once H3K56 is deacetylated around Tf elements by Hst4, Ku and condensin are efficiently loaded onto Tf elements through Abp1 and rebuild Tf clustering at centromeres (Figure 7F).
We find that the ATR kinase is required for disassembly of Tf clustering at centromeres upon DNA damage, and this DNA damage-response of the genome organization is mediated by the destruction of Hst4 HDAC. H3K56 acetylation through the destruction of Hst4 upon DNA damage can disassemble Tf clustering at centromeres due to the dissociation of Ku and condensin from Tf elements. In addition, the destruction of Hst4 upon DNA damage is compromised in pkuΔ and abp1Δ cells, where condensin-mediated genome organization is diminished. It has been shown that Hst4 is preferentially recruited to Tf elements, probably through Abp1 (Durand-Dubief et al., 2007; Figure S6A). Therefore, our current hypothesis is that Tf clustering at centromeres contributes to enrich Hst4 molecules in the subnuclear domain, and a concentration of Hst4 molecules in physical proximity might facilitate their efficient destruction mediated by the ATR kinase.
Ku diffusion occurs upon DNA damage and requires H3K56 acetylation (Figure 7D). This Ku diffusion also occurs in budding yeast (Martin et al., 1999). Upon DNA damage, Hst4 is down-regulated through the ATR kinase, leading to H3K56 acetylation. H3K56 acetylation dissociates Ku from retrotransposons, which at least partially accounts for the Ku diffusion (Figures 6E and 7D). We also find that NHEJ efficiency is enhanced and reduced in hst4Δ and rtt109Δ cells, respectively. It is important to note that NHEJ still occurs at reduced efficiency in rtt109Δ cells, in which H3K56 acetylation is abolished, indicating that H3K56 acetylation facilitates NHEJ but is not essential. We hypothesize that Ku diffusion helps efficiently detect DNA breakages dispersed throughout the fission yeast genome. To support this hypothesis, Ku is known to directly recognize DNA breakage (Martin et al., 1999).
How epigenetic mechanisms govern genomic associations remains largely uncharacterized. As a sole example, cohesin is recruited to CTCF binding sites through its interaction with CTCF and mediates associations among genes and their regulatory elements (Hadjur et al., 2009; Hou et al., 2010; Nativio et al., 2009). At the mouse H19/Igf2 locus, CTCF binding to chromatin is inhibited by DNA methylation at the imprinting control region, resulting in allele-specific conformation of this locus (Hark et al., 2000; Kurukuti et al., 2006; Nativio et al., 2009). Therefore, studies in our and other laboratories demonstrate that the structural maintenance of chromosomes (SMC) complexes such as condensin and cohesin, which are essential for mitotic chromosome condensation and sister chromatid cohesin, respectively, both function in global genome organization during interphase. More importantly, condensin and cohesin are recruited by DNA-associating factors such as Ku and CTCF, respectively, and binding of these factors to chromatin is governed by epigenetic regulations such as H3K56 acetylation and DNA methylation. Therefore, a general concept that emerges is that epigenetic mechanisms control global genome organization through changing affinity of the SMC recruiters to chromatin.
EXPERIMENTAL PROCEDURES
Strain construction and culture conditions
Pku70, Pku80, Abp1, Cut14, Hst4, and Rtt109 proteins were tagged with Myc, Flag, or Pk at the C-termini of their proteins, and pku70, pku80, lig4, and bqt4 genes were deleted using a PCR-based module method (Bähler et al., 1998). Strains carrying abp1Δ, clr3Δ, sir2Δ, rtt109Δ, gcn5Δ, hat1Δ, mst2Δ, elp3Δ, hos2Δ, rap1Δ, and rad3Δ were obtained from S. pombe Haploid Deletion Mutant Set ver. 2.0 (Bioneer). Strain constructions were performed using conventional genetic crosses. Yeast cells were cultured in yeast-extract adenine (YEA) medium at 30°C, unless otherwise noted.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (Iwasaki et al., 2010), with slight modification. Chromatin was sheared by Bioruptor (Diagenode). Tagged proteins were purified by rabbit polyclonal anti-Myc (Novus Biologicals) or mouse monoclonal anti-Pk (Serotec), and protein G coupled Dynabeads (Invitrogen).
Immunoprecipitation
The immunoprecipitation (IP) experiment was carried out as previously described (Iwasaki et al., 2010).
Nonhomologous end joining assay
Procedure for the NHEJ assay in fission yeast was previously established (Manolis et al., 2001; Miyoshi et al., 2003). Plasmid pAL19 was digested by PvuII-HF (New England Biolabs). Logarithmically growing cells were transformed with 1 μg of linear or circular pAL19 using electroporation. Transformed cells were incubated on AA-leu+1/10 YEA plates at 30°C. NHEJ efficiency was calculated as the percentage of the number of LEU2 colonies arising from cells transformed with the linear plasmid divided by that of colonies transformed with the undigested plasmid, and normalized by the average value in wild type.
Supplementary Material
HIGHLIGHTS.
Retrotransposon clustering is mediated by Ku, condensin, and CENP-B.
Histone H3K56Ac regulates localization of Ku and condensin at retrotransposons.
Retrotransposon clustering is released during S phase and upon DNA damage.
ATR mediates disassembly of retrotransposon clustering upon DNA damage.
ACKNOWLEDGEMENTS
We would like to thank Joel Huberman for helpful discussion; Robin Allshire and the Yeast Genetic Resource Center (YGRC) for fission yeast strains; Antony Carr, Robert Martienssen, and Mikel Zaratiegui for plasmids; Osami Niwa for an antibody; the Wistar Institute Imaging Facility for microscopic analysis; and the Wistar Institute Genomics and Bioinformatics Facilities for high-throughput sequencing and genomic data analyses. We also thank Gerd Blobel and Louise Showe for critically reading the manuscripts and Mea Fuller for editorial assistance. This work was supported by the National Institutes of Health grant CA010815, the NIH Director’s New Innovator Award Program DP2-OD004348, the G. Harold & Leila Y. Mathers Foundation, the V Foundation, the Edward Mallinckrodt Jr. Foundation, and the Wistar Pilot Project Funds to K.N. This work was also supported by NIH grant GM077604 to E.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, Supplemental Experimental Procedures, and Supplemental References.
ACCESSION NUMBERS
Data described in this paper were deposited into the NCBI SRA database with accession number SRP015264.
REFERENCES
- Bähler J, Wu J, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 2009;187:413–427. doi: 10.1083/jcb.200902122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu. Rev. Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Sinha I, Fagerstrom-Billai F, Bonilla C, Wright A, Grunstein M, Ekwall K. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J. 2007;26:2477–2488. doi: 10.1038/sj.emboj.7601690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar D, Kamakaka RT. Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryotic Cell. 2008;7:800–813. doi: 10.1128/EC.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Botsios S, Donaldson AD. Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. J. Cell Biol. 2008;183:641–651. doi: 10.1083/jcb.200806065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Sadaie M, Kanoh J, Ishikawa F. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J. Biol. Chem. 2003;278:1924–1931. doi: 10.1074/jbc.M208813200. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, Grewal SI, Tanaka K, Murakami Y. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters J, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Physical Review E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Zhang W. Identifying network communities with a high resolution. Physical Review E. 2008;77:016104. doi: 10.1103/PhysRevE.77.016104. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Bahler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J. Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaminy S, Newcomb B, Kim J, Gatbonton T, Foss E, Simon J, Bedalov A. Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J Biol Chem. 2007;282:37805–37814. doi: 10.1074/jbc.M706384200. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce B, Miller KM, Driscoll R, Masumoto H, Jackson SP, Kouzarides T, Verreault A, Arcangioli B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bähler J, Arcangioli B, Martienssen RA. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.