Abstract
Recent discovery of biological function of endothelial cell-specific chemotaxic regulator (ECSCR), previously known as endothelial cell-specific molecule 2 (ECSM2), in modulating endothelial cell migration, apoptosis, and angiogenesis, has made it an attractive molecule in vascular research. Thus, identification of splice variants of ECSCR could provide new strategies for better understanding its roles in health and disease. In this study, we performed a series of blast searches on the human EST database with known ECSCR cDNA sequence (Variant 1), and identified additional three splice variants (Variants 2–4). When examining the ECSCR gene in the human genome assemblies, we found a large unknown region between Exons 9 and 11. By PCR amplification and sequencing, we partially mapped Exon 10 within this previously unknown region of the ECSCR gene. Taken together, in addition to previously reported human ECSCR, we identified three novel full-length splice variants potentially encoding different protein isoforms. We further defined a total of twelve exons and nearly all exon-intron boundaries of the gene, of which only eight are annotated in current public databases. Our work provides new information on gene structure and alternative splicing of the human ECSCR, which may imply its functional complexity. This undoubtedly opens new opportunities for future investigation of the biological and pathological significance of these ECSCR splice variants.
Keywords: ECSCR (ECSM2), alternative splicing, data mining, EST alignment, sequence homology, exon-intron boundary
1. Introduction
Alternative splicing of pre-mRNA is a common posttranscriptional process utilized by eukaryotic organisms to generate multiple transcript variants from a single gene (Black, 2003; Maniatis and Tasic, 2002). Large-scale analyses of available expressed sequence tag (EST) and mRNA datasets have estimated that 35–90% of human genes can undergo alternative splicing (Brett, et al., 2000; Brett, et al., 2002; Johnson, et al., 2003; Kan, et al., 2001; Mironov, et al., 1999; Modrek, et al., 2001; Pan, et al., 2008; Wang, et al., 2008b). Many mRNA variants appear to be regulated in a tissue-specific manner (Wang, et al., 2008b) and protein isoforms may differ in function and/or subcellular localization (Kriventseva, et al., 2003; Resch, et al., 2004; Stamm, et al., 2005). Thus, alternative splicing is considered as a major mechanism contributing to protein diversity and proteome complexity (Graveley, 2001; Maniatis and Tasic, 2002; Resch, et al., 2004). Further, mounting evidence indicates that alternative splicing is not only important for normal development and differentiation (Grabowski, 2011; Jiang, et al., 2000; Lopez, 1998; Suzuki, et al., 2011), but also implicated in numerous diseases including cancer (Carstens, et al., 2000; Faustino and Cooper, 2003; Garcia-Blanco, et al., 2004; Hui, et al., 2004; Krawczak, et al., 1992; Nair, 2009; Prinos, et al., 2011; Saxena, et al., 2002).
Endothelial cell-specific chemotaxis receptor (ECSCR) (Verissimo, et al., 2009), previously known as endothelial cell-specific molecule 2 (ECSM2) (Armstrong, et al., 2008; Ma, et al., 2009), was initially identified a decade ago by in silico cloning of novel endothelial cell-specific genes (Huminiecki and Bicknell, 2000). However, its biological and cellular functions have only recently begun to be understood. We and others have independently shown that ECSCR is preferentially expressed in endothelial cells and vasculature, and is critically involved in cell migration, angiogenesis, and apoptosis (Armstrong, et al., 2008; Ikeda, et al., 2009; Ma, et al., 2009; Verma, et al., 2010). However, the precise roles of ECSCR in regulating endothelial cell functions such as motility (inhibitory or stimulatory) and angiogenesis (pro-angiogenic or anti-angiogenic) remain controversial (Armstrong, et al., 2008; Ikeda, et al., 2009; Ma, et al., 2009; Verissimo, et al., 2009; Verma, et al., 2010), and further detailed investigation is needed. Recently, we have uncovered that ECSCR is a novel endothelial cell junctional protein and promotes cell-cell adhesion (Shi, et al., 2011). Furthermore, we have demonstrated that ECSCR can attenuate basic fibroblast growth factor (bFGF)-driven cell migration through the FGF receptor (EGFR)-extracellular regulated kinase (ERK)-focal adhesion kinase (FAK) pathway, suggesting that ECSCR could be an important role in coordinating receptor tyrosine kinase (RTK)-, integrin-, and cell-cell junction-mediated signaling (Shi, et al., 2011).
The multifaceted roles of ECSCR suggested by these recent studies prompted us to further explore the possibility of alternative splicing. Given the wealth of information stored in EST databases, alignment of EST sequences to the genome and/or to other ESTs/cDNAs has been a popular and powerful means to predict alternative splicing of gene transcripts (Brett, et al., 2000; Clark and Thanaraj, 2002; Florea, 2006; Gupta, et al., 2004; Hui, et al., 2004; Mironov, et al., 1999; Modrek, et al., 2001; Wang, et al., 2008a). Very recently, we have reported the molecular characterization of two alternative splicing variants of the mouse ECSCR (Wu, et al., 2012). In this study, by employing EST-based data mining, bioinformatics, and molecular biology approaches, we identified three novel full-length mRNA variants of the human ECSCR that are predicted to encode different protein products. Moreover, we identified two novel exons (previously thought as introns), which are only included in some ECSCR isoforms. Finally, we defined nearly all exon-intron boundaries and corrected the sequence and structure of the ECSCR gene. Our work provides new information on gene structure and alternative splicing of the human ECSCR, which may imply its functional complexity.
2. Materials and methods
2.1. Data source
The human (Homo sapiens) EST database (dbEST) and the human genome database are from the NCBI website (www.ncbi.nlm.nih.gov). The human dbEST was released May 2011 and contains more than eight million human ESTs. The three assemblies of human chromosome 5 (AC_000137, Homo sapiens chromosome 5, alternate assembly HuRef, whole genome shotgun sequence; AC_000048, Homo sapiens chromosome 5, alternate assembly Hs_Celera, whole genome shotgun sequence; and NC_000005, Homo sapiens chromosome 5, GRCh37.p2 primary reference assembly) were used in this study.
2.2. Bioinformatics tools
Software products including Megablast (Zhang, et al., 2000), ClustalW2 (Larkin, et al., 2007) (www.ebi.ac.uk/Tools/msa/clustalw2), ExPASy (web.expasy.org), SOSUIsignal (Gomi, et al., 2004) and SOSUI engine (version 1.11) (Hirokawa, et al., 1998) (bp.nuap.nagoya-u.ac.jp/sosui) were used.
2.3. EST clones and sequencing correction
Human (Homo sapiens) EST clones BI834795 (dbEST ID: 9654133), AA187059 (dbEST ID: 827161), and BF526332 (dbEST ID: 6962415) were purchased from Open Biosystems, Inc. (Huntsville, Alabama, USA). Each of these EST clones was bi-directionally sequenced using forward and reverse sequencing primers on the respective vector (pCMV-SPORT6 or pBluescript SK−).
2.4. Genomic DNA extraction
Human kidney epithelial cells (HEK293) were from ATCC (Manassas, VA) and grown in DMEM supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin (all from Mediatech, Manassas, VA). Cells were harvested by scraping in cold PBS and collected by centrifugation for 10 min at 4C. The cell pellets were washed twice with cold PBS and resuspended in TE buffer (10 mM Tris-HCl, 1mM EDTA, pH 8.0) containing 1% SDS in a centrifuge tube, mixed well by shaking gently, and incubated at room temperature for 10 min. The genomic DNA was extracted with an equal volume of phenol/chloroform/isoamylalcohol (25:24:1) for three times, followed by precipitation in 1/10 volume of sodium acetate (pH 5.2) and 3 volumes of isopropanol. The DNA pellets were washed with 70% ethanol, dried, and dissolved in TE buffer.
2.5. Polymerase chain reaction (PCR)
Genomic DNA from HEK293 cells was used as templates and primer pairs for PCRs are shown in Figure S4. Amplification was performed using Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA). Thermal cycling conditions comprised an initial denaturation step at 94°C for 5 min, followed by 35 cycles of denaturing at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, and a final extension step at 72°C for 10 min. The PCR products were examined by agarose gel electrophoresis, purified, and bi-directionally sequenced.
3. Results
The strategies used to identify novel ECSCR splice variants and to define all exon-intron boundaries in this study are outlined in Figure 1.
Figure 1.
Flow chart depicting computational and experimental approaches for identification of novel human ECSCR splice variants and determination of exon-intron boundaries.
3.1. Initial identification of potential novel human ECSCR splice variants
To identify potential splice variants of human ECSCR, we first used the 1027bp of human ECSCR mRNA/cDNA (GenBank accession number NM_001077693) as a query sequence to search the NCBI database of human ESTs (dbEST, released May 2011 and containing more than eight million human ESTs) with Megablast. As shown in Figure 2, we obtained more than 100 hits, of which 18 ESTs have a unique missing DNA fragment (Figure 2, horizontal lines in grey between the two black arrows) compared to the query sequence. We also noticed that two ESTs (AA187059 and BF526332) harbor an insertion of DNA sequence at different positions (Figure 2, two short vertical lines in black, as indicated by arrows). We thus chose the three representative ESTs with GenBank accession numbers of BI834795, BF526332, and AA187059, respectively, for further analysis. Bi-directionally (forward and reverse) sequencing of the three EST clones identified many errors in their original sequences deposited in the GenBank (data not shown).
Figure 2. Blast results showing the initial discovery of novel ECSCR splice variants.
A blast search on the dbEST was performed using the 1027bp human ECSCR mRNA/cDNA (GenBank accession number NM_001077693) as a query sequence. The two dotted vertical lines (in cyan) indicate the boundaries for a missing fragment (138bp) present in 18 ESTs, suggesting a relatively common splice variant. If counting the numbers of all ESTs partially or fully covering the region between the two dotted lines, this ECSCR variant form represents approximately one third of the total ESTs identified (18 out of 54). Note that the second transcript from the bottom has a longer gap. This is due to sequencing errors in the original EST, given the fact that its sequence contains multiple unknown nucleotides in this region. The short black vertical lines present in two blast hits in the middle of this map (EST clones AA187059 and BF526332) indicate that the two ESTs contain additional DNA insertions in their transcripts. They were later defined as novel Exon 2 and Exon 8 in this study.
3.2. Identification of Variant 2 containing a 138bp deletion compared to ECSCR (Variant 1)
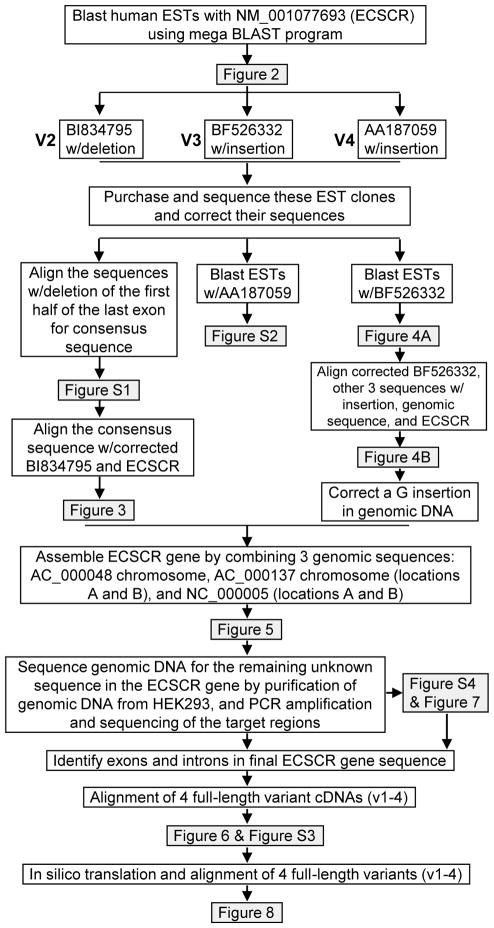
It is known that EST clones frequently contain sequencing errors. To minimize the influence of sequencing errors on the result, we sought to generate a consensus sequence. We extracted available sequences of the 17 out of 18 ESTs containing the unique missing DNA fragment from the NCBI database and then aligned them with the ClustalW2 program. The results indicated that they are most likely identical sequences (Supplemental Figure 1). The single nucleotide variations could be due to sequencing errors in the original ESTs deposited in the GenBank database or single-nucleotide polymorphism (SNP) within coding sequences of human ECSCR gene. Alignment of the consensus sequence (529bp, generated from Supplemental Figure 1), corrected BI834795 sequence (871bp), and the human ECSCR cDNA sequence (1027bp), revealed that the missing DNA fragment contains 138 nucleotides (Figure 3). This 138bp sequence corresponds to the unique missing DNA fragment present in the 18 ESTs shown in Figure 2. Thus, this relatively common, novel splice variant was named Variant 2 in distinction from the known human ECSCR (Variant 1). To map this 138bp deletion on the human ECSCR gene, we performed a blast search with this sequence against the three human genome assemblies (AC_000048, AC_000137, and NC_00005) focusing on the region of ECSCR. We found that the 138bp deletion is actually located within the first half of the last exon of the human ECSCR gene. Such a deletion can lead to a slight difference in the C-terminus of deduced amino acid sequences between Variants 1 and 2 (The stop codon TAA is highlighted in Figure 3).
Figure 3. Splicing out the 138bp DNA fragment (Exon 12A) in Variant 2.
Alignment of the 529bp EST consensus sequence (generated from Figure S1), corrected sequence of EST clone BI834795 (denoted as BI834795_CR), and full-length human ECSCR mRNA/cDNA (GenBank accession number NM_001077693) reveals the actual nucleotide sequences of the unique missing DNA fragment (138bp) in the 18 ESTs (identified in Figure 2). Thus, ECSCR Variant 2 represented here by BI834795 has a skipped exon (138bp). This exon was later named Exon 12A in this study. The start codon (ATG) and stop codon (TAA) in ECSCR and BI834795 are shaded.
3.3. Identification of Variants 3 containing a 108bp insertion and a 138bp deletion compared to ECSCR (Variant 1)
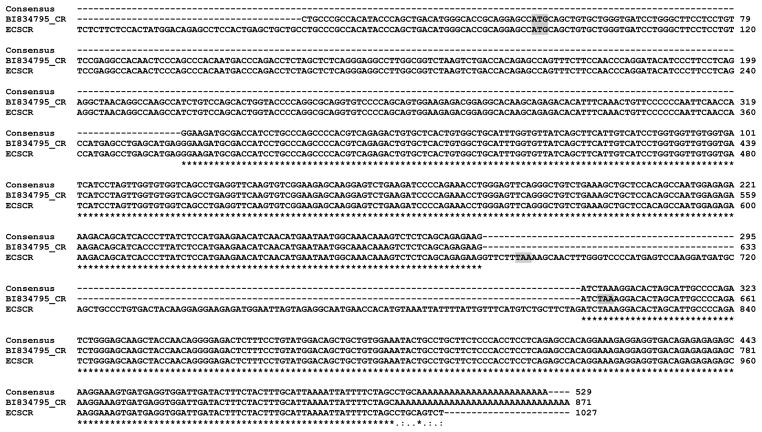
We next sought to obtain more information on the insertion present in the EST clone BF526332 (identified in Figure 2). By employing a similar strategy, we performed a blast search on the dbETS using BF526332 as a query sequence. The results confirmed that BF526332 indeed contains an insertion compared to ECSCR (Variant 1) (Figure 4). Notably, the other three EST clones (BP397865, AA304184, and BE177183) contain partial sequences of this insertion (Figure 4, fragments indicated by blue arrows). Multiple sequence alignment of these three ESTs, corrected BF526332, the ECSCR (Variant 1) cDNA, and the ECSCR gene (GenomicEx8) indicated that the exact length of this insertion is 108bp. Further mapping this 108bp insertion on the human ECSCR gene revealed that it is located within a region that was previously considered as an intron (Figure 4, lower panel). The extra single nucleotide present in the genomic sequence only (Figure 4, red arrow) could be a sequencing error. This novel splice variant, which is represented by BF526332 and at least other three ESTs, was designated by the name of Variant 3.
Figure 4. Blast results showing the identification of Variant 3.
As noted in Figure 2, the EST clone BF526332 harbors an insertion (later named Exon 8). To confirm that this is not a random event, a blast search on the dbEST was performed using BF526332 as a query sequence. At least three additional EST clones (BP397865, AA304184, and BE177183) were identified to contain this inserted region (indicated by blue arrows), suggesting that it is a novel variant (Variant 3). Nucleotide sequence alignment of ECSCR (Variant 1), genomic region corresponding to Exon 8 (GenomicEx8), corrected BF526332 (BF526332_CR) and three ESTs (BP397865, AA304184, and BE177183) shows that Variant 3 contains an insertion of 108bp (Exon 8) highlighted in grey, which is not present in Variant 1. The red error indicates a sequencing error in the genomic DNA.
3.4. Identification of Variants 4 containing an 119bp insertion compared to ECSCR (Variant 1)
Similarly, we performed a blast search on the dbEST using AA187059 (identified in Figure 2) as a query sequence. Our results indicated that the EST clone AA187059 indeed harbors an insertion compared to ECSCR (Variant 1) (Supplemental Figure 2). Alignment of corrected AA187059 sequences, ECSCR mRNA/cDNA (1027bp), and the ECSCR gene revealed that the length of this insertion is 119bp. Similar to the 108bp insertion described above, this insertion is also located in a region that was previously determined as an intron at the N-terminus of the ECSCR gene (data not shown).
3.5. Determination of exon-intron boundaries of the human ECSCR gene and identification of two novel exons (Exon 2 and 8)
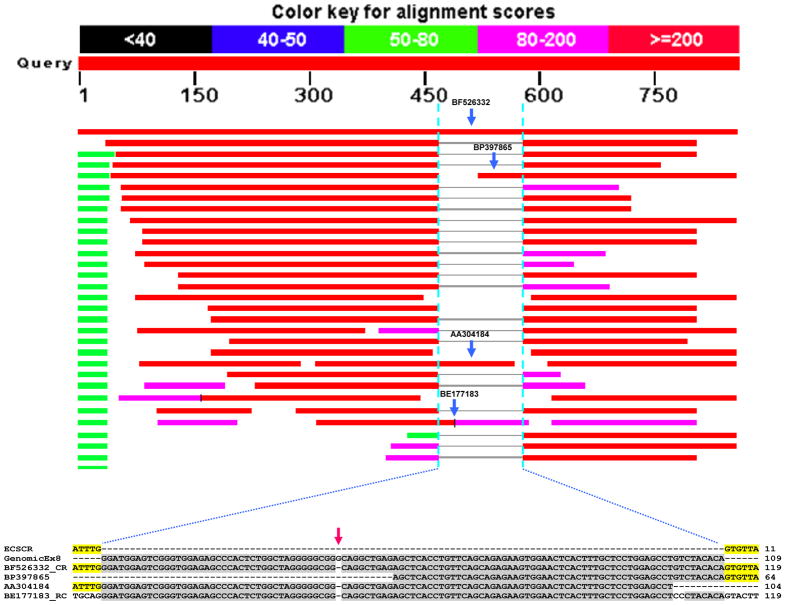
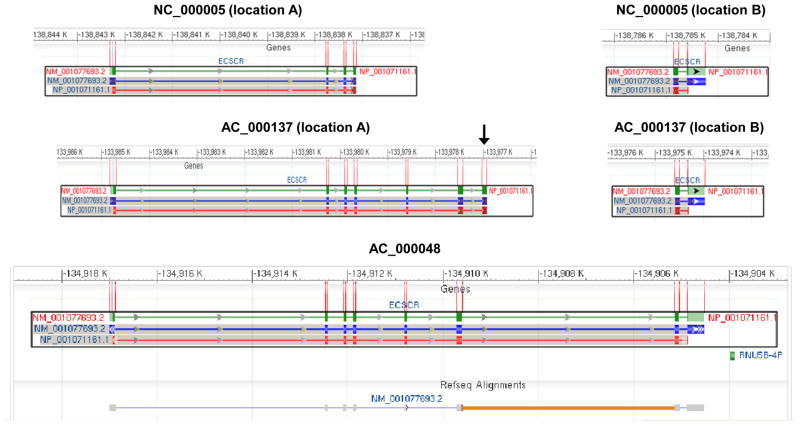
To define the exon-intron boundaries of the human ECSCR gene, we examined the human ECSCR gene structure on the three assemblies of human chromosome 5 (AC_000137, Homo sapiens chromosome 5, alternate assembly HuRef, whole genome shotgun sequence; AC_000048, Homo sapiens chromosome 5, alternate assembly Hs_Celera, whole genome shotgun sequence; and NC_000005, Homo sapiens chromosome 5, GRCh37.p2 primary reference assembly), which are available on the NCBI website. As shown in Figure 5, a total of eight exons are annotated on the AC_000048 chromosome. However, in this assembly, the ECSCR gene contains a large, un-sequenced “intron” between the original 6th and 7th exons (Figure 5, lower panel, indicated by an orange line). The ECSCR gene is also mapped on AC_000005 chromosome and AC_000137 chromosome, respectively. In both cases, the gene is split into two parts (location A and location B) (Figure 5, upper and middle panels), which together do not cover the entire gene. Notably, the N-terminus of the ECSCR gene on AC_000137 chromosome (location A) contains an exon (Figure 5, middle panel, indicated by a black arrow), which is not identified on the AC_000048 chromosome (Figure 5, lower panel).
Figure 5. Original gene structure of human ECSCR.
The gene structures shown here are the mapping results on the three assemblies of human chromosome 5 (AC_000137, Homo sapiens chromosome 5, alternate assembly HuRef, whole genome shotgun sequence; AC_000048, Homo sapiens chromosome 5, alternate assembly Hs_Celera, whole genome shotgun sequence; and NC_000005, Homo sapiens chromosome 5, GRCh37.p2 primary reference assembly) that are available in the NCBI databases. Green bars (the first row in each panel) represent the ECSCR gene and blue bars (the second row) show exons. The protein product is in the third row with red bars as translated gene products. Note that the last exon only encodes for two amino acid residues before the stop codon TAA (see ECSCR in Figure 3). On the AC_000048 assembly, a total of eight exons are initially defined. However, the ECSCR gene contains a large, un-sequenced region (thought as a large intron), which is indicated by the orange line. Also, note that the gene is split into two parts (location A and location B) on either AC_000137 or NC_000005 assembly, which together do not cover the entire gene. The black arrow indicates the extra exon identified only on the AC_000137 assembly (location A) but not on the AC_000048 assembly.
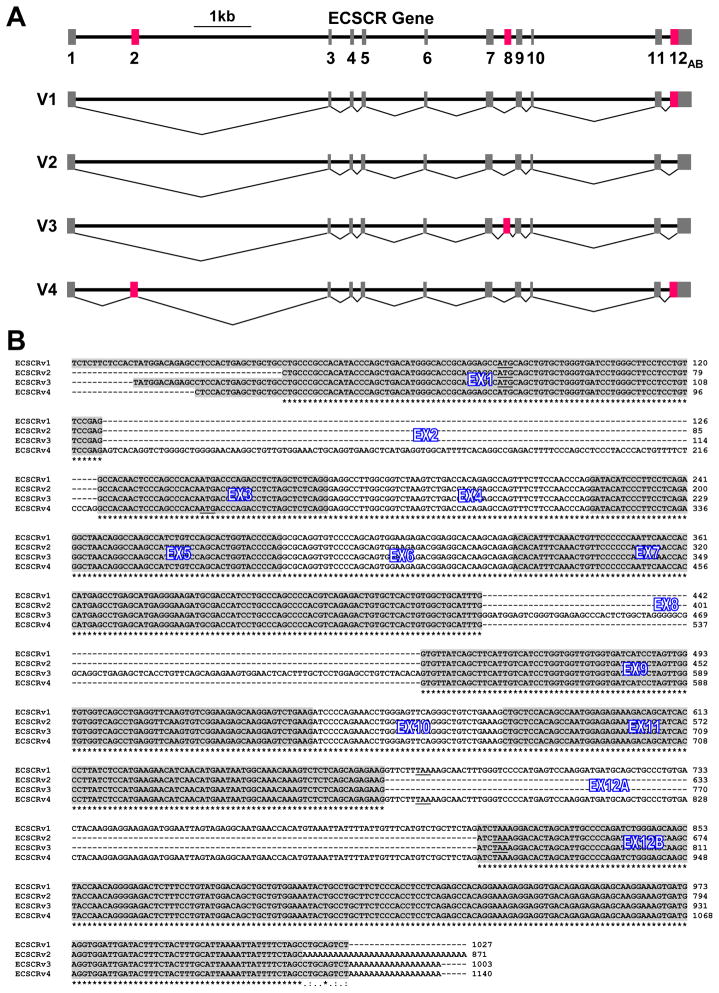
As outlined above, compared to ECSCR (Variant 1), Variant 2 has a 138bp deletion whereas Variant 4 harbors a 119bp insertion. In contrast, Variant 3 contains a 108bp insertion and the same 138bp deletion as in Variant 2. Considering all known exons that have been included in AC_000048 chromosome, AC_000137 chromosome (location A and location B), and NC_000005 chromosome (location A and location B) (Figure 5), we assembled the ECSCR gene by combining the three genomic sequences and aligning the full sequences of Variants 1–4 against them. By employing this strategy, we identified a total of 12 exons, of which Exons 2 and 8 are novel (Figure 6A). In addition, we found that Exon 12 (the last exon of the ECSCR gene) is split into Exon 12A and Exon 12B due to the 138bp deletion present in Variants 2 and 3 (Figure 6A). The sequence alignment results of the transcripts (containing exons only) of Variants 1–4 are shown in Figure 6B. As expected, the two newly identified exons (Exon 2 and Exon 8) are located within the regions that were previously thought as introns of the human ECSCR gene (Supplemental Figure 3). This was further confirmed by PCRs using genomic DNA extracted from human embryonic kidney (HEK) 293 cells as templates followed by bi-directional sequencing of the PCR products. The primers used in these PCRs are displayed in Supplemental Figure 4. As expected, PCRs with primer pairs Ex2L/Ex2R and Ex8L/Ex8R resulted in specific products of ~650bp (containing Exon 2) and ~500bp (containing Exon 8), respectively (Figure 7A). In particular, the single nucleotide sequencing error in the original ECSCR gene sequence (Figure 4B, the red arrow) was confirmed by our sequencing results of the ~500bp PCR product containing Exon 8 (Figure 7B).
Figure 6. Reconstruction of the human ECSCR gene based on the data presented in this study.
(A) New ECSCR gene structure and alternative splicing events. Exons are shown by closed boxes. Red boxes indicate the newly identified exons whose splicing results in four splice variants of human ECSCR. Variant 1 is the originally known ECSCR, the most common isoform. Variant 2 is a second common isoform with skipped Exon 12A compared to Variant 1. Variant 3 is an uncommon isoform including the novel Exon 8. Variant 4 is a rare isoform including the novel Exon 2. (B) Alignment of the complete nucleotide sequences of the four full-length ECSCR variants. A total of twelve exons (Exons 1–11, Exon 12A and Exon 12B) are marked. The start codon (ATG) and stop codon (TAA) for each splice variant are underlined. See text for details.
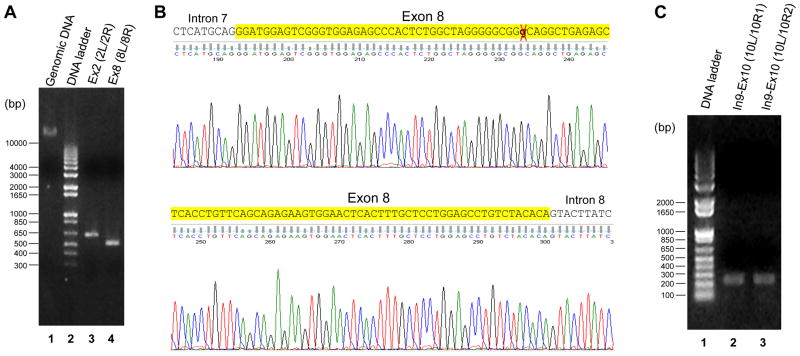
Figure 7. Experimental validation of exons 2, 8, and 10.
(A) PCR results of the gene fragments containing Exon 2 and Exon 8. The primer pairs used in PCRs are shown in Figure S4. As revealed by agarose gel electrophoresis, the targeting sizes of specific PCR products are ~650bp (containing Exon 2, lane 3) and ~500bp (containing Exon 8, lane 4), respectively. The genomic DNA extracted from HEK293 cells (lane 1) was used as templates in the PCRs. The sizes of DNA ladder are also shown (lane 2). (B) Representative sequencing results of the PCR products. The PCR product containing Exon 8 (lane 4 in Panel A) was sequenced. Comparison results confirm that a single nucleotide (G) in the original ECSCR gene (marked by a red cross) is indeed a sequencing error. Also see Figure 4B. (C) PCR results of the gene fragments spanning the junction of Exon 9 – Intron 9, the entire Intron 9, and the junction of Intron 9 – Exon 10. The primer pairs used in PCRs are shown in Figure S4. The targeting sizes of specific PCR products using primer pairs Ex10L/Ex10R1 and Ex10L/Ex10R2 are ~250bp and ~240bp, respectively.
In an attempt to map Exon 10, we performed a blast search on all human genome assemblies. However, we failed to identify its exact locus. Since there exists a large un-sequenced “intron” in the original ECSCR gene on the AC_000048 chromosome (Figure 5, orange line), we reasoned whether Exon 10 could be located within this unknown region. Thus, we designed primers on Exons 9, 10 and 11 and used different combinations of primers for PCRs. We obtained two specific DNA products of ~250bp and ~240bp (Figure 7C) from PCRs using primer pairs Ex10L/Ex10R1 and Ex10L/Ex10R2 (Supplemental Figure 4), respectively. Sequencing of these two PCR fragments revealed that both contain the region spanning the junction of Exon 9-Intron 9, the entire Intron 9, and the junction of Intron 9-Exon 10. The sequences of these regions have been updated in Supplemental Figure 3. However, we were not able to obtain specific products from PCRs using the primer pairs on Exon 9/Exon 11 and Exon 10/Exon 11 (data not shown). One possibility is that this previously un-sequenced region (Figure 5, orange line) contains extremely high GC contents or other secondary structures, which resulted in the failure of PCRs. Nevertheless, our results showed that Exon 10 is indeed located after Intron 9. The sequences of Exon 9 through Exon 10 have been obtained and validated.
3.6. Predicted protein sequences of the four full-length ECSCR variants (Variants 1–4)
Sequencing results of the ESTs BI834795 (Variant 2), BF526332 (Variant 3), and AA187059 (Variant 4) and their alignment with ECSCR (Variant 1) showed that the three EST clones contain full-length cDNAs (Figure 6B). We thus performed in silico translation of these cDNAs. As shown in Figure 8, the four variants share a very high amino acid homology. The slight differences between Variants 1 and 2 are the last two residues at their C-termini (Figure 8), which are resulted from the alternative 3′ splicing event (skipping of Exon 12A in Variant 2, see Figure 6). Notably, compared to the other three variants, Variant 3 has an extra 36 amino acid segment (Figure 8) due to the inclusion of Exon 8 (Figure 6). Interestingly, this insertion occurs in the beginning of the transmembrane domain. Variant 4 is the only one of the four splice variants that contains Exon 2 (Figure 6). There are two possibile in silico translations of Variant 4 transcript. If using the open reading frame (ORF) with the original start codon of ATG in Exon 1 like the other three variants, the ORF would end with the stop codon of TGA in the middle of Exon 3 (Figure 6), resulting in a predicted protein product of a total of 67 amino acids with no apparent homology with Variants 1–3 (data not shown). Alternatively, the translation of Variant 4 can start from the first ATG in Exon 3 and ends at the first stop codon of TAA in Exon 12A (Figure 6). This leads to the absence of the signal peptide in the N-terminus of the hypothetical protein of Variant 4 (Figure 8).
Figure 8. Predicted protein sequences of the four full-length ECSCR variants.
The putative signal peptide is boxed. The predicted single transmembrane domain is underlined. The arrows indicate where alternative splicing occurs in the four ECSCR variants. All identical amino acids among the four variants are highlighted. See text for details.
4. Discussion
Recently, with experimental validation of ECSCR as an endothelial cell-specific, cell-cell junctional protein and discovery of its important roles in cell-cell adhesion, motility, angiogenesis, and apoptosis (Armstrong, et al., 2008; Ikeda, et al., 2009; Ma, et al., 2009; Shi, et al., 2011; Verma, et al., 2010), ECSCR has been an interesting molecule in vascular biology research. Complete information regarding its transcriptional and posttranscriptional regulation is urgently needed to further understand the precise function of ECSCR. Alternative splicing contributes significantly to the complexity of the human transcriptome and proteome, and is a common mechanism of posttranscriptional regulation, which occurs in averagely one-half of the human genes (Brett, et al., 2002; Johnson, et al., 2003; Modrek, et al., 2001). Very recently, we have identified and characterized two alternative splicing variants of the mouse ECSCR (Wu, et al., 2012). However, the alternative splicing events of the human ECSCR gene are yet to be investigated. Complete sequencing of the human genome and public availability of EST databases have made this possible. In this study, we combine computational and experimental methods to identify splice variants of ECSCR, define nearly all exon-intron boundaries, and update the information on the human ECSCR gene.
It is known that a gene in higher eukaryotes consists of exons and introns. The removal of introns and the joining of exons by RNA splicing is an essential step in generation of mature, functional RNA product of a gene. Alternative splicing events can assemble different combinations of exons to produce mRNA variants with distinct protein coding potentials. From a gene structure standpoint, alternative splicing events can be classified into five common types, which include exon skipping/inclusion, alternative 3′ splice sites, alternative 5′ splice sites, mutually exclusive exons, and intron retention (Cartegni, et al., 2002). As for the human ECSCR gene, we found that Exon 12A (the first half of the last exon) is spliced out in Variants 2 and 3 whereas the intact Exon 12 (including 12A and 12B) are retained in Variants 1 and 4. This represents the type of alternative 3′ splice sites. We also showed that Exon 2 and Exon 8 are present only in Variant 4 and Variant 3, respectively, which are good examples of exon skipping/inclusion events. Since ESTs are generally derived from mature spliced mRNA populations, our data obtained in this study through combined methods of EST-based data mining, PCR, and sequencing demonstrates the mRNA diversity from the single ECSCR gene.
Alignment of predicted amino acid sequences from the four full-length transcript variants of ECSCR indicates a high homology among them (Figure 8). For Variant 4, inclusion of 119bp Exon 2 in its transcript leads to a shift of the ORF and translation ending at the stop codon of TGA in the middle of Exon 3 if the same start codon (ATG) in Exon 1 is used as in Variants 1–3 (Figure 6B). Alternatively, utilization of the second ATG in Exon 3 results in a putative protein short of the signal peptide consisting of first 27 amino acids at the N-terminus. In this case, Variant 4 is identical to the mature ECSCR (Variant 1). Due to the inclusion of 108bp Exon 8, Variant 3 shows the least similarity to others. It is noted that the only difference between Variant 1 and Variant 2 occurs within the last two amino acid residues that are resulted from the alternative 3′ splicing event (intact Exon 12 versus Exon 12B only). This implies that the function of ECSCR Variants 1 and 2 could be very similar. This is somewhat surprising. Of all the blast hits biased towards the 3′ ends of ECSCR transcripts, approximately two-thirds of the ESTs resemble Variant 1 (ECSCR) containing the intact Exon 12 and the remaining one-third are similar to Variant 2 in which the Exon 12A is skipped (Figure 2). Because ESTs generally originate from random sequencing efforts, the number of ESTs corresponding to a specific variant in the dbEST is likely representative of its expression level. Thus, it is reasonable to conclude that Variants 1 and 2 are the two common isoforms of ECSCR. If this is true, one may ask why the two variants share such a high homology (nearly 98% of amino acid identity) and what the biological and functional significance is. There are several explanations. Firstly, the coding sequences for the last two residues in Variant 1 and Variant 2 correspond to the first 6 nucleotides in Exon 12A and the first 3 nucleotides in Exon 12B (Figure 6B). Thus, the major difference between the two transcript variants is located within alternative 3′ untranslated regions (UTRs). In fact, in addition to the above-mentioned five common types of alternative splicing events, alternative 5′ and 3′ UTRs are often encountered (Florea, 2006). However, they are generally harder to identify and characterize because of the intrinsic difficulty in determining the gene boundaries (Florea, 2006). The alternative 5′ and 3′ UTRs could play a role in translational control and development (Kuersten and Goodwin, 2003; Wilkie, et al., 2003). Secondly, it has been long appreciated that the alternative transcripts containing premature termination codons are targets of nonsense-mediated mRNA decay (NMD), a surveillance mechanism that selectively degrades nonsense mRNAs (Buchwald, et al., 2010; Ishigaki, et al., 2001; Kim, et al., 2001; Lewis, et al., 2003; Lykke-Andersen, 2001; Lykke-Andersen, et al., 2001; Nagy and Maquat, 1998). As already noted, splicing out Exon 12A results in the removal of the first stop codon, which makes the second stop codon in the same ORF in Exon 12B effective. Thus, it will be interesting to speculate the possibility of NMD for ECSCR, which deserves further investigation in the future. Thirdly, although the predicted protein sequences of Variants 1 and 2 are highly similar, their transcripts may represent different tissue origins, cell types, subcellular localizations, developmental stages, and disease specificity. Indeed, previous studies have demonstrated the spatial and temporal expression patterns of ECSCR (Armstrong, et al., 2008; Ikeda, et al., 2009; Ma, et al., 2009; Shi, et al., 2011; Verma, et al., 2010), in which the potential splice variants of ECSCR were not distinguished. Given the past research has been focused on the expression of ECSCR (Variant 1) either heterologously or endogenously, identification of the three novel full-length ECSCR isoforms (Variants 2–4) in our current study undoubtedly introduces a new dimension of studying the ECSCR gene regulation.
Although the wealth information in the dbEST and alignment of EST sequences to the human genome assemblies provide a useful, effective tool to unravel alternative transcripts, it is important to point out that there are several limitations of the EST-based methods. Firstly, lower expressed splice variant(s) may be underscored because a transcript with a higher expression level obviously has a better chance of inclusion in the dbEST. In some cases, minor splice forms can be functionally important (Geerlings, et al., 2003; Hao, et al., 1994; Wieder, et al., 1997). Secondly, alternative splicing can be highly tissue, cell type, and/or developmental stage specific and thereby detection of such splice variants really depends on the tissue type, timing and conditions when ESTs are sampled. Lower expressed variants especially have a tendency to be tissue specific (Modrek and Lee, 2003). Furthermore, many ESTs are sampled from tumor libraries. However, a predominant splice form can be expressed in normal tissue instead (Nair, 2009; Xu and Lee, 2003). Thus, the frequency of the ESTs for a specific splice variant could be biased. Thirdly, as most ESTs are generated from the 5′ or the 3′ termini of the transcript, ESTs are biased towards underrepresentation of splice forms involving exons that are in the middle of long transcripts (Johnson, et al., 2003). Lastly, the inherent problem of error-prone sequences in ESTs can lead to false positive predictions of splice variants (Modrek and Lee, 2002; Sorek and Safer, 2003). Indeed, we re-sequenced the three ESTs representing ECSCR Variants 2–4 and found many errors in their original sequences. One simple and useful method to lower the number of EST sequencing errors is to generate a consensus sequence from multiple transcripts (Gupta, et al., 2004), as used in our study (Supplemental Figure 1). Nevertheless, to overcome these potential problems, other strategies such as non-EST based methods (Hiller, et al., 2005; Sorek, et al., 2004) should be considered in the future.
In summary, in this study, using combined computational and experimental approaches we identified at least three novel splice variants (Variants 2–4) and two previously unknown exons (Exons 2 and 8) of the human ECSCR gene. We defined nearly all exon-intron boundaries and mapped a total of 12 exons on the ECSCR gene, of which only eight have been previously annotated in public databases. To our knowledge, this is the first demonstration of alternative splicing of human ECSCR gene. The existence of multiple splice variants implies that they may contribute to the multifaceted functions of ECSCR. Our study provides the molecular foundation for future exploiting the roles of ECSCR in health and disease that likely involve transcriptional, posttranscriptional, and translational regulation.
Supplementary Material
Sequences of the 18 ESTs (identified from Figure 2) with the unique missing fragment (138bp Exon 12A) were extracted from the NCBI database. A 529bp consensus sequence was generated using multiple sequence alignment of the 17 ESTs (one of the 18 ESTs failed to align with others possibly due to high frequency of sequencing errors in this EST clone).
As already noted in Figure 2, the EST clone AA187059 harbors an insertion (later named Exon 2) compared to ECSCR (Variant 1). To confirm this, AA187059 was used as a query sequence to blast the dbEST. Note that all EST hits do not have this insertion (Exon 2) except the query sequence itself.
All exons are highlighted in blue or yellow. The start codon (ATG) in Exon 1 (for Variants 1–3), the start codon (ATG) in Exon 3 (for Variant 4), the stop codon (TAA) in Exon 12A (for Variants 1 and 4), and the stop codon (TAA) in Exon 12B (for Variants 2 and 3) are underlined.
(A) Locations of PCR primers on the ECSCR gene. (B) Sequences of PCR primers.
Highlights.
We identify four human ECSCR splice variants (Variants 1–4) in this study.
We define all twelve exons and exon-intron boundaries of human ECSCR gene.
Exons 2 and 8 are novel and have not been previously annotated in public databases.
Our data provides new information on ECSCR gene structure and alternative splicing.
Acknowledgments
This work was supported by a St. Joseph’s Foundation (SJF) Startup Fund (to Y.H.), a Catholic Healthcare West (now Dignity Health) Intellectual Innovation Network (CHWiin) SEED Fund (to Y.H.), and in part by American Heart Association (AHA) Beginning Grant-in-Aid (to Y.H.) and National Institute of Health (R01GM085237, to Y.C.). J. Lu was partially supported by a China Scholarship Council (CSC) Studentship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong LJ, Heath VL, Sanderson S, Kaur S, Beesley JF, Herbert JM, Legg JA, Poulsom R, Bicknell R. ECSM2, an endothelial specific filamin a binding protein that mediates chemotaxis. Arterioscler Thromb Vasc Biol. 2008;28:1640–1646. doi: 10.1161/ATVBAHA.108.162511. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Brett D, Hanke J, Lehmann G, Haase S, Delbrück S, Krueger S, Reich J, Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- Brett D, Pospisil H, Valcárcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci U S A. 2010;107:10050–10055. doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens RP, Wagner EJ, Garcia-Blanco MA. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Clark F, Thanaraj TA. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet. 2002;11:451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Florea L. Bioinformatics of alternative splicing and its regulation. Brief Bioinform. 2006;7:55–69. doi: 10.1093/bib/bbk005. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Geerlings TH, Faber AW, Bister MD, Vos JC, Raué HA. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae. J Biol Chem. 2003;278:22537–22545. doi: 10.1074/jbc.M300759200. [DOI] [PubMed] [Google Scholar]
- Gomi M, Sonoyama M, Mitaku S. High performance system for signal peptide prediction: SOSUIsignal. Chem-Bio Info J. 2004;4:142–147. [Google Scholar]
- Grabowski P. Alternative splicing takes shape during neuronal development. Curr Opin Genet Dev. 2011;21:388–394. doi: 10.1016/j.gde.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Gupta S, Zink D, Korn B, Vingron M, Haas SA. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics. 2004;20:2579–2585. doi: 10.1093/bioinformatics/bth288. [DOI] [PubMed] [Google Scholar]
- Hao H, Tyshenko MG, Walker VK. Dihydrofolate reductase of Drosophila. Cloning and expression of a gene with a rare transcript J Biol Chem. 1994;269:15179–15185. [PubMed] [Google Scholar]
- Hiller M, Huse K, Platzer M, Backofen R. Non-EST based prediction of exon skipping and intron retention events using Pfam information. Nucleic Acids Res. 2005;33:5611–5621. doi: 10.1093/nar/gki870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hui L, Zhang X, Wu X, Lin Z, Wang Q, Li Y, Hu G. Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene. 2004;23:3013–3023. doi: 10.1038/sj.onc.1207362. [DOI] [PubMed] [Google Scholar]
- Huminiecki L, Bicknell R. In silico cloning of novel endothelial-specific genes. Genome Res. 2000;10:1796–1806. doi: 10.1101/gr.150700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nakano R, Uraoka M, Nakagawa Y, Koide M, Katsume A, Minamino K, Yamada E, Yamada H, Quertermous T, Matsubara H. Identification of ARIA regulating endothelial apoptosis and angiogenesis by modulating proteasomal degradation of cIAP-1 and cIAP-2. Proc Natl Acad Sci U S A. 2009;106:8227–8232. doi: 10.1073/pnas.0806780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Cote J, Kwon JM, Goate AM, Wu JY. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol Cell Biol. 2000;20:4036–4048. doi: 10.1128/mcb.20.11.4036-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kan Z, Rouchka EC, Gish WR, States DJ. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, Koch I, Apweiler R, Vingron M, Bork P, Gelfand MS, Sunyaev S. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AJ. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. mRNA quality control: Marking the message for life or death. Curr Biol. 2001;11:R88–91. doi: 10.1016/s0960-9822(01)00036-7. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Ma F, Zhang D, Yang H, Sun H, Wu W, Gan Y, Balducci J, Wei Y, Zhao X, Huang Y. Endothelial cell-specific molecule 2 (ECSM2) modulates actin remodeling and epidermal growth factor receptor signaling. Genes Cells. 2009;14:281–293. doi: 10.1111/j.1365-2443.2008.01267.x. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Nair TM. On selecting mRNA isoform features for profiling prostate cancer. Comput Biol Chem. 2009;33:421–428. doi: 10.1016/j.compbiolchem.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Prinos P, Garneau D, Lucier JF, Gendron D, Couture S, Boivin M, Brosseau JP, Lapointe E, Thibault P, Durand M, Tremblay K, Gervais-Bird J, Nwilati H, Klinck R, Chabot B, Perreault JP, Wellinger RJ, Elela SA. Alternative splicing of SYK regulates mitosis and cell survival. Nat Struct Mol Biol. 2011;18:673–679. doi: 10.1038/nsmb.2040. [DOI] [PubMed] [Google Scholar]
- Resch A, Xing Y, Modrek B, Gorlick M, Riley R, Lee C. Assessing the impact of alternative splicing on domain interactions in the human proteome. J Proteome Res. 2004;3:76–83. doi: 10.1021/pr034064v. [DOI] [PubMed] [Google Scholar]
- Saxena S, Szabo CI, Chopin S, Barjhoux L, Sinilnikova O, Lenoir G, Goldgar DE, Bhatanager D. BRCA1 and BRCA2 in Indian breast cancer patients. Hum Mutat. 2002;20:473–474. doi: 10.1002/humu.9082. [DOI] [PubMed] [Google Scholar]
- Shi C, Lu J, Wu W, Ma F, Georges J, Huang H, Balducci J, Chang Y, Huang Y. Endothelial cell-specific molecule 2 (ECSM2) localizes to cell-cell junctions and modulates bFGF-directed cell migration via the ERK-FAK pathway. PLoS One. 2011;6:e21482. doi: 10.1371/journal.pone.0021482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Safer HM. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 2003;31:1067–1074. doi: 10.1093/nar/gkg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Shemesh R, Cohen Y, Basechess O, Ast G, Shamir R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Osaki K, Sano K, Alam AH, Nakamura Y, Ishigaki Y, Kawahara K, Tsukahara T. Comprehensive analysis of alternative splicing and functionality in neuronal differentiation of P19 cells. PLoS One. 2011;6:e16880. doi: 10.1371/journal.pone.0016880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo AR, MHJ, Heath VL, Legg JA, Sheldon H, Andre M, Swain RK, Bicknell R. Functionally defining the endothelial transcriptome, from Robo4 to ECSCR. Biochem Soc Trans. 2009;37:1214–1217. doi: 10.1042/BST0371214. [DOI] [PubMed] [Google Scholar]
- Verma A, Bhattacharya R, Remadevi I, Li K, Pramanik K, Samant GV, Horswill M, Chun CZ, Zhao B, Wang E, Miao RQ, Mukhopadhyay D, Ramchandran R, Wilkinson GA. Endothelial cell-specific chemotaxis receptor (ecscr) promotes angioblast migration during vasculogenesis and enhances VEGF receptor sensitivity. Blood. 2010;115:4614–4622. doi: 10.1182/blood-2009-10-248856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, O’Toole M, Brendel V, Young ND. Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes. BMC Plant Biol. 2008a;8:17. doi: 10.1186/1471-2229-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008b;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieder R, Wang H, Shirke S, Wang Q, Menzel T, Feirt N, Jakubowski AA, Gabrilove JL. Low level expression of basic FGF upregulates Bcl-2 and delays apoptosis, but high intracellular levels are required to induce transformation in NIH 3T3 cells. Growth Factors. 1997;15:41–60. doi: 10.3109/08977199709002111. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Wu W, Shi C, Ma F, Balducci J, Huang H, Ji HL, Chang Y, Huang Y. Structural and Functional Characterization of Two Alternative Splicing Variants of Mouse Endothelial Cell-Specific Chemotaxis Regulator (ECSCR) Int J Mol Sci. 2012;13:4920–4936. doi: 10.3390/ijms13044920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of the 18 ESTs (identified from Figure 2) with the unique missing fragment (138bp Exon 12A) were extracted from the NCBI database. A 529bp consensus sequence was generated using multiple sequence alignment of the 17 ESTs (one of the 18 ESTs failed to align with others possibly due to high frequency of sequencing errors in this EST clone).
As already noted in Figure 2, the EST clone AA187059 harbors an insertion (later named Exon 2) compared to ECSCR (Variant 1). To confirm this, AA187059 was used as a query sequence to blast the dbEST. Note that all EST hits do not have this insertion (Exon 2) except the query sequence itself.
All exons are highlighted in blue or yellow. The start codon (ATG) in Exon 1 (for Variants 1–3), the start codon (ATG) in Exon 3 (for Variant 4), the stop codon (TAA) in Exon 12A (for Variants 1 and 4), and the stop codon (TAA) in Exon 12B (for Variants 2 and 3) are underlined.
(A) Locations of PCR primers on the ECSCR gene. (B) Sequences of PCR primers.