Abstract
Leptin is a circulating protein which regulates dietary intake through binding the leptin receptor. Numerous labs have used known structures and mutagenesis to study this binding process in common animal models (human, mouse and rat). Understanding this binding process in other vertebrate species will allow for a better understanding of leptin and leptin receptor function. The binding site between leptin and leptin receptor is highly conserved in mammals as confirmed through sequence alignments mapped onto structures of both leptin and leptin receptor. More variation in this interaction is found in lizard and frog sequences. Using our models, we show that the avian leptin sequences have far less variation in the binding site than does the leptin receptor. This analysis further suggests that avian leptins are artifactual. In fish, gene duplication events have led to the expression of multiple leptin proteins. These multiple leptin proteins have variation in the regions interacting with leptin receptor. In zebrafish and the Japanese rice fish, we propose that leptin A has a higher binding energy than does B. Differing binding energies are evidence of either divergent functions, different binding confirmations, or other protein partners of leptin B.
Keywords: Leptin, leptin receptor, protein interaction, bioinformatics
1 Introduction
Leptin is one of the most studied molecules in endocrinology with tens of thousands of publications. The leptin protein is coded by the obese (ob) gene [45] which shares high homology in mammalian species. Leptin’s (Lep) role in appetite and adipose regulation is now well supported. Initial studies in mice and humans suggested the endocrine functions of Lep in adipose storage, with support through injections of recombinant Lep in the ob/ob mouse model [17], and a correlation between human serum Lep concentration and body fat [5]. Leptin RNA is expressed at high levels in adipocytes [28], while the Leptin receptor (LepR) is expressed at high levels in the hypothalamus [41, 25, 35], T-cells [27], and vascular endothelial cells [36]. A model has emerged with dietary intake and fat stores regulating the production of Lep, which then enters the circulatory system where it binds to LepR in multiple tissues eliciting response in the brain, immune system and vasculature. The Lep pathway also influences numerous other physiological processes, including body temperature, energy regulation, immune, reproductive and development [8, 2, 10, 33, reviewed in 15].
The great majority of leptin studies have been conducted using mammalian models, most notably the mouse, rat, and human. The aim of these studies has been to build a model of leptin function, and to uncover its pleiotropic effects. The quest for Lep (or leptin-like) sequences in other taxa, however, has been far less studied. Among mammalian Leps, there is generally high sequence conservation (including armadillo, rabbit, bat, skunk, raccoon and whale) [9]. In phocid seals, containing higher variation than most mammals, Hammond et al. show that the sequence variation does not fall in the interaction site with LepR, but may undergo positive selection for electrostatics [18]. Considerable effort has gone into characterizing Lep in birds [1, 40] yielding conservation levels far higher when comparing to mouse than those of other mammals, leading to debate over accuracy and reproducibility of the studies [34, 14]. Using structures, models and sequences Denver et al., provided an analysis of evolution of Leptin in not only mammals but also fish, lizards and frogs [7]. In fish, two separate Lep transcripts are found expressed primarily in liver with believed roles in reproduction, food intake and lesser known role in fat metabolism [reviewed in 6]. It is thought that both of these Lep may bind the same LepR, however this has not been verified experimentally.
In humans, Lep is a 167 amino acid protein that takes a globular fold involving hydrophobic packing between four helices as can be seen in the known structure, PDB 1ax8 [44]. Lep binds to a membrane protein, LepR, composed of four cytokine receptor homologous domains (CRH), an Ig-like domain, a transmembrane segment, and a C-terminal cytoplasmic domain in the long isoform. Multiple shorter isoforms are found with some of these domains missing. Initial work on identifying the leptin binding domain (LBD) of LepR showed amino acids 323–640 in binding [13], which consists of the Ig-like and the second CRH (CRH2) domains. Binding of Lep to the LepR induces intracellular signal transduction through the JAK/STAT pathway [42] through phosphorylation of Tyr986 and Tyr1141 of LepR [3]. Detailed molecular interactions between Lep and LepR have been addressed by various groups through use of site directed mutagenesis and modeling approaches [20, 29, 30, 31], and more recently through the structure of the CRH2 of LepR, PDB 3v6o [4]. It is proposed that Lep binds tightly to the CRH2 domain, while also interacting with a second LepR through the Ig-like domain.
Several studies shown above have investigated the evolution of Leptin in multiple species, including some that model molecular interactions of Lep with LepR through structures and mutagenesis. What is currently lacking is the integration of those molecular models and structures with a comparative evolutionary approach. This study addresses the evolution on both Lep and LepR, through combining the known structures for both, the known mutagenic data for Lep and LepR, docking predictions between them and finally phylogenetic and sequence comparisons mapped onto the interaction of the two proteins. This approach reveals a highly conserved interaction between the CRH2 of LepR with Lep, with higher levels of variation in this interaction in fish. Fish contain multiple Lep proteins expressed from multiple Ob genes, resulting from gene duplications. This suggests reduced evolutionary pressure on fish Lep, which may allow for interaction with multiple (unidentified) receptors and/or multiple binding confirmations of Lep to the LepR in fishes.
2 Methods
2.1 Sequences and structures of Lep and LepR
Far fewer LepR sequences are determined than Lep, therefore we began by identifying all sequences available for the LepR, followed by obtaining the sequence of Lep for those species (Table S1). All species with known LepR sequences but not Lep (Anser anser, Dromaius novaehollandiae, Taeniopygia guttata, Oryzias melastigma) were removed from analysis. Functional data for Lep and LepR were obtained through either Uniprot (http://www.uniprot.org/) or from published literature. For sequence alignments, ClustalW2 was used with default settings of the EBI (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Phylogenetic analysis was performed on leptin protein sequences included in the study (Table S1) along with additional representative leptin sequences obtained from GenBank. Two alignments representing fish and terrestrial vertebrates were constructed and maximum likelihood analysis performed using MEGA5 [39] (http://www.megasoftware.net/) with default evolutionary assumptions using the JTT matrix-based model [22].
2.2 Structural docking of Lep and LepR
Known structures for human Lep (PDB: 1ax8) and LepR (PDB: 3v6o) were downloaded from the PDB, reduced to a single structure taking the first of the models, and all water molecules removed. Each structure was placed into a simulation square, filled with water to a density of 0.998g/mL and energy minimized with Amber03 force field [11] multiple times. Amino acids revealed to be highly or functionally conserved, known mutagenic functionality, post translational modifications and known natural variants in humans were manually shown on the structures of each. Using the known data, and amino acid properties, Lep was manually docked to the CRH2 domain of LepR. This was then placed into a new simulation box (x,y,z=94,81,55), water added and five energy minimizations performed. All structures were visualized, energy minimized, and imaged with YASARA structure.
2.3 Sequence divergence on structure
To determine the sequence/structure relationship over multiple species we mapped sequence variation for each species onto the docked structure. As both structures are of human, human Lep and LepR were used as the reference to be compared to. The sequence for each species was aligned with the human, exported as a fasta file, and color gradient applied from grey (conserved) to red (divergent) with the fasta file in YASARA. Amino acids found with variation on Lep and LepR within 10Å were considered for mutual evolution allowing for flexibility, dynamics and error in docking.
2.4 Lep binding energies for multiple species
To determine how amino acids affect the binding of Lep to LepR, amino acids identified to differ between human and other species in the interaction site of Lep to LepR were swapped from the human to that found in the species on both Lep and LepR. The interaction was then energy minimized as in methods section 2.2 for each species. Water was then removed and cell boundary applied. Binding energy was then calculated for Lep using YASARA.
2.5 Creation of models for fish Lep and LepR
As fish Lep have little homology with human Lep, we modeled the three fish containing two Lep molecules to address potential differences in interaction with LepR of the species. Threading models were created for one of the two Lep and the LepR using LOMETS [43]. The top three models for Lep and LepR of each species were aligned to the human docked Lep/LepR using the MUSTANG algorithm [23]. The model with the lowest root-mean squared deviation (RMSD) to the human was used for all further studies. The docked Lep and LepR for each species were then energy minimized five times. For the second Lep of the species containing two Lep, homology modeling was performed on the already docked threaded models by swapping those amino acids recognized in sequence and structures to vary in the active site followed by five rounds of energy minimization. Energy calculations (sum of all energy components) and structural variation (averaged carbon alpha RMSD) were measured with YASARA.
3 Results
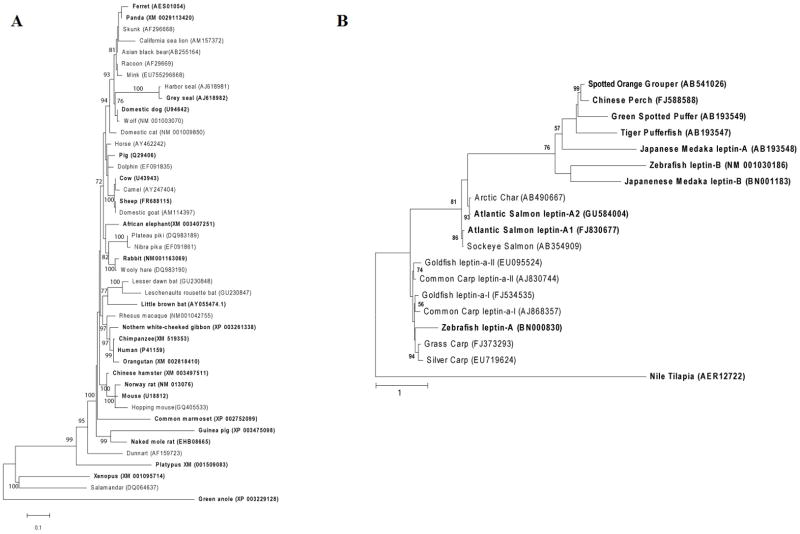
A total of 35 species have both Lep and LepR sequences available in partial or full that cover the interaction between the two (Table S1). These sequences consist of mostly mammals (24 total), a frog, a lizard, six fish and three birds. Phylogenetic analysis of terrestrial vertebrates included 44 leptin sequences excluding avian sequences which are not thought to be valid sequence (see section 4.3). Lep protein phylogeny (Figure 1A) of terrestrial vertebrates reveals a typical phylogeny of mammals but a growing number of divergent sequences compared to what was known just a few years ago. In addition to the doubling of non-mammalian representatives, the addition of seal, bat, marmoset and guinea pig sequences reported in the last five years have greatly expanded our appreciation for the variation possible among terrestrial vertebrates. A total of 19 known sequences of fish Lep were aligned and a phylogeny generated (Figure 1B). This poorly resolved phylogeny shows the high divergence and complicated evolution of fish Leps. The deeply divergent A and B orthologs of Lep have been demonstrated to occur in some fish lineages (Zebrafish and Pufferfish) while the A form is thought to have experienced a duplication events much more recently in some lineages (Salmon, Goldfish and common Carp). The function and origin of each Lep protein in fish has not been determined. The recently released Nile tilapia sequence is the most divergent fish leptin yet found and our phylogeny does nothing to resolve its relationships with other fish leptin proteins.
Figure 1.
A) Evolutionary history of 45 terrestrial vertebrate leptin protein sequences (215 total positions) was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The tree with the highest log likelihood (−4376.6338) is shown. Numbers at nodes reflect support as a percentage of 500 bootstrap replicates with no numbers representing less than 70% support. Initial tree(s) for the heuristic search were obtained by maximum parsimony method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Taxa in bold were used in the binding site analyses. B) Evolutionary history of 19 fish leptin protein sequences (223 total positions) was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The unrooted tree with the highest log likelihood (−4704.7164) is shown. Initial tree(s) for the heuristic search were obtained by maximum parsimony method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers at nodes reflect support from 500 bootstrap replicates with no number representing less than 50% bootstrap support. Leptin proteins are indicated as they have been identified in the literature. Taxa in bold were used in the binding site analyses.
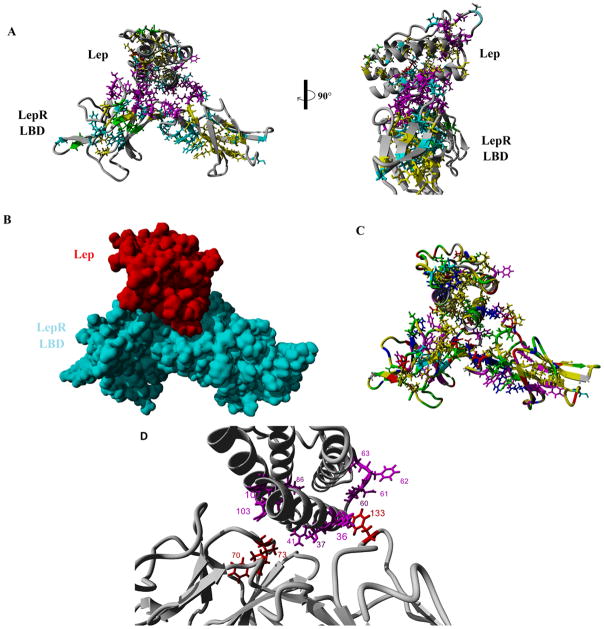
Literature and Uniprot analysis of known functional residues revealed many sites on both Lep (Table 1) and LepR (Table 2). Using the numbering of the human Lep (Figure S1) and LepR (Figure S2), sequence alignments reveal amino acids that are highly conserved (cyan), functionally conserved (yellow), found as a natural variants (red), posttranslationally modified (green), or has known mutagenic functionality (magenta). Because of the increased divergence of Lep in fish, we chose to not include them in the initial analysis of sequence conservation for Lep. Twenty one amino acids are highly conserved in Lep, with a conserved disulfide bridge, and 26 functionally conserved amino acids. Of the known mutagenic data (22 amino acids) that lead to altered protein function, ten amino acids are conserved in the sequence alignments (amino acids 36, 37, 41, 60, 61, 62, 63, 86, 103, and 107). Additionally the known natural variant at amino acid 105 associated with obesity and hypogonadism [37] is conserved. Mapping these functional properties onto the known structure of Lep reveals a conserved hydrophobic core between the four helices with the disulfide bridge shown in the front on the left half and the right side on the 90 degree rotation of Figure 2A. Two functional sites are found on the surface of Lep, 1 and 2 (Figure 2A).
Table 1.
Functional data on Leptin.
| AA | Species | Original AA | AA in human Leptin | Human | Importance | Reference |
|---|---|---|---|---|---|---|
| Mutational results | ||||||
| 15 | Human | Lys | 36 | Lys | Mutation to Ser decreased binding | [31] |
| 16 | Human | Thr | 37 | Thr | Mutation to Asn decreased binding | [31] |
| 20 | Human | Arg | 41 | Arg | Mutation to Asn decreased binding | [31] |
| 20 | Human | Arg | 41 | Arg | Mutation to Asn decreased signaling | [31] |
| 28 | Human | Gln | 49 | Gln | Mutation to Ser increased signaling | [31] |
| 29/30/31 | Human | Ser/Val/Ser | 50/51/52 | Ser/Val/Ser | Mutations to Gln/Gln/Asn decreased signaling | [31] |
| 34/35 | Human | Gln/Arg | 55/56 | Gln/Arg | Mutations to Ser/Ser decreased signaling | [31] |
| 39/40/41 | Human | Leu/Asp/Phe | 60/61/62 | Leu/Asp/Phe | Mutation to Ala/Ala/Ala decreased FRET activation with receptor | [29] |
| 39/40/41 | Ovine | Leu/Asp/Phe | 60/61/62 | Mutation to Ala/Ala/Ala decreased FRET activation with receptor | [29] | |
| 39/40 | Human | Leu/Asp | 60/61 | Mutations to Ala/Ala served as antagonist blocking leptin response | [29] | |
| 41/42 | Human | Phe/Ile | 61/62 | Mutations to Ala/Ala served as antagonist blocking leptin response | [29] | |
| 39/40/41 | Human | Leu/Asp/Phe | 60/61/62 | Mutations to Ala/Ala/Ala served as antagonist blocking leptin response | [29] | |
| 39/40/41/42 | Human | Leu/Asp/Phe/Ile | 60/61/62/63 | Mutations to Ala/Ala/Ala/Ala served as antagonist blocking leptin response | [29] | |
| 41 | Human | Phe | 62 | Phe | Mutation to Ser decreased signaling | [31] |
| 75 | Human | Gln | 96 | Gln | Mutation to Ser increased signaling | [31] |
| 75 | Human | Gln | 96 | Gln | Mutation to Ser decreased binding | [31] |
| 82/85 | Human | Asn/Asp | 103/106 | Asn/Asp | Mutation to Ser/Ser decreased binding | [31] |
| 86 | Human | Leu | 107 | Leu | Mutation to Ala decreased binding | [31] |
| 86 | Mouse | Leu | 107 | Leu | Mutation to N or Q decrease activation | [20] |
| 115 | Human | Glu | 136 | Glu | Mutations to Ser decreased signaling | [31] |
| 117 | Human | Ser | 138 | Ser | Mutations to Gln decreased signaling | [31] |
| 120/121 | Human | Ser/Thr | 141/142 | Ser/Thr | Mutation to Ala decreased signaling, strong antagonist of leptin receptor | [31] |
| 120/121 | Human | Ser/Thr | 141/142 | Mutations to Ala/Ala served as antagonist blocking leptin response | [29] | |
| 122 | Human | Glu | 143 | Glu | Mutations to Ser decreased signaling | [31] |
| 138/139/142 | Human | Gln/Gln/Val | 159/160/161 | Gln/Gln/Val | Mutations to Ser/Ser/Ala decreased signaling | [31] |
| Post-translational modifications | ||||||
| 117/167 | Cys | 117/167 | Cys | Disulfide bridge | [44] | |
| Natural Variants | ||||||
| 105 | Human | Arg | 105 | Arg | Change to Trp associated with obesity and hypogonadism | [37] |
Table 2.
Functional data on the Leptin binding domain (LBD) of the Leptin receptor.
| AA in LBD (431–633 of human) | Species | Original AA | AA in Human LepR | Human | Importance | Reference |
|---|---|---|---|---|---|---|
| Mutational results | ||||||
| 11 | Chicken | Tyr | 441 | Tyr | Mut to Ala decreases binding | [30] |
| 11/70 | Chicken | Tyr/Phe | 441/500 | Tyr/Phe | Mut to Ala abolished binding | [30] |
| 38 | Chicken | Arg | 468 | Arg | Mut to Ala decreases binding | [30] |
| 38/39/40 | Chicken | Arg/Ser/Lys | 468/469/470 | Arg/Ser/Ser | Mut to Ala decreases binding | [30] |
| 70 | Chicken | Phe | 500 | Phe | Mut to Ala abolished binding | [30] |
| 70 | Chicken | Phe | 500 | Phe | Contact-surface analysis showed high importance to interaction | [30] |
| 73 | Mouse | Ile | 503 | Ile | Mut to A or S reduced activation around 25% | [20] |
| 73/74 | Chicken | Val/Phe | 503/504 | Ile/Phe | Mut to Ala abolished binding | [30] |
| 73/74/75/76 | Chicken | Val/Phe/Leu/Leu | 503/504/505/506 | Ile/Phe/Leu/Leu | Mut to Ala abolished binding | [30] |
| 74 | Chicken | Phe | 504 | Phe | Contact-surface analysis showed high importance to interaction | [30] |
| 74 | Mouse | Phe | 504 | Phe | Mut to A or S reduced activation around 25% | [20] |
| 75 | Mouse | Leu | 505 | Leu | Mut to A or S reduced activation around 25% | [20] |
| 75/76 | Chicken | Leu/Leu | 505/506 | Leu/Leu | Mut to Ala abolished binding | [30] |
| 76 | Chicken | Leu | 506 | Leu | Contact-surface analysis showed high importance to interaction | [30] |
| 76 | Mouse | Leu | 506 | Leu | Mut to A reduced activation around 25% | [20] |
| 77 | Mouse | Ser | 507 | Ser | Mut to A reduced activation around 25% | [20] |
| 100 | Mouse | Leu | 530 | Leu | Mut to A reduce signal transduction around 50% | [20] |
| 100 | Chicken | Val | 530 | Leu | Mut to Ala decreases binding | [30] |
| 102 | Mouse | Asp | 532 | Asp | Mut to A reduce signal transduction around 50% | [20] |
| 102 | Chicken | Ala | 532 | Asp | Mut to Asp decreased binding (humanizes chicken LepR) | [30] |
| 102/103 | Chicken | Ala/Asp | 532/533 | Asp/Ser | Mut to Ala abolished binding | [30] |
| 103 | Mouse | Ser | 533 | Ser | Mut to A reduce signal transduction around 75% | [20] |
| 132/133 | Chicken | Val/Phe | 562/563 | Val/Phe | Mut to Ala decreases binding | [30] |
| 133 | Chicken | Phe | 563 | Phe | Mut to Ala decreases binding | [30] |
| 135 | Mouse | Glu | 565 | Glu | Mut to A reduce signal transduction around 50% | [20] |
| 136 | Mouse | Asn | 566 | Asn | Mut to A reduce signal transduction around 50% | [20] |
| 137 | Mouse | Asn | 567 | Asn | Mut to A reduce signal transduction around 50% | [20] |
| 164 | Mouse | Lys | 594 | Lys | Mut to A reduce signal transduction around 50% | [20] |
| 185 | Mouse | Arg | 615 | Arg | Mut to A reduce signal transduction around 75% | [20] |
| 187 | Mouse | Asp | 617 | Asp | Mut to A reduced activation around 25% | [20] |
| Post-translational modifications | ||||||
| 6/17 | Human | Cys | 436/447 | Cys | Disulfide bridge | PDB: 3V6o [4] |
| 43/98 | Human | Cys | 473/528 | Cys | Disulfide bridge | [19] |
| 58/68 | Human | Cys | 488/498 | Cys | Disulfide bridge | PDB: 3V6o [4] |
| 58/68 | Human | Cys | 488/498 | Cys | Disulfide bridge | [19] |
| 86 | Human | Asn | 516 | Asn | N-linked(GlcNAc…) | [19] |
| 194 | Human | Asn | 624 | Asn | N-linked(GlcNAc…) | [19] |
| Natural Variants | ||||||
| 73 | Human | Ile | 503 | Ile | Change to Val | NCBI |
Figure 2.
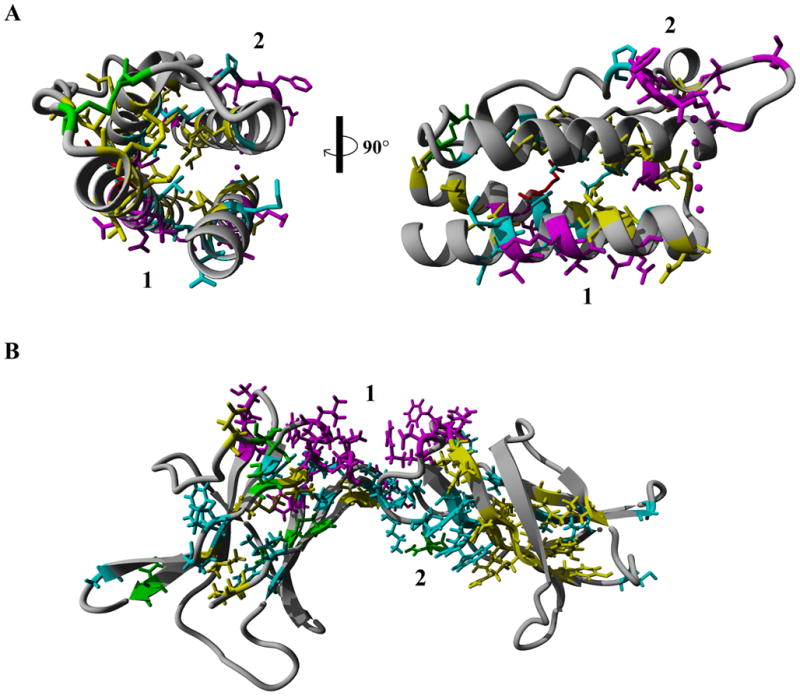
Sequence conservation shown on structures of either leptin (A) or the leptin receptor (B) with the multiple conserved sites. The first conserved site (1) of each represents the leptin/leptin receptor interface.
The LepR sequence alignments, including fish, had 32 conserved amino acids and 25 functionally conserved. Two disulfide bridges are conserved, with an additional Asn (amino acid 194) known to be N-glycosylated [19] highly conserved. Of the known mutations (21 amino acids), three are found as conserved (amino acids 70, 73, and 133). Conserved and mutated amino acids shown on the structure of LepR, reveal highly conserved packing of the beta sheets while also having two conserved binding sites, 1 and 2 (Figure 2B).
Docking of Lep and LepR is believed to be through the first identified site, 1, of each in figure 2. Multiple loops of the LepR interact with helices A and C of Lep (Figure 3A–B). Coloring the docking based on amino acid properties, reveals a hydrophobic packing (yellow) with several salt bridges (red and blue) and hydrogen bonding through hydrophilic (green) amino acids stabilizing the interaction (Figure 3C). Highly conserved known mutagenic data (Lep= 36, 37, 41, 60, 61, 62, 63, 86, 103, and 107; LepR= 70, 73, and 133) mostly fall into this binding pocket between Lep and LepR (Figure 3D).
Figure 3.
A) Functional docking of Lep and LepR from figure 2. B) Van der Waals surface of each. C) Amino acid properties (Blue=Arg, His, Lys; Red=Asp, Glu; Yellow= Ala, Val, Ile, Leu, Met, Pro; Magenta= Phe, Tyr, Trp, Green= Ser, Thr, Asn, Gln; Cyan= Cys). D) Conserved functional amino acids shown on the docking.
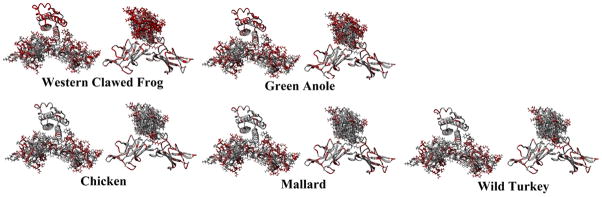
Taking this docking and looking at each individual species we can see if any amino acid variations are found in Lep and LepR that may stabilize binding and suggest a co-evolutionary change (Figure 4–5). A breakdown of the amino acids that vary can be seen in table 3, with amino acids with significantly altered functional groups shown in red. Species closely related to human have few to no variations in the regions interacting between Lep and LepR, while some mammals have increasing variation (Figure 4). Binding energies are similar as human for all species except the grey seal, chicken, platypus and the green anole (table 3). The three bird species have high variation in the LepR, but not in Lep; yet both Lep and LepR vary significantly in lizard and frog (Figure 5).
Figure 4.
Mammalian Leptin and Leptin receptor variations from human sequence. For each species the figure to the left shows the amino acids on the receptor while on the right shows leptin amino acids. Those in red differ from human.
Figure 5.
Non-mammalian Leptin and Leptin receptor variations from human sequence. For each species the figure to the left shows the amino acids on the receptor while on the right shows leptin amino acids. Those in red differ from human.
Table 3.
Amino acid variations in the docking of Lep and LepR for multiple species compared to human
| Lep Amino acid variations in the binding with LepR | LepR Amino acid variations in the binding with Lep | Binding Energy (KJ/mol) | |
|---|---|---|---|
| Human | - | 1979.66 | |
| Sumatran orangutan | - | - | 1979.66 |
| Common Chimpanzee | - | - | 1979.66 |
| Northern White-Cheeked Gibbon | - | - | 1979.66 |
| Rhesus macaque | D29S, V110L | Y11H, L186S | 1648.24 |
| Crab-Eating Macaque | D29S, V110L | Y11H, L186S | 1648.24 |
| Giant Panda | V110L, F113S | L100V | 1976.03 |
| Horse | V110L, F113S | L100V, L186Q | 1960.17 |
| Sheep | V110L, F113A | L100I | 1976.91 |
| Cattle | V110L, F113A | L100I, R185S | 1968.73 |
| Little Brown Bat | R92G, V110L, F113S | L100V, V132I | 2199.13 |
| Ferret | incomplete sequence | L100V | - |
| European Rabbit | N43S, V110L, F113S | - | 1975.9 |
| African elephant | N43S, V110L, F113A | L100V | 1969.57 |
| Pig | K26R, N43S, V110L, F113S | L100I | 1977.58 |
| Grey Seal | K26R, E102A, D106A, V110L, F113S | L100V, V132I | 1404.81 |
| Common Marmoset | K26R, T40A, R92G, D106A, V110L, F113A | Y11S, S40R, D102N, K130N, N136T | 2500.01 |
| Dog | T40A, V110L, F113S | L100V | 1968.61 |
| Chinese hamster | V110L, F113S | - | 1976.03 |
| Naked Mole rat | T40S, E102D, D106N, V110M, F113S | Q71K, N137K | 2217.13 |
| Guinea Pig | V27L, T33F, T40N, N43R, R92S, D106S, V110M, F113S | L76F, L100T, S102A, K130T, V132S, P134S, N137Q | 1711.5 |
| Rat | R92Q, N99H, V110L | - | 2211.09 |
| Mouse | R92Q, V110L | - | 2186.01 |
| Platypus | V27I, D29A, N43I, R92P, D106S, V110L, F113T | F74Y, L100V, K130R, V132T, N137H | 1477.96 |
| Mallard | R92Q, N99D, V110L | S40K, L41I, F74S, L100V, D102A, S103D, K130N, P134T, E135N, N136D, N137D, R185A | 2146.29 |
| Chicken | K26I, V27F, R92Q, V110L | R38M, S40K, L41I, L100V, D102A, S103D, K130N, P134T, E135N, N136D, N137D, R185A | 1055.16 |
| Wild Turkey | K26I, V27F, R92Q, V110L | S40K, L41I, L100V, D102A, S103D, K130N, P134A, E135N,N136D, N137D, R185A | 1685.26 |
| Green anole | V27T, D29A, T33M, K36R, T40S, N43Q,R92E, N99Y, D106S, V110L, F113A | Y11N, T13Q, L41V, F74Y, D102K, K130N, V132E, E135K, N136Y, N137D, R185L, L186T | 1403.94 |
| Western Clawed Frog | K26R, D29N, T33M, K36R, N40Q, R92E, N99S, D106S | Y11N, L12Q, T13K, Q71E, F74H, L76V, D102I, K130R, V132A, F133L, E135S, N136T, N137D, L186T | 1869.87 |
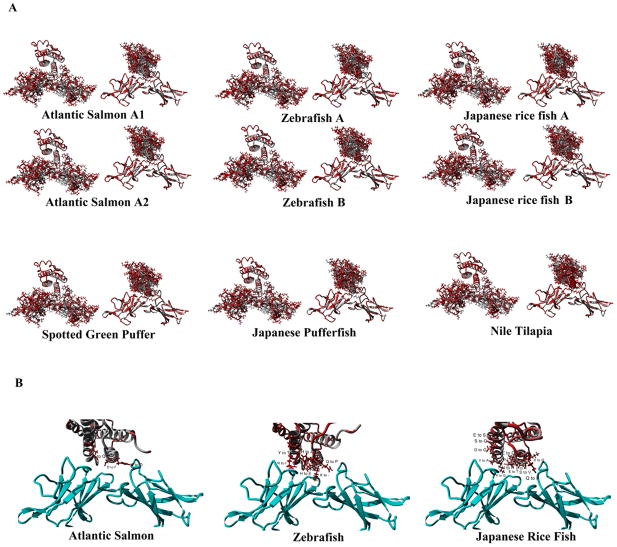
Fish Leps have some conservation in the interior of the four helixes and the disulfide bridge while containing minimal conservation on the surface (Figure 6A), including the region that interacts with LepR. Additionally, few amino acids are conserved in the LBD of LepR. Three species (Salmon, Zebrafish, and Japanese Rice Fish) that have two known Lep proteins also have known LepR sequences allowing us to address which has a higher probability of binding to the LepR of the species. Clearly, all three species have variation at the interface of Lep/LepR interaction (Figure 6B). In Salmon, three amino acids differ between the presumably recently diverged A-1 and A-2 Lep proteins with H/D, D/F, L/Q differences (Figure S5A). Zebrafish A and B proteins show more differences with H/A, Q/P, H/E, K/-, N/-, L/I, R/I, and Y/T changes (Figure S5B). The Japanese rice fish contains the most differences with 13, Q/I, E/Q, S/V, N/H, D/N, I/L, K/Q, E/T, K/Q, V/A, D/G, S/Q, E/S (Figure S5E).
Figure 6.
A) Fish Leptin and Leptin receptor variations from human sequence. For each species the figure to the left shows the amino acids on the receptor while on the right shows leptin amino acids. Those in red differ from human. B) Differences between the two forms of Lep in the three species of fish with two known and the LepR.
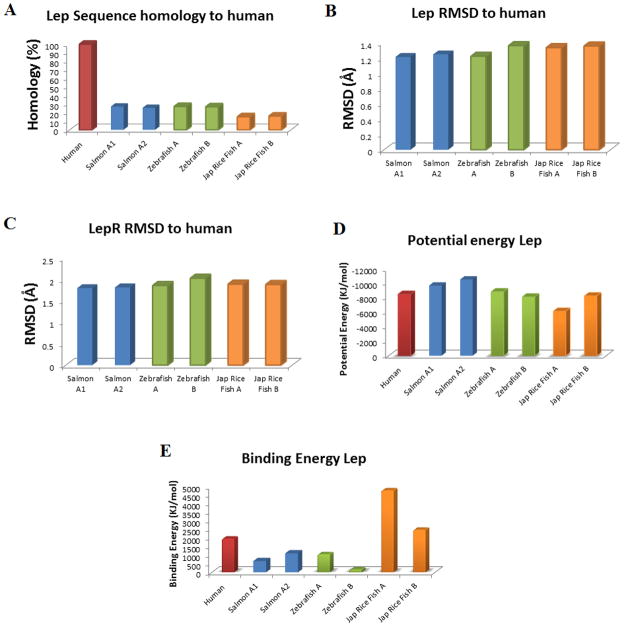
Sequence homology to human is 27/26% for LepA1/LepA2 of Salmon, 27% for both LepA and LepB of Zebrafish and 15/16% for LepA/LepB of the Japanese rice fish (Figure 7A). Docking of Lep to LepR for each species gave a RMSD of 1.2 to 1.4 for all models of Lep and 1.8–2 for LepR when compared with the human structures (Figure 7B–C). All Lep structures provided potential energies similar to that of human (Figure 7D), while the binding energies were different (Figure 7E). For our docking of human Lep to LepR there was a binding energy of 1975kJ/mol. In Salmon the A2 form of Lep had a higher binding with 1115 kJ/mol compared to the A1 form with 662 kJ/mol. Both the A forms for Zebrafish and the Japanese rice fish had higher binding (1056/4792 kJ/mol respectively), with the B forms significantly lower (158/2485 kJ/mol).
Figure 7.
A) Sequence homology of multiple fish Lep with human Lep. B–C)RMSD of each Lep to human for either Lep (B) or LepR (C). D–E) Potential energy (D) and binding energy (E) of Lep when bound to the LepR for human and fish with two known Lep.
4 Discussion
Lep and LepR sequences are highly conserved among mammals; however, Lep exhibits considerable primary protein sequence variation among other vertebrates (especially in fish). Despite this, structural analyses of Lep have suggested high functional homology. Eighteen years after its discovery in mouse, we now have leptin and leptin receptor representatives from all major branches of the vertebrate phylogeny (with the caveat of bird Lep) with coverage of considerable phylogenetic breadth of mammals (Figure 1A). The addition of the LepR protein structure [4] allows us to examine the evolution of these interactions through a comparative perspective. We find that the interaction described by Carpenter et al. [4] between human Lep and LepR reflects a strongly conserved interaction from fish to mammals.
4.1 Conserved sites on Lep and LepR
For both Lep and LepR, the first identified conserved site is where the two proteins have been proposed to interact. In Lep a second site is found, which is believed to be the location that contacts the Ig-like domain of a second LepR, inducing dimerization and potential higher order interactions such as the 2:4 (Lep:LepR) hexameric structure [32]. For the LepR, the second conserved site has not been characterized to the best of our knowledge. This site is composed of D9, K106, Q139, Q141, R143, Q180, R182, W192, W195, N194 all of which surround amino acid N194 which is known to be N-glycosylated [19]. Due to the number of charged amino acids and the two tryptophans, we propose that this site serves as a docking location for another protein, which may be regulated through the N-glycosylation of amino acid 194 or perform the N-glycosylation. Future mutagenic and biochemical experiments need to be performed on this site to show its importance in physiology. This site may serve for proper leptin signaling however, it is too preliminary to speculate as to what proteins may bind to this site and how it functions. In this paper we have focused on the CRH2 domain of LepR. Future analysis can be performed on the IG-like and other domains, with interest into domains that are contained in the multiple isoforms of the LepR.
4.2 Highly conserved terrestrial vertebrate Lep and LepR binding domains
Terrestrial vertebrate Lep and LepR binding domains are highly conserved. Variation in primary sequences among terrestrial vertebrates is reflected in some variation in the binding domains of Lep and LepR. In most cases that variation can be explained by the co-evolution of sequences to maintain the high stability of this interaction in evolutionary history. For example, in both macaque species of Lep there is a D29S change while on LepR there is a L186S change. The distance from the carbon beta of each amino acid is 8.3Å. The Asp (29) of Lep appears to interact with Lys (26) of Lep and Asn (137) of LepR. The Leu (186) of LepR changed to Ser may stabilize the loss of one of the two oxygen groups of D29S at the end of the amino acid. Additionally, in platypus, green anole and Xenopus, amino acid 29 of Lep is found to vary (Ala, Ala, and Asn respectively) with an amino acid within 10Å on LepR covariant (N137H, N137D, N137D). Amino acid 33 of Lep is also found close to this site in which Guinea pig, green anole and Xenopus are variant from human (T33F, T33M, T33M). Guinea pig, possessing the most divergent mammalian Lep/LepR sequences, also contains variation at amino acid 137 (N137Q) on LepR. It is likely that these amino acid variations found on both Lep and LepR aid in stabilizing the interaction between Lep and LepR and thus a possible site of coevolution. In both the green anole and Xenopus other amino acids are found in close proximity to each other and are possibly linked in evolution (K36R of Lep to E135K/S of LepR and N99Y/S of Lep to F74Y/H). The binding energy between Lep and LepR is not altered for most of the species in this paper, further supporting the proper interaction between Lep and LepR. This would suggest also that Lep would preferentially bind to the LepR CRH2 domain. In some species, such as the grey seal, platypus, and the green anole (all containing a decreased binding energy), the role of other domains or an alternative conformation may stabilize binding. In addition the interaction with more than one LepR through the Ig-like domain interacting with the site 2 on Lep (figure 2) may stabilize binding in these species.
4.3 Further evidence against Lep in birds
Docking analysis can speak to the debate regarding the cloning and identification of the leptin gene in chickens and other birds. This debate stems from an attempt to rectify the scarcity of non-mammalian leptin sequences through the identification and characterization of the first known avian leptin. Taouis, et al. [40] published the first avian-isolated leptin data in 1998 during their study of lipogenesis in chickens. Sequence was amplified using specially designed primers based on known mouse leptin sequences and their results yielded a product that apparently demonstrated 97%, 96% and 83% coding sequence similarity to mice, rats and humans, respectively. After the publication of this discovery, labs in Scotland and Israel sought to verify this sequence. Using up to 14 designed primers (as well as the primers published by Taouis, et al. [40]) and freshly isolated DNA and reverse-transcribed RNA sources from different varieties of chickens and various tissues sources, these labs attempted to amplify avian leptin [14]. The combined techniques of RT-PCR, northern hybridization and Southern hybridization (all with valid mammalian controls) all failed to detect the leptin sequence from either freshly isolated DNA or RNA in tissues from any variety of chicken [14]. Finally, the Taouis reported sequence could not be identified in the solved chicken genome [34]. These results have led many in the field to question not only the existence of leptin in birds, but the high sequence similarity between the published bird sequences to mice. Whereas high sequence similarities of that nature are expected between related mammals, it is atypical when comparing such divergent lineages as mammals and birds (especially in a gene that is so divergent within taxa). Since the original publication of the chicken leptin sequence, there have been no successful attempts to isolate the leptin gene in any bird even in labs experienced in the identification of avian homologue sequences [12, 38]. With three genomes of birds (chicken, turkey and zebra finch) and no Lep identified in any of these, Lep is either nonexistent or found in poorly resolved regions of the genome.
It can be seen in our sequence alignments that there are several amino acids found in all three bird species that are not found in other species. However, these are located in the first few amino acids before the splicing gap on leptin. After this, the sequences share 100% homology with mouse. Modeling of the interaction of Lep with LepR, clearly demonstrates an increased divergence in the LepR where Lep should bind, with no corresponding Lep variation to account for this (table 3). If the bird Lep sequences were true, binding may be reduced to the LepR and thus forces would be diminished in conserving the amino acid sequence seen in birds. As the chicken Lep binding energy in table 3 shows a 50% lower binding energy, it adds further evidence that this sequence is artifactual. Therefore, these data lend strong support to the prior work that concluded that these sequences are not from birds, but most likely represent contamination in cloning. The turkey and mallard sequences are database entries without peer-reviewed publications, and as such their validity is difficult to judge. Given that they show much lower rates of evolution than the bird receptor sequences, and much lower rates of evolution than any other vertebrate Lep, we assert they are likely artifactual even though they were sequenced from a separate lab than the chicken Lep.
4.4 Fish Lep and LepR interactions suggest possible divergent functionality of the multiple Lep proteins
Typically gene orthologues are identified via enough sequence similarity so that portions of sequence can be recognized in a multiple alignment, but also enough divergence to indicate the evolutionary distance between the two species. This approach worked well to identify leptin in all mammalian species investigated, but failed for other vertebrates. Although Londraville’s group [21] identified a leptin-like protein in fishes, it was not until Kurokawa’s lab identified leptin via gene-synteny in the Fugu genome [24] that a leptin gene was definitively established in fish. Fugu Lep is only 13% identical to human leptin, and sequence is highly variable (20–25%) even among fish species (Fig. 1B and Fig 7). In addition, most fish express two Lep proteins resulting from gene duplication, whereas mammals are only known to express one [6]. Despite their extreme sequence divergence, threading algorithms predict similar tertiary structures for all vertebrate Leps [16, 6, figures 4 and 5), and leptin receptors are considerably more conserved [26]. Which of the Lep proteins in fish is the common ancestor of mammalian leptin is debatable. Although each fish species has been proposed to have only two proteins, the two are not necessarily orthologous among species (e.g. Salmon A-I and A-II are clearly not orthologous to Zebrafish A and B, Figure 1B). Difficulties in assigning sequence homology and the lack of thorough sampling in fish result in a tentative and unresolved phylogeny of fish Lep proteins (Figure 1B). For example, the single existing partial sequence of Nile Tilapia only shows 10% amino acid homology while having a predicted high structural homology.
It can be speculated that the multiple Lep found in fish may bind through altered hydrophobic interactions in a temperature dependent manner. As there is a high variation in the binding interface of Lep to LepR between fish and mammals (Figure 6A), the role of hydrophobic interactions may aid in the temperature differences in fish not experienced by other vertebrates. In Salmon there is a loss of hydrophobic amino acids on LepR comparing human to either the A1 or A2 form of leptin (Figure S6). In Zebrafish there is a greater number of hydrophobic amino acids relative to human (Figure S7) suggesting more temperature controlled interactions. There is also a variation between the Zebrafish A and B forms and Salmon A1 and A2 forms in hydrophobic amino acids. Based on binding energies, we suggest that the multiple forms of Lep in Salmon, Zebrafish and the Japanese rice fish are functionally different (note that the binding energy of Zebrafish and medaka Lep A is an order of magnitude higher than Lep B in Figure 7). Duplication events in the fish Lep could allow for either divergence or neofunctionality, with different protein affinities to either LepR or other unidentified proteins that interact with Lep. This could allow for selective binding of either A or B Lep in different molecular environments based on temperature, other molecular interactions or relative concentrations of the respective Lep protein.
5 Conclusions
The interaction between Lep and LepR is conserved in terrestrial vertebrates, with an increased variation in fish. Mammalian Lep and LepR are highly conserved with few variations. Previously published and debated sequences of chicken Lep would have an altered binding to the LepR, which serves as an additional argument against this sequences being accurate. In fish the multiple Lep proteins, resulting from gene duplication, have variation found in the binding between Lep with LepR, which are predicted to have different binding energies. The Lep gene duplication may allow Lep in fish to bind in different confirmations or to additional proteins.
Supplementary Material
S1 Sequence alignments of Lep
S2 Sequence alignments of LepR
S3 Active site variations of Lep shown on sequence alignments
S4 Active site variations of LepR shown on sequence alignments
S5 Fish Sequence alignments to human
S6 Amino acid properties on Lep-LepR of Salmon
S7 Amino acid properties on Lep-LepR of Zebrafish
Highlights.
Interaction between leptin and the leptin receptor is conserved in land vertebrates.
Avian leptin sequences appear artifactual as evolution rates of Lep do not match LepR.
Fish contain multiple amino acid variations in the interaction of Lep to LepR.
Predicted binding for Lep are stronger for A vs. B proteins in zebrafish.
Acknowledgments
We thank Amy Milsted for support throughout the project. JWP is funded by an AHA predoctoral fellowship. JWP, HB and DLC have bioinformatics funds from the Ohio board of regents Choose Ohio First. Partially supported by NIH DK079282-01A1 to RLL.
Abbreviations
- Lep
Leptin
- LepR
Leptin receptor
- CRH
cytokine receptor homologous domains
- LBD
Leptin binding domain
- RMSD
root-mean squared deviation
Footnotes
Contributions: JWP performed all modeling, sequence alignment comparisons, and wrote the manuscript. HB and RJD performed phylogenetics and advised on sequence comparisons. DLC and RLL advised on fish leptin and helped in analysis of understanding of sequence to structure relationship in fish. All authors contributed to manuscript preparation and approve the final draft.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ashwell CM, Czerwinski SM, Brocht DM, McMurtry JP. Hormonal regulation of leptin expression in broiler chickens. Am J Physiol. 1999;276:R226–32. doi: 10.1152/ajpregu.1999.276.1.R226. [DOI] [PubMed] [Google Scholar]
- 2.Belgardt BF, Brüning JC. CNS leptin and insulin action in the control of energy homeostasis. Ann N Y Acad Sci. 2010;1212:97–113. doi: 10.1111/j.1749-6632.2010.05799.x. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter LR, Farruggella TJ, Symes A, Karow ML, Yancopoulos GD, Stahl N. Enhancing leptin response by preventing SH2-containing phosphatase 2 interaction with Ob receptor. Proc Natl Acad Sci USA. 1998;95:6061–6. doi: 10.1073/pnas.95.11.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpenter B, Hemsworth GR, Wu Z, Maamra M, Strasburger CJ, Ross RJ, Artymiuk PJ. Structure of the human obesity receptor leptin-binding domain reveals the mechanism of leptin antagonism by a monoclonal antibody. Structure. 2012;20:487–97. doi: 10.1016/j.str.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Considine RV, Sinha MK, Heiman ML, Kriauciunas A, Stephens TW, Nyce MR, et al. Serum immunoreactive-leptin concentrations in normal-weight and obese humans. N Engl J Med. 1996;334:292–5. doi: 10.1056/NEJM199602013340503. [DOI] [PubMed] [Google Scholar]
- 6.Copeland DL, Duff RJ, Liu Q, Prokop J, Londraville RL. Leptin in teleost fishes: an argument for comparative study. Front Physiol. 2011;2:26. doi: 10.3389/fphys.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denver RJ, Bonett RM, Boorse GC. Evolution of leptin structure and function. Neuroendocrinology. 2011;94:21–38. doi: 10.1159/000328435. [DOI] [PubMed] [Google Scholar]
- 8.Donato J, Jr, Cravo RM, Frazão R, Elias CF. Hypothalamic sites of leptin action linking metabolism and reproduction. Neuroendocrinology. 2011;93:9–18. doi: 10.1159/000322472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon C, Drouin G, Trudeau VL, Moon TW. Molecular evolution of leptin. Gen Comp Endocrinol. 2001;124:188–98. doi: 10.1006/gcen.2001.7701. [DOI] [PubMed] [Google Scholar]
- 10.Driessler F, Baldock PA. Hypothalamic regulation of bone. J Mol Endocrinol. 2010;45:175–81. doi: 10.1677/JME-10-0015. [DOI] [PubMed] [Google Scholar]
- 11.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 12.Dunn IC, Chen Y, Hook C, Sharp PJ, Sang HM. Characterization of the chicken preprogonadotrophin-releasing hormone-I gene. J Mol Endocrinol. 1993;11:19–29. doi: 10.1677/jme.0.0110019. [DOI] [PubMed] [Google Scholar]
- 13.Fong TM, Huang RC, Tota MR, Mao C, Smith T, Varnerin J, et al. Localization of Leptin binding domain in the leptin receptor. Mol Pharmacol. 1998;53:234–40. doi: 10.1124/mol.53.2.234. [DOI] [PubMed] [Google Scholar]
- 14.Friedman-Einat M, Boswell T, Horev G, Girishvarma G, Dunn IC, Talbot RT, Sharp PJ. The chicken leptin gene: has it been cloned? Gen Comp Endocrinol. 1999;115:354–63. doi: 10.1006/gcen.1999.7322. [DOI] [PubMed] [Google Scholar]
- 15.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–70. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 16.Gorissen M, Bernier NJ, Nabuurs SB, Flik G, Huising MO. Two divergent leptin paralogues in zebrafish (Danio rerio) that originate early in teleostean evolution. J Endocrinol. 2009;201:329–39. doi: 10.1677/JOE-09-0034. [DOI] [PubMed] [Google Scholar]
- 17.Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–6. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 18.Hammond JA, Hauton C, Bennett KA, Hall AJ. Phocid seal leptin: tertiary structure and hydrophobic receptor binding site preservation during distinct leptin gene evolution. PLoS One. 2012;7:e35395. doi: 10.1371/journal.pone.0035395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haniu M, Arakawa T, Bures EJ, Young Y, Hui JO, Rohde MF, et al. Human leptin receptor. Determination of disulfide structure and N-glycosylation sites of the extracellular domain. J Biol Chem. 1998;273:28691–9. doi: 10.1074/jbc.273.44.28691. [DOI] [PubMed] [Google Scholar]
- 20.Iserentant H, Peelman F, Defeau D, Vandekerckhove J, Zabeau L, Tavernier J. Mapping of the interface between leptin and the leptin receptor CRH2 domain. J Cell Sci. 2005;118:2519–27. doi: 10.1242/jcs.02386. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RM, Johnson TM, Londraville RL. Evidence for leptin expression in fishes. J Exp Zool. 2000;286:718–24. doi: 10.1002/(sici)1097-010x(20000601)286:7<718::aid-jez6>3.0.co;2-i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 23.Konagurthu AS, Whisstock JC, Stickey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–74. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 24.Kurokawa T, Uji S, Suzuki T. Identification of cDNA coding for a homologue to mammalian leptin from pufferfish, Takifugu rubripes. Peptides. 2005;26:745–50. doi: 10.1016/j.peptides.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, Lee JI, Friedman JM. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–5. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Chen Y, Copeland D, Ball H, Duff RJ, Rockich B, Londraville RL. Expression of leptin receptor gene in developing and adult zebrafish. Gen Comp Endocrinol. 2010;166:346–55. doi: 10.1016/j.ygcen.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 28.Maffei M, Fei H, Lee GH, Dani C, Leroy P, Zhang Y, et al. Increased expression in adipocytes of ob RNA in mice with lesions of the hypothalamus and with mutations at the db locus. Proc Natl Acad Sci USA. 1995;92:6957–60. doi: 10.1073/pnas.92.15.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niv-Spector L, Gonen-Berger D, Gourdou I, Biener E, Gussakovsky EE, Benomar Y, et al. Identification of the hydrophobic strand in the A-B loop of leptin as a major binding site II: implications for large-scale preparation of potent recombinant human and ovine leptin antagonists. Biochem J. 2005;391:221–30. doi: 10.1042/BJ20050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niv-Spector L, Raver N, Friedman-Einat M, Grosclaude J, Gussakovsky EE, Livnah O, Gertler A. Mapping leptin-interacting sites in recombinant leptin-binding domain (LBD) subcloned from chicken leptin receptor. Biochem J. 2005;390:475–84. doi: 10.1042/BJ20050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peelman F, Beneden KV, Zabeau L, Iserentant H, Ulrichts P, Defeau D, et al. Mapping of the leptin binding sites and design of a leptin antagonist. J Biol Chem. 2004;279:41038–46. doi: 10.1074/jbc.M404962200. [DOI] [PubMed] [Google Scholar]
- 32.Peelman F, Iserentant H, Smet AS, Vandekerckhove J, Zabeau L, Tavernier J. Mapping of binding site III in the leptin receptor and modeling of a hexameric leptin-leptin receptor complex. J Biol Chem. 2006;281:15496–504. doi: 10.1074/jbc.M512622200. [DOI] [PubMed] [Google Scholar]
- 33.Pénicaud L. The neural feedback loop between the brain and adipose tissues. Endocr Dev. 2010;19:84–92. doi: 10.1159/000316900. [DOI] [PubMed] [Google Scholar]
- 34.Pitel F, Monbrun C, Gellin J, Vignal A. The chicken LEP (OB) gene has not been mapped. Anim Genet. 2000;31:281. doi: 10.1046/j.1365-2052.2000.00610.x. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz MW, Seeley RJ, Campfield LA, Burn P, Baskin DG. Identification of targets of leptin action in rat hypothalamus. J Clin Invest. 1996;98:1101–6. doi: 10.1172/JCI118891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sierra-Honigmann MR, Nath AK, Murakami C, Garcia-Cardena G, Papapetropoulos A, Sessa WC, et al. Biologic action of leptin as an angiogenic factor. Science. 1998;281:1683–5. doi: 10.1126/science.281.5383.1683. [DOI] [PubMed] [Google Scholar]
- 37.Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD. A leptin missense mutation associated with hypogonadism and morbid obesity. Nat Genet. 1998;18:213–5. doi: 10.1038/ng0398-213. [DOI] [PubMed] [Google Scholar]
- 38.Talbot RT, Dunn IC, Wilson PW, Sang HM, Sharp PJ. Evidence for alternative splicing of the chicken vasoactive intestinal polypeptide gene transcript. J Mol Endocrinol. 1995;15:81–91. doi: 10.1677/jme.0.0150081. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taouis M, Chen JW, Daviaud C, Dupont J, Derouet M, Simon J. Cloning the chicken leptin gene. Gene. 1998;208:239–42. doi: 10.1016/s0378-1119(97)00670-7. [DOI] [PubMed] [Google Scholar]
- 41.Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–71. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 42.Vaisse C, Halaas JL, Horvath CM, Darnell JE, Stoffel M, Friedman JM. Leptin activation of Stat2 in the hypothalamus of wildtype and ob/ob mice but not db/db mice. Nat Genet. 1996;14:95–7. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 43.Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–82. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, Basinski MB, Beals JM, Briggs SL, Churgay LM, Clawson DK, et al. Crystal structure of the obese protein leptin-E100. Nature. 1997;387:206–9. doi: 10.1038/387206a0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–32. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 Sequence alignments of Lep
S2 Sequence alignments of LepR
S3 Active site variations of Lep shown on sequence alignments
S4 Active site variations of LepR shown on sequence alignments
S5 Fish Sequence alignments to human
S6 Amino acid properties on Lep-LepR of Salmon
S7 Amino acid properties on Lep-LepR of Zebrafish