Summary
A key feature of RNA polymerase II preinitiation complexes (PICs) is their ability to coordinate transcription initiation with chromatin modification and remodeling. To understand how this coordination is achieved, we employed extensive proteomic and mechanistic analyses to study the composition and assembly of PICs in HeLa and mouse embryonic stem (ES) cell nuclear extracts. Strikingly, most of the machinery necessary for transcription initiation on chromatin is part of the PIC. The PIC is nearly identical between ES and HeLa cells, and contains two major co-activator complexes, Mediator and SAGA. Genomewide analysis of Mediator reveals a close correlation with Pol II, TBP and mRNA levels implying a major role in PIC assembly. Moreover, Mediator coordinates assembly of the Pol II initiation factors and chromatin machinery into a PIC in vitro, while SAGA acts after PIC assembly to allow transcription on chromatin.
Keywords: Preinitiation complex, Mediator, SAGA, Embryonic stem cell, HeLa cell, MuDPIT
Introduction
Genes transcribed by RNA polymerase II (Pol II) contain chromatin modifications that facilitate initiation and early elongation (Li et al., 2007, Wang et al., 2009, Wang et al., 2008, Agalioti et al., 2002, Li et al., 2008). These modifications often bind to ATP-dependent remodeling proteins such as the bromodomain-containing SWI/SNF and the chromodomain-containing CHD1, which mobilize nucleosomes to allow binding of the Pol II machinery (Hargreaves et al., 2011). Our groups are interested in how the assembly and function of the Pol II preinitiation complex (PIC) is coordinated with the chromatin modification and remodeling events at a promoter. To address this issue and to study its mechanism, we have recreated transcription on naked DNA and chromatin templates, and captured the resulting PICs using templates immobilized on magnetic beads (Lin et al., 2011, Black et al., 2006, Johnson et al., 2002). This approach permits a detailed examination of PIC composition and function.
In our initial studies on immobilized naked DNA templates, we found that the activator GAL4-VP16 formed a complex with the TFIID and Mediator co-activators that was necessary for efficient recruitment of Pol II and the general transcription factors (GTFs) (Johnson et al., 2002) in broad agreement with the view of these co-activators as bridging factors (D'alessio et al., 2009, Naar et al., 2001, Kornberg, 2005). On chromatin, we found that the histone acetyltransferase p300 acts in concert with the Mediator co-activator very early in PIC assembly and prior to the recruitment of the GTFs (Black et al., 2006, Black et al., 2008). The p300-mediated acetylation of itself and chromatin led to p300 dissociation and allowed binding of the TFIID complex to Mediator, which facilitated assembly of the PIC.
Our early studies employed immunoblotting to identify factors thought to be involved in PIC assembly and function. To provide a more detailed understanding, we employed Multidimensional Protein Identification Technology (MuDPIT) to detect factors captured by the immobilized template on unmodified and H3K4-trimethylated (H3K4me3) chromatin templates (Lin et al., 2011). Our analysis revealed that a wide range of protein complexes involved in chromatin modification and remodeling were recruited to the PIC along with Mediator, TFIID and Pol II. Importantly, SAGA, a well-studied H3 histone acetyltransferase in yeast and mammals, and a major co-activator in yeast (Baker et al., 2007, Nagy et al., 2007), was typically among the highest abundance factors. Moreover, numerous Pol II elongation complexes such as PAF and the CDK9-containing super-elongation complex (SEC) were detected (Smith et al., 2011).
After initiation, Pol II pauses 30–50 bp downstream of the transcription startsite (TSS) through the action of DSIF and NELF. The SEC, which is recruited by Mediator (Takahashi et al., 2011), plays a post-initiation role by phosphorylating DSIF and NELF, thereby releasing the paused Pol II. Since many genes in vivo contain paused Pol II (Nechaev et al., 2011), our work suggests that in these cases, Mediator’s initial role may be in releasing the pause after which it establishes a PIC to allow multiple rounds of transcription.
The results of our initial proteomic study raised several important questions. First, are the chromatin modification/remodeling and Pol II elongation factors detected in our chromatin study components of PICs in vivo and in vitro? This bears on whether chromatin factors are general features of the PIC, whose recruitment is controlled by activator or a major co-activator. Second, are PICs formed in HeLa cells representative of PICs in other cell types like ES cells, which are emerging as an exciting area of biological interest. Finally, given the abundance of SAGA, what is its role in PIC assembly and function? In S. cerevisiae, SAGA replaces TFIID at TATA box-containing promoters (Bhaumik et al., 2002, Lee et al., 2000, Huisinga et al., 2004), while TFIID is employed at TATA-less promoters (Basehoar et al., 2004). These findings were a surprise at the time because SAGA was thought to be simply a histone acetyltransferase and deubiquitinase.
To address these questions, we employed MuDPIT to determine the composition of GAL4-VP16-stimulated PICs in vitro on DNA templates using transcriptionally active extracts from HeLa and mouse ES cells. We then compared the DNA PICs with the composition of our chromatin PICs. We found them to be highly similar indicating that the chromatin modification and remodeling machinery is an inherent component of PICs. We then compared the in vitro PICs to Mediator-associated factors from HeLa and ES cell nuclei isolated at a low salt concentration, where PIC components remain associated with chromatin (Dignam et al., 1983). The composition of the native PICs was remarkably similar to in vitro PICs. Finally, we delineated the roles of SAGA and Mediator. Our data suggest that the coordinated binding of most chromatin and Pol II elongation factors, which act near the startsite, is largely due to the Mediator co-activator. Importantly, we found that for the GAL4-VP16 activator, SAGA is not a co-activator in the traditional sense but functions independent of PIC assembly to allow transcription on chromatin templates.
Results and Discussion
Mediator and SAGA Co-Activators Are Abundant in PICs from HeLa and ES Cells
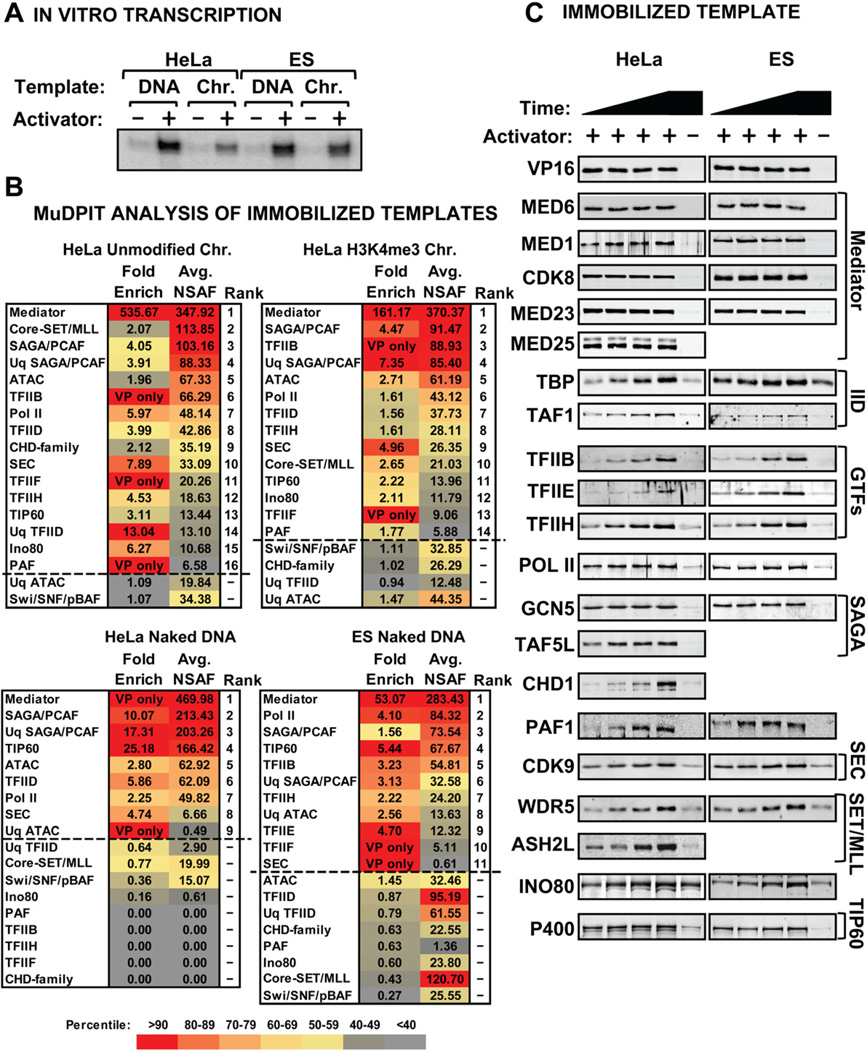
To study PIC assembly in HeLa versus mouse ES cells, we prepared nuclear extracts and compared their transcriptional activities using the model activator GAL4-VP16 along with a GAL4-responsive template. Figure 1A shows that ES cell nuclear extracts are transcriptionally active and responsive to the activator (Lin et al., 2011). We then captured the transcriptionally competent PICs using templates immobilized on magnetic beads (Lin et al., 2012). We compared unmodified chromatin, H3K4me3 chromatin, and naked DNA for HeLa nuclear extract, and naked DNA for the E14 ES cell nuclear extract. The associated factors were analyzed by MuDPIT and expressed in units of NSAF (normalized spectral abundance factor) (Washburn et al., 2001). Individual transcription-related proteins were sorted into complexes by an R-based program termed “MS Sort,” which was developed in-house (see Supplemental information). The complexes were then ranked according to activator-inducibility and overall abundance. Because TFIID and SAGA share certain core subunits, we employed a “unique” average NSAF to identify if each complex was definitively present. For cases in which unique subunit coverage of closely related complexes was too low, we relied on shared subunits or related family members to derive a score for the family of complexes (i.e., the SET1a/b and MLL1–4 complexes, and the CHD family of proteins) (Nagy et al., 2007, Eissenberg et al., 2010, Marfella et al., 2007). Figure 1B lists the activator inducibility and rank abundance of the captured PICs. Proteins that were enriched more than 1.5-fold following addition of GAL4-VP16 were designated as activator-stimulated. Proteins recruited from dialyzed nuclear extracts accurately reflect the preinitiation state because no nucleotides were added and similar recruitment data were obtained in the presence of α-amanitin and Apyrase (Supplemental Fig. 1).
Figure 1. PIC Analysis in HeLa and Mouse ES Cell Nuclear Extracts.
(A) In vitro transcription from HeLa extract and mouse ES cell nuclear extract on naked DNA and chromatin templates. (B) MuDPIT Analysis of PICs formed on naked DNA and chromatin templates in HeLa and ES cell extracts in the presence and absence of GAL4-VP16. The primary data were analyzed by MS Sort (See Supplemental Table 1) and the resulting activator-enriched (1.5-fold or greater; above dotted line) protein complexes were scored by average NSAF abundance. Average unique NSAFs (Uq-) for some complexes are also shown as described in the text (see supplementary information for detailed description of analysis). The fold enrichment and average NSAF values are color-coded according to the percentile of proteins shown within each experimental condition (e.g. both fold enrichment and NSAF values for Mediator are above the 90th percentile of proteins shown in the chart for each experimental condition and are therefore colored red (see key)). (C) Immunoblots representing known PIC components assembled during a 54-min time course on naked DNA templates using either HeLa (left) or mouse ES cell (right) nuclear extract.
Mediator was the most abundant activator-stimulated factor on both DNA and chromatin templates in both ES and HeLa cell extracts (Fig. 1B, Supplemental Table 1). Mediator is nearly identical between mouse and humans (Conaway et al., 2011) and 94% of the subunits were detected. SAGA was typically the second most abundant co-activator in both HeLa and ES cell PICs and, like Mediator, found on both DNA and chromatin templates (Fig. 1B). SAGA contains the GCN5 HAT and a histone H2B monoubiquitination-specific protease submodule (Nagy et al., 2007). The abundance of SAGA on naked DNA templates was surprising. Most of our work and that of others in mammalian systems suggested that Mediator and TFIID play the central roles in assembly of transcriptionally active PICs by recruiting the GTFs (Johnson et al., 2003, Johnson et al., 2002, Thomas et al., 2006). Although TFIID was detected in both ES and HeLa cell PICs, it was less abundant than SAGA and was less activator-responsive in ES cell extracts. Collectively, these data pointed to the possibility of a role for SAGA in PIC function.
Pol II and GTFs were also detected by MuDPIT in HeLa and in ES cell PICs on naked DNA and chromatin. TFIIB recruits Pol II (Kettenberger et al., 2004, Chen et al., 2003, Liu et al., 2010), while TFIIE and TFIIH represent the final steps in PIC assembly (Carey, 2012, Grunberg et al., 2012). Factors known to facilitate Pol II elongation such as the CDK9-containing SEC, PAF, and TFIIF were also found in PICs on DNA and chromatin. CDK9 promotes the release of Pol II paused by DSIF and NELF downstream of the TSS (Smith et al., 2011). PAF serves as a platform for binding of Pol II (Jaehning, 2010) but also associates with chromatin factors including the SET1 complex (Smith et al., 2011) and CHD1 (Warner et al., 2007). Indeed, the core WDR5-RBBP5-ASH2L-DPY30 (WRAD) submodule from the SET1 trimethylase complexes was also detected (Supplemental Table 1). The SWI/SNF complex was abundant on chromatin and DNA templates but not activator-stimulated while the Ino80 chromatin-remodeling complex was modestly activator-stimulated (Fig. 1B).
Due to the crude nature of HeLa and ES cell extracts, low abundance proteins typically fall outside the dynamic range of detection. To compensate, we performed immunoblotting of select unique subunits of a protein identified by MuDPIT to validate the relative amounts in a DNA versus a chromatin PIC. Analysis of DNA PICs revealed that subunits from each of the complexes were present in both HeLa and ES cell PICs and their activator-inducibility and binding profiles in a time course were nearly identical among the two extracts (Fig. 1C). Finally, immunoblotting of PICs on chromatin showed that most factors detected by MuDPIT were present and activator inducible on chromatin as on DNA (Supplemental Fig. 1).
Taken together, our data suggest a model of the mammalian PIC where an activator stimulates recruitment of GTFs and co-activators including Mediator and TFIID, along with numerous chromatin remodeling and modifying enzymes. The observation that numerous chromatin and early Pol II elongation factors are detected in activator-stimulated PICs alongside Pol II and the GTFs in HeLa and ES cells suggests that they are important components of all mammalian PICs. This conclusion suggests that the major co-activators controlling assembly of the GTFs also control chromatin events associated with initiation. To further address this issue, we examined whether Mediator and SAGA associate with PIC components on native chromatin isolated from HeLa and ES cells to determine roles they play in PIC assembly and transcription in vitro.
The Roles of Mediator and SAGA in PIC Assembly
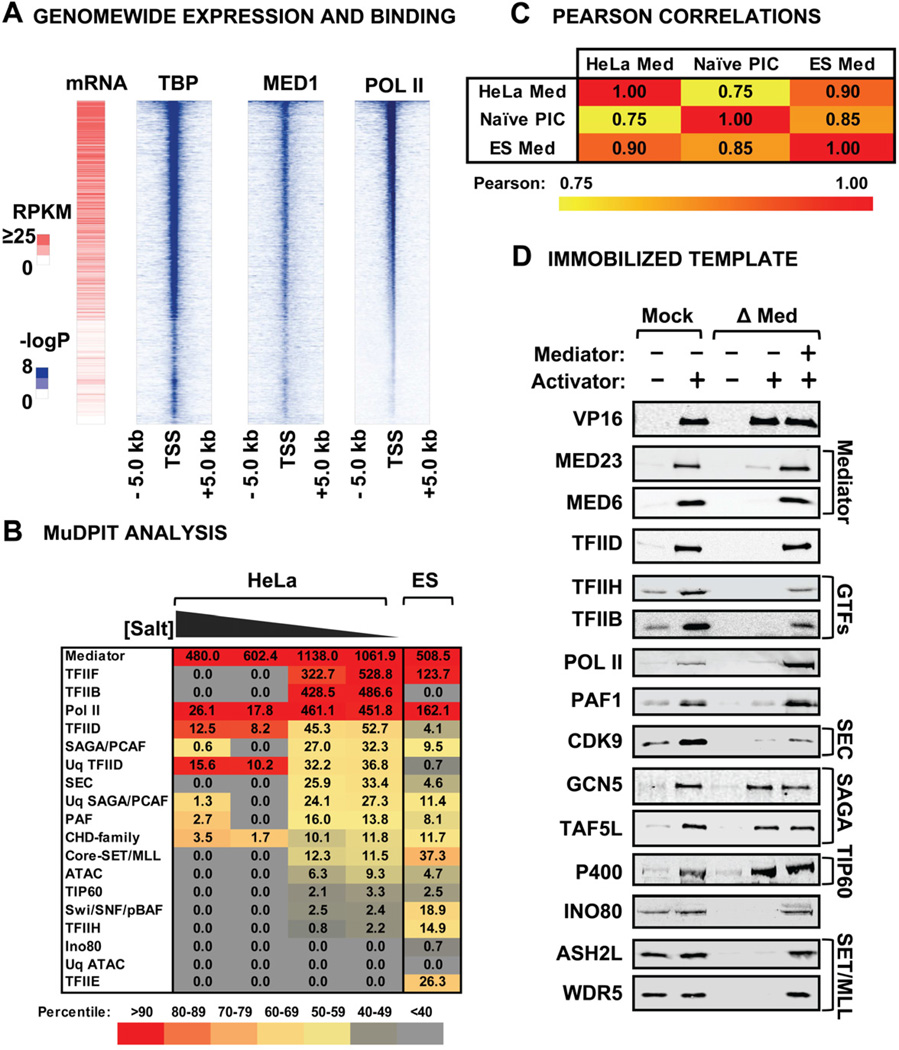
Figure 2A shows a heat map of Mediator distribution in ES cells generated from published genomewide datasets sorted by Pol II abundance (Kagey et al., 2010, Shen et al., 2012). The data reveal a remarkable similarity among the relative amounts of Mediator, Pol II and TBP bound to a given gene and a close correlation with mRNA levels. This observation, along with Mediator’s abundance in PICs formed in vitro, suggests that Mediator is common to many Pol II PICs in vivo, where it likely acts both in PIC assembly and release of paused Pol II (Fig. 2A). If true, then isolation of Mediator from native chromatin, as opposed to soluble nuclear extracts, should be representative of PIC composition in vivo. We isolated Med29-tagged Mediator under different stringencies from sonicated HeLa and ES cell nuclei isolated at low salt under conditions in which the transcriptional machinery is not solubilized (Dignam et al., 1983). MuDPIT revealed that many of the proteins found in an in vitro-assembled PIC co-purified with Mediator from HeLa and ES cell chromatin (Fig. 2B). However, unlike the in vitro assembled PIC, the most abundant Mediator-associated proteins from cellular chromatin are TFIIF, TFIIB, and Pol II in HeLa and TFIIF and Pol II in ES cells (Fig. 2B)(Supplemental Table 2). The association of TFIIF and Pol II with the Mediator has been noted (Liu et al., 2008) and TFIIB is known to associate with Pol II when it is engaged at the TSS (Kostrewa et al., 2009). TFIID is the next most abundant complex in HeLa cells. TFIIH is also detected in the HeLa and ES cell preparations and TFIIE in the ES, albeit at lower abundance. These observations are consistent with current models where TFIID, Mediator and Pol II cooperatively bind TFIIB, and nucleate the assembly of other GTFs at the promoter. Other factors such as PAF, SEC, and DSIF were identified [Reviewed in (Nechaev et al., 2011)] along with the chromatin modifying enzymes SAGA, SET1 and TIP60 (Fig. 2B)(Supplemental Table 2).
Figure 2. Mediator Composition and Function in PIC Assembly.
(A) Genomewide analysis of Mediator, TBP and Pol II distribution in ES cells. Genes were ranked by average binding of Pol II (-log (P-val)) within a 10-kb window surrounding the transcription start site of mouse promoters with a significant enrichment for any of the three factors (p-val <10−5). Expression data (RPKM) were plotted consequently for the same ranking order. (B) Chart depicting average NSAFs of complexes detected in MuDPIT analysis of purified FLAG-Med29 from sonicated HeLa and ES cell nuclei at 100, 200, 300 and 500 mM KCl (indicated as a gradient). ES cell Mediator was isolated from nuclei at 420 mM KCl. After MuDPIT and MS Sort, average NSAFs for proteins were ranked by abundance relative to the HeLa 100 mM dataset. Values are color-coded according to percentile rank within all detected complexes. (C) Pearson correlation comparison of Mediator-associated factors in unmodified chromatin and PICs assembled in vitro. (D) Immunoblots of PIC assembly comparing the recruitment of Mediator-associated protein complexes identified by MuDPIT in Mock- or Mediator-depleted HeLa nuclear extracts. Pure Mediator was added back to depleted extracts to rescue binding.
A Pearson correlation comparison of the factors bound to Mediator revealed a strong similarity among HeLa and ES cell chromatin and GAL4-VP16-stimulated PICs (Fig. 2C). We conclude that the factors recruited by GAL4-VP16 in vitro are similar to those recruited by cellular activators in Mediator-associated PICs in vivo. Importantly, an immobilized template MuDPIT analysis of ATF6α, on the HSPA5 promoter, revealed a similar set of factors as those found in our PIC study (Sela et al., 2012).
Mediator plays a central role in recruitment of many new PIC factors. Immobilized template assays in Mediator-depleted extracts revealed that Mediator is necessary for efficient recruitment of TFIID, the GTFs, the CDK9 subunit of PTEFb/SEC in agreement with a previous study (Takahashi et al., 2011), and the PAF1, SET1/MLL and Ino80 complexes. The addition of pure Mediator to the depleted extract restored binding of the affected factors, which were recruited from the depleted extract because they are not found in highly purified Mediator (Fig. 2D and Supplemental Fig. 2). Interestingly, despite previous work suggesting a TRRAP subunit-independent interaction between Mediator and SAGA, Mediator was not necessary for recruitment of SAGA or TIP60 (Fig. 2D) (Liu et al., 2008). If SAGA were a bona fide co-activator for GAL4-VP16 it should associate with GTFs or Mediator, or be important for some other aspect of PIC assembly.
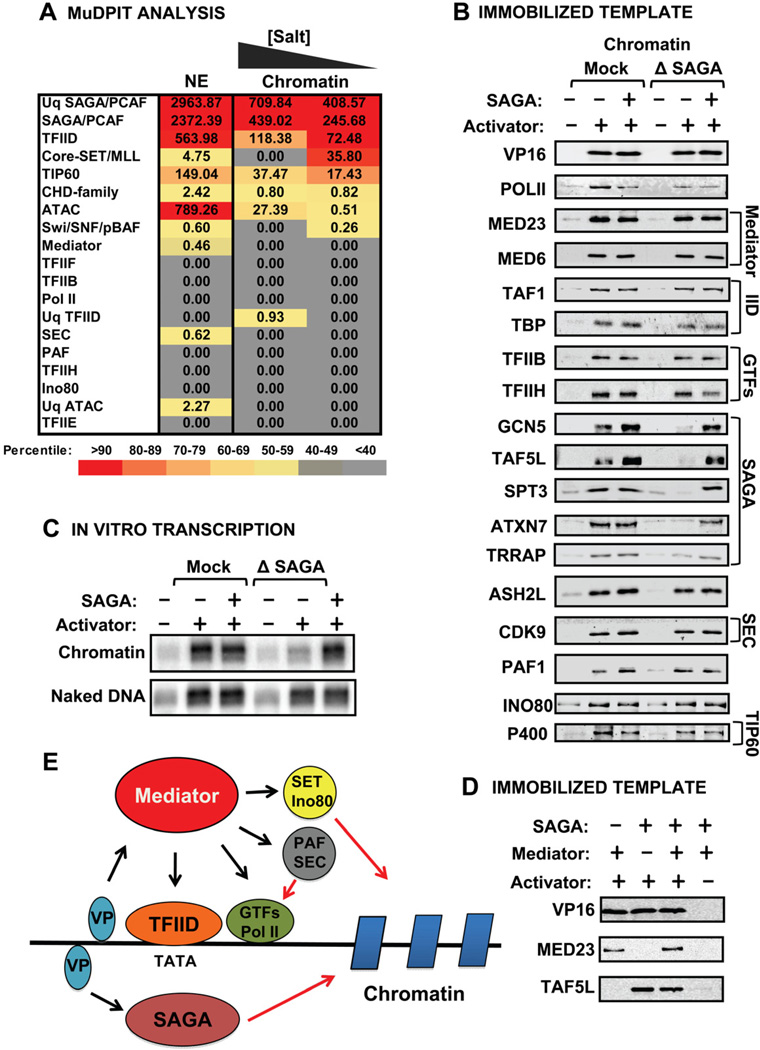
Interestingly, unlike Mediator, Spt3-tagged SAGA (Martinez et al., 1998) isolated from chromatin did not contain any other factors in abundance except for the chromatin remodeler NuRD (Fig. 3A) (Supplemental Table 3). Indeed, many of the factors detected by MS Sort were not unique, i.e., the TFIID subunits detected were shared by SAGA. To our knowledge, SAGA has not been examined by ChIP-Seq in ES cells but studies in human cell lines suggest that it binds only a subset of genes and in a somewhat exclusive manner with another GCN5-containing complex called ATAC (Krebs et al., 2011).
Figure 3. SAGA Composition and Role in PIC Assembly and Function.
(A) Chart depicting average NSAFs of complexes detected in MuDPIT analysis of FLAG-Spt3 from sonicated HeLa nuclei at 150 and 300 mM KCl and from soluble HeLa nuclear extract at 450 mM. Values are color-coded according to percentile rank within detected complexes. (B) Immunoblots of PIC assembly comparing the recruitment of complexes identified by MuDPIT in Mock- or SAGA-depleted HeLa nuclear extracts plus or minus pure SAGA. Different modules of SAGA were detected using antibodies against the TRRAP, HAT/core (GCN5), SPT (SPT3), Deubiquitination (ATXN7), and TAF (TAF5L) modules. (C) In vitro transcription in SAGA-depleted extracts on naked DNA and unmodified chromatin templates plus or minus pure SAGA. A 2-fold longer exposure of transcription on chromatin is shown. (D) Immunoblots of immobilized template recruitment experiments with GAL4-VP16 and pure Mediator and/or SAGA. (E) Model showing that activators recruit Mediator and SAGA independently. Mediator recruits the indicated factors. SAGA functions on chromatin.
To further analyze SAGA’s role in the PIC, it was depleted from HeLa extracts using GCN5 and TAF5L antibodies (Supplemental Fig. 2). Surprisingly, we found that SAGA depletion does not significantly affect GAL4-VP16-mediated PIC assembly on chromatin (Fig. 3B) or DNA templates (data not shown). This result coupled with our proteomics data argues that SAGA is not linked directly to PIC assembly and is therefore not a co-activator in the traditional sense, i.e., a bridging factor. Moreover, the depletion of SAGA has virtually no effect on naked DNA transcription (Fig. 3C). Remarkably, chromatin transcription was strongly and reproducibly diminished by depletion and restored with pure SAGA. We conclude that SAGA does not affect PIC assembly but is required for efficient transcription of chromatin templates. It is unclear whether SAGA directly influences PIC assembly in yeast. Spt3 depletion does not affect TBP or TFIIA binding in PICs from yeast extracts (Warfield et al., 2004) but SAGA depletion abolishes activated transcription on naked DNA templates (Fishburn et al., 2005), which differs from our results in HeLa. Also, Tra1 mutants display some defects in recruitment of Pol II in vivo by ChIP assays (Knutson et al., 2011) as do mutations that disrupt the Spt3-TBP interface (Mohibullah et al., 2008).
Previous studies had suggested a role of Mediator in SAGA recruitment to promoters (Liu et al., 2008). However, Figure 3D shows that SAGA and Mediator bind directly to GAL4-VP16 both independently and together. Moreover, Mediator recruitment is not affected by prebound SAGA, or vice versa (Supplemental Fig. 3). We conclude that SAGA is able to join the GAL4-VP16-assembled PIC independent of Mediator.
In summary, we determined that the primary architecture of the activator-stimulated PIC consists of high-affinity interactions between Pol II and the GTFs, driven by the Mediator. Mediator also plays an essential role in recruitment of several key chromatin modifying and remodeling complexes like SET1, CHD1, and Ino80 along with two major Pol II elongation complexes, PAF1 and SEC (Fig. 3E). Therefore, the activator-stimulated PIC contains most key factors thought to control promoter accessibility within chromatin and early Pol II elongation.
Importantly, most of the interactions are found in PICs formed on naked DNA suggesting that promoter-bound chromatin factors are targeted to the PIC not by chromatin but by interactions with Mediator, by direct interactions with activator, or through complex networks of interactions linked to both activator and Mediator. Despite SAGA’s relative abundance, its binding is directed by GAL4-VP16 and it functions after PICs are assembled (Fig. 3E).
Experimental Procedures
Recruitment and In Vitro Transcription Assays
G5E4T (Johnson et al., 2003) was assembled into chromatin by salt dilution and immobilized as described previously (Steger et al., 1997). The immobilized template and in vitro transcription assays and the corresponding scaled-up reactions for MuDPIT were performed as described previously (Lin et al., 2012, Lin et al., 2011).
Extract and Protein Preparation
HeLa and mouse ES cell E14 nuclear extract, and GAL4-VP16 were prepared as described (Dignam et al., 1983, Tantin et al., 1996). Immunodepletion of Mediator was performed as described (Lin et al., 2011). SAGA depletion was performed with GCN5 (Santa Cruz) and TAF5L (Sigma) antibodies. Mediator was purified as described previously (Sato et al., 2003). In parallel, Mediator was isolated from the chromatin of the low salt nuclear fraction at the salt concentrations described in the legend of Fig. 2. FLAG-Med29 was generated from the V6.5 ES cell line (Beard et al., 2006) bearing a Doxycycline inducible FLAG-Med29 murine cDNA. SAGA was purified from nuclear extract of cells expressing FLAG-Spt3 as described (Martinez et al., 1998) and from the chromatin of the low salt nuclear fraction using conditions described in the legend of Fig. 3.
Genomewide Analysis
Datasets used were TBP-GSE22303, MED1-GSE22557, and RNA Pol II-GSM723019 from the GEO database (www.ncbi.nlm.nih.gov/geo/). The expression levels for all annotated genes were determined using the GSM881355 GEO data set. All data analysis was performed as described previously (Ferrari et al., 2012)
MuDPIT Analysis of Immobilized Templates and Purified Proteins
MS/MS spectra were collected and normalized spectral abundance factor (NSAF) values were calculated as described previously (Lin et al., 2011). Average NSAFs for each complex and unique NSAFs were calculated by MS Sort by adding NSAFs for each subunit, and dividing by the total number of subunits.
Supplementary Material
Highlights.
The composition of mammalian PICs was determined by proteomic analysis
A wide range of chromatin modifying/remodeling factors are recruited to PICs
Mediator coordinates binding of Pol II initiation, elongation and chromatin machineries
SAGA acts after PIC assembly to make chromatin templates transcriptionally competent
Acknowledgements
This work was supported by NIH grants GM074701, GM089778 and GM099134 to M.C., J.W., and K.P., respectively. R.F. and S.K.K. were supported by a California Institute for Regenerative Medicine grant. M.C. wrote the paper; X.F.C., J.J.L., L.L and C.H. performed the experiments; J.W. and A.V. performed MuDPIT; R.S. developed MS Sort; K.P. directed ES growth and scaleup; R.F. and S.K. performed genomewide analyses; X.F.C., L.L., J.J.L., A.M. and M.C. performed data analysis. We thank Joan and Ron Conaway for Flag-Med29 and Ernest Martinez for FLAG-Spt3 HeLa cell lines. We thank Steve Hahn for discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agalioti T, Chen G, et al. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, et al. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, et al. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Choi JE, et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Black JC, Mosley A, et al. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. PICking apart Pol II initiation. Nat Struct Mol Biol. 2012;19:737–738. doi: 10.1038/nsmb.2349. [DOI] [PubMed] [Google Scholar]
- Chen HT, Hahn S. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol Cell. 2003;12:437–447. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. Origins and activity of the Mediator complex. Semin Cell Dev Biol. 2011;22:729–734. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'alessio JA, Wright KJ, et al. Shifting players and paradigms in cell-specific transcription. Mol Cell. 2009;36:924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, et al. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Su T, et al. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, et al. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Grunberg S, Warfield L, et al. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Carey M. Assembly of a mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr Biol. 2003;13:772–777. doi: 10.1016/s0960-9822(03)00283-5. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Wang J, et al. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, et al. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Knutson BA, Hahn S. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol Cell Biol. 2011;31:818–831. doi: 10.1128/MCB.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- Krebs AR, Karmodiya K, et al. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Carey M. In vitro transcription and immobilized template analysis of preinitiation complexes. Curr Protoc Mol Biol. 2012;Chapter 12(Unit 12):14. doi: 10.1002/0471142727.mb1214s97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Lehmann LW, et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 2011;25:2198–2209. doi: 10.1101/gad.17554711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, et al. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Vorontchikhina M, et al. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Kundu TK, et al. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, et al. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as coactivators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, et al. A Mammalian Homolog of Drosophila melanogaster Transcriptional Coactivator Intersex Is a Subunit of the Mammalian Mediator Complex. Journal of Biological Chemistry. 2003;278:49671–49674. doi: 10.1074/jbc.C300444200. [DOI] [PubMed] [Google Scholar]
- Sela D, Chen L, et al. Endoplasmic Reticulum Stress-responsive Transcription Factor ATF6alpha Directs Recruitment of the Mediator of RNA Polymerase II Transcription and Multiple Histone Acetyltransferase Complexes. J Biol Chem. 2012;287:23035–23045. doi: 10.1074/jbc.M112.369504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, et al. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Owen-Hughes T, et al. Analysis of Transcription Factor-Mediated Remodeling of Nucleosomal Arrays in a Purified System. Methods. 1997;12:276–285. doi: 10.1006/meth.1997.0479. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D, Chi T, et al. Biochemical mechanism of transcriptional activation by GAL4-VP16. Methods Enzymol. 1996;274:133–149. doi: 10.1016/s0076-6879(96)74013-2. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Wang Z, Schones DE, et al. Characterization of human epigenomes. Curr Opin Genet Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Ranish JA, et al. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 2004;18:1022–1034. doi: 10.1101/gad.1192204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, et al. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, et al. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.