Abstract
The Peg3 gene is expressed only from the paternally inherited allele located on proximal mouse chromosome 7. The PEG3 protein encoded by this imprinted gene is predicted to bind DNA based on its multiple zinc finger motifs and nuclear localization. In the current study, we demonstrated PEG3’s DNA-binding ability by characterizing its binding motif and target genes. We successfully identified target regions bound by PEG3 from mouse brain extracts using chromatin immunoprecipitation analysis. PEG3 was demonstrated to bind these candidate regions through the consensus DNA-binding motif AGTnnCnnnTGGCT. In vitro promoter assays established that PEG3 controls the expression of a given gene through this motif. Consistent with these observations, the transcriptional levels of a subset of the target genes are also affected in a mutant mouse model with reduced levels of PEG3 protein. Overall, these results confirm PEG3 as a DNA-binding protein controlling specific target genes that are involved in distinct cellular functions.
Keywords: Peg3, genomic imprinting, DNA-binding motif
INTRODUCTION
Genomic imprinting is a gene dosage-controlling mechanism that restricts transcription from one inherited allele (either maternal or paternal) through epigenetic mechanisms [1, 2]. Many imprinted genes encode cellular ligands and receptors for signaling pathways, cytoplasmic enzymatic modulators, and membrane transporters that are ultimately responsible for embryonic growth and development [3, 4]. Several imprinted genes, however, are predicted to function as DNA-binding transcription factors based on specific protein motifs within their open reading frames such as the helix-loop-helix motif and the Cys2His2 zinc finger motif. Among these imprinted genes, only Klf14, Wt1, and Zac1, all containing Cys2His2 zinc fingers, have been proven to bind DNA [5-7]. Interestingly, many genes bound by ZAC1 are known imprinted genes, such as H19/Igf2, Grb10, and Cdkn1c. This set of genes exhibits similar expression level fluctuations in response to multiple extrinsic and intrinsic changes [7]. This data and other recent publications have proposed the existence of an ‘imprinting network,’ whereby transcription of many imprinted genes is co-regulated to accomplish a similar goal, typically controlling growth and metabolic rates in both fetal and adult eutherian animals [8-10].
Another imprinted gene, Peg3, is predicted to contain twelve Cys2His2 Kruppel-type zinc finger protein domains with the capability of binding to DNA [11-13]. PEG3 has also been demonstrated to be present in the nucleus at equal or slightly greater amounts than in the cytoplasm [14]. Earlier studies, however, have focused on PEG3’s cytoplasmic interactions and functional roles. PEG3’s interaction with SIAH1A induces p53-mediated cell apoptosis in multiple cell types [15, 16]. PEG3 was also demonstrated to inhibit Wnt signaling by promoting β-catenin degradation through the same pathway [17]. In addition, PEG3’s interaction with TRAF2 regulates the TNF responsive pathway by activating NFκβ [18]. Its connection to both pathways was demonstrated in vivo to be involved in skeletal muscle growth and homeostasis with alterations creating increased muscle atrophy [19, 20]. Furthermore, a mouse knockout model targeting Peg3 demonstrated that it is responsible for diverse phenotypic outcomes such as altered/reduced maternal offspring-rearing behavior, olfactory alterations that decrease male mating, low birth weight, alterations in fat tissue synthesis and storage, problems regulating core body temperature, and lower metabolic activity [21-24]. Although many independent observations have suggested significant roles for PEG3 in diverse cellular pathways, the molecular mechanisms by which PEG3 is involved in these pathways remain unexplored.
In the current study, we tested PEG3’s functionality as a predicted DNA-binding transcription factor. Initially, we identified and characterized a subset of PEG3’s target genes and their DNA-binding motif. PEG3 was also shown to regulate the expression of Pgm2l1 through the binding of the identified motif. Ultimately, this data reveals a new and important function for PEG3 that may be the molecular mechanism responsible for a subset of the Peg3 phenotypic effects in the animal knockout model.
RESULTS
Identification of genomic regions bound by PEG3
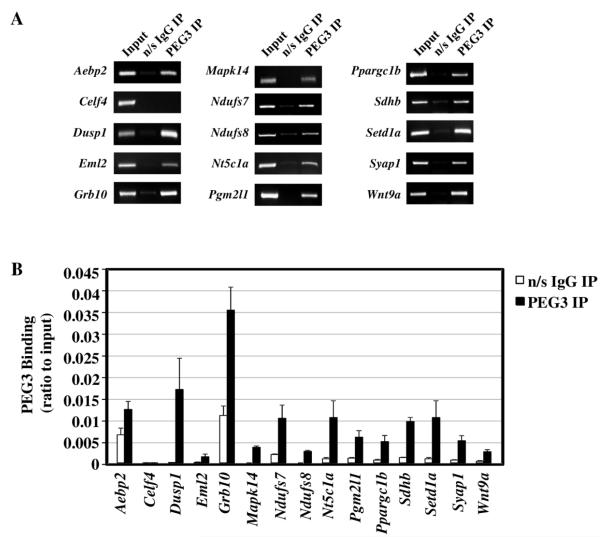
Peg3 is predicted to encode a DNA-binding protein based on its zinc finger domains and nuclear localization [11]. To initially test this prediction and identify candidate loci, we used a ChIP (Chromatin Immunoprecipitation) based strategy. For ChIP experiments, we generated polyclonal antibodies against mouse PEG3 as described in the Materials and Methods section. The purified antibody was demonstrated to recognize mouse PEG3 protein almost exclusively (Supplemental Figure 1). This same antibody has also been successfully used in our previously published studies [25]. For actual ChIP experiments, brain extracts from a 3-month-old male mouse were first immunoprecipitated with the PEG3 antibody, and the precipitated DNA was sequenced using a NGS (Next Generation Sequencing) protocol. PEG3 sequencing experiments generated a small number of reads (3,336,046 million filtered PEG3 IP sequencing reads normalized to Input) that had an average read length of 134 bp (data not shown). In order to demonstrate that PEG3 binds to specific genomic regions, a subset of these candidate genomic regions was chosen for analysis by ChIP followed by either PCR-gel imaging or qPCR (Figure 1A,B). The chosen regions were also located near genes that either control mitochondria functions (Ndufs7, Ndufs8, Ppargc1b, Sdhb), tissue development (Dusp1, Mapk14, Wnt9a), or are uniquely interesting (Aebp2, Eml2, Grb10, Syap1, Nt5c1a, Setd1a). Complete gene names are listed in Supplemental Table 1. Pgm2l1 was especially chosen because PEG3 bound to its promoter region, a typical location for gene regulation, which was useful for performing multiple follow-up experiments. All of these genomic regions showed higher levels of enrichment by IP with the PEG3 antibody when compared to those with the control antibody, confirming the in vivo binding of PEG3 to these regions. One selected region from the ChIP-Seq data, located in the intron of Celf4, was negative for PEG3 binding based on the low signal levels from the PEG3 antibody IP. Overall, the data demonstrate that the PEG3 protein is capable of binding specific DNA regions across the mouse genome. Further analysis of more specific tissue subtypes will allow for the delineation of PEG3’s tissue specific binding regions.
Figure 1. Chromatin Immunoprecipitation (ChIP) with PEG3 antibody.
A subset of genomic regions was demonstrated to be bound by PEG3 through both (A) regular PCR and (B) qPCR. For regular PCR, the entire PCR reaction for the control (n/s IgG) and PEG3 ChIP or 2/5 of the Input DNA were separated by agarose gel electrophoresis and visualized with ethidium bromide. For qPCR, each target region was amplified using DNA immunoprecipitated from either n/s IgG (open bars) or PEG3 antibody (closed bars). The values on the Y-axis represent values relative to those of the Input DNA. Amplifications were repeated in triplicate and the subsequent S.D. is shown.
Prediction of a potential DNA-binding motif for PEG3
Cys2His2 Kruppel-type zinc finger domains generally recognize a specific, although sometimes degenerate, nucleic acid sequence motif [13]. To identify a DNA-binding motif for the PEG3 protein, the program Multiple Em for Motif Elicitation (MEME) was used [26]. Initially, all peak regions identified by ChIP-Seq were analyzed using the large dataset ‘ChIP’ version of MEME. This analysis identified a non-specific T-rich motif in about 40% of the peak regions. To derive a more defined consensus sequence for experimental studies, all fourteen verified regions from Figure 1 were used for motif analysis. Given that the motifs produced by this analysis were extremely degenerate, the motif was refined by using four regions that had a p-value less than or equal to 1.0e-5 associated with them (Figure 2). Analysis of these five regions produced a motif that is 15 bp in length with conserved positions at nucleotides 2, 11, 12, and 14 (A, T, G, and C respectively). The motif also demonstrated specificity for six other positions as they are restricted to one of two nucleotide bases. The most conserved nucleotide bases are located at the outer edges of the motif with the middle portion being less selective. The overall length, specificity, and pattern of this predicted motif was particularly attractive given that PEG3 contains a cluster of five zinc finger domains near its C-terminus that are well conserved among mammalian species [27, 28]. Previous research suggests that when these DNA-binding domains are present in tandem arrays they typically bind DNA at a ratio of three base pairs per zinc finger [13]. Thus, the predicted 15 bp motif with small clusters of conserved bases is a reasonable motif for a protein containing a tandem array of five zinc fingers.
Figure 2. DNA motif analysis and prediction.
The MEME program was used to predict a DNA-binding motif for PEG3. Both the predicted sequence logo and the consensus sequence are shown. The locations of this motif in five validated regions of PEG3 binding, along with the program generated p-value, are shown with their associated gene name.
Characterization of a DNA-binding motif for PEG3
The predicted DNA-binding motif of PEG3 was further analyzed by EMSA using the Pgm2l1-bound region as a representative motif given that it is located in the promoter region and is highly conserved among mammalian species (Supplemental Figure 2). A 39-bp region of the Pgm2l1 promoter was labeled with [γ-32P]ATP and used as an experimental probe (P, Figure 3). The Pgm2l1 probe was found to be bound by a prominent protein complex (C) in mouse brain nuclear extract. This protein complex was shifted specifically by the PEG3 antibody (S) but not by either non-specific IgG or YY1 antibodies indicating that this complex likely contains the PEG3 protein. The binding specificity of the PEG3 complex was further analyzed with a series of competition assays. First, the binding of the PEG3 complex was abolished by incubation with an excessive amount (100X) of unlabeled Pgm2l1 probe. Second, the binding was also inhibited by the competitor oligo containing the Ndufs8 WT sequence. This competitor sequence contains a small region that shares sequence identity with the initial 15-bp long motif found within the Pgm2l1 probe, as depicted in Figure 3. Similar analysis was also performed using the Syap1-bound region as the radiolabeled probe and the Pgm2l1 sequence as a competitor (Supplemental Figure 3). This data suggests that the 15-bp-long region of the Pgm2l1 probe is most likely responsible for the binding to the PEG3 complex. Third, three mutated forms of the Pgm2l1 probe were generated as competitors to further define the critical bases for PEG3 binding. A duplex oligo (Mutant 12) containing a stretch of 15 As in place of the PEG3 binding motif did not compete against the labeled Pgm2l1 probe, demonstrating that the 15-bp-long region is required for the binding. This competition was further repeated with two additional competitors: Mutant F3 (CAGT to AAAA) and Mutant B4 (TGTC to AAAA). Mutant F3 competed well against the labeled probe, indicating the 4 bp (CAGT) on the 5′-side of the motif may be less critical for PEG3 binding. In contrast, Mutant B4 did not compete well, confirming that this 4 bp region (TGTC) at the 3′-end of the motif is the most critical region for PEG3 binding.
Figure 3. Electromobility shift assay (EMSA) to validate the predicted DNA-binding motif of PEG3.
The sequences for the 32P -labeled probe and all of the unlabeled competitors are provided in the bottom boxes. The red-colored nucleotide bases in the right box indicate the nucleotide bases mutated to A. The labels for the gel picture are as follows: P, 32P -labeled probe; C, complex formed between probe and proteins in the nuclear extract; n/s, non-specific complex formation between probe and antibody; S, supershifted complex composed of probe, PEG3 protein, and PEG3 antibody.
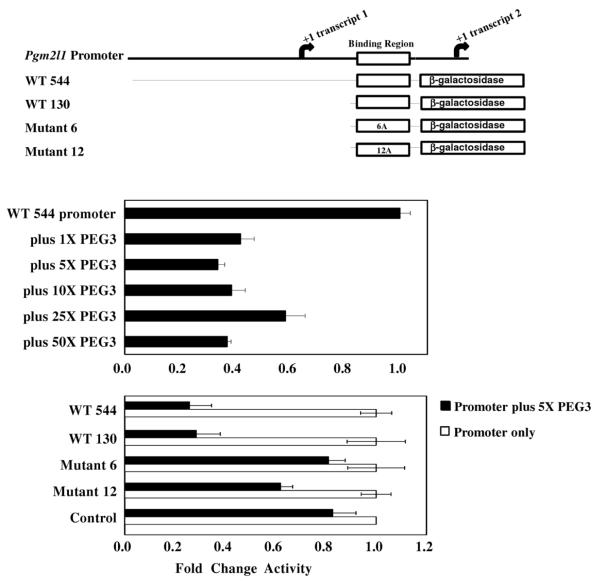
PEG3’s ability to bind the predicted motif was also tested using promoter assays (Figure 4). A series of DNA fragments derived from the promoter of Pgm2l1 were used to drive the expression of a promoterless β-galactosidase cassette. These fragments include an intact 544-bp promoter region of Pgm2l1 (WT 544) and a smaller promoter region of 130 bp containing the predicted PEG3 binding motif (WT 130). The 15-bp motif, located within the 130 bp region, was additionally mutated with either 6 As (Mutant 6) or 12 As (Mutant 12). For the in vitro assays, these reporter constructs were co-transfected into Neuro2a cells with an expression vector containing the full-length PEG3 protein. The transfection of the WT 544 reporter yielded detectable levels of β-galactosidase activity, confirming the promoter activity of the 544-bp region of Pgm2l1. In the presence of varying amounts of the full-length mouse PEG3 expression vector, the WT 544 reporter demonstrated similar levels of reduction in the β-galactosidase activity, demonstrating that PEG3 has a repressive effect on the Pgm2l1 promoter. This repressive effect on the WT 544 promoter by the PEG3 protein was also observed with the WT 130 construct. The repressive effect on WT 130 demonstrates that the PEG3 binding motif is located within this region of the Pgm2l1 promoter. When the PEG3 motif was partially or completely mutated, Mutants 6 and 12, this repressive effect was lessened or completely abolished, further confirming that the repressive effect is mediated through the 15-bp PEG3 motif. This assay was also repeated with an independent control reporter containing the Celf4 sequence, which lacks the binding motif of PEG3, and was negative for PEG3 binding (Control in Figure 4). This control reporter did not yield a repressive effect as seen in the WT 544 reporter, demonstrating that the repressive effect by the PEG3 protein is specific for some of PEG3-bound sequences. Overall, the data demonstrate that PEG3 binds the predicted DNA motif in a sequence specific manner, and also that the PEG3 protein may function as a repressor with regard to the Pgm2l1 promoter.
Figure 4. β-galactosidase promoter assay to demonstrate PEG3 binding through its identified motif.
(Top panel) The assays used fragments from the Pgm2l1 promoter both with and without the DNA-binding motif of PEG3 (indicated with an open box). The promoter constructs 6A and 12A contain mutated forms of the DNA-binding motif having either 6 or 12 adenosines (A)s that are changed from the original sequence of the PEG3 binding motif, respectively. (Middle panel) The β-galactosidase activity driven by the WT 544 promoter was measured in either the absence or presence of increasing concentrations of PEG3 protein. All data were normalized to the activity of the WT 544 promoter in the absence of PEG3 protein. (Bottom panel) Different regions of the Pgm2l1 promoter were tested for activity in the absence (white bars) or presence (black bars) of 5X PEG3 protein. For each promoter construct, data was normalized to the promoter activity in the absence of PEG3. All promoters gave similar amounts of activity in the absence of PEG3.
Expression level changes in PEG3 target genes in a mutant model of Peg3
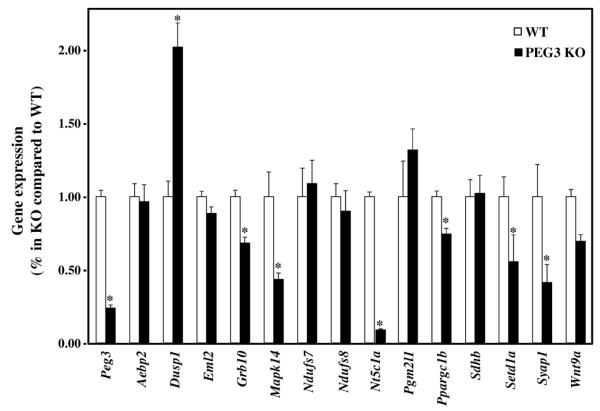
The results from the β-galactosidase assay suggest that one potential function of PEG3 may be as a transcriptional repressor. To determine the functional consequences of PEG3 binding, we analyzed the expression levels of PEG3 bound genes in a mutant mouse model with low levels of Peg3 expression. This mutant model has a 2.5-kb genomic deletion in the 1st intron of the Peg3 gene, a region which is known to harbor several regulatory elements for Peg3 transcription [29]. For this series of analyses, two sets of RNA were individually isolated from neonatal brains (KO and WT), reverse-transcribed, and used for qRT-PCR analyses (Figure 5). As expected, Peg3 displayed an approximately 75% reduction in its expression level in the KO relative to WT. A gene expression survey of the validated PEG3 bound genes (from Figure 1) demonstrated multiple responses in which gene expression was either reduced (Grb10, Mapk14, Nt5c1a, Ppargc1b, Setd1a, Syap1, Wnt9a), increased (Dusp1, Pgm2l1), or remained the same (Aebp2, Eml2, Ndufs8, Sdhb). This varied result could be due to several factors including the heterogeneity of the tissue used (total brain), diverse functional nature of the genes chosen, or the possibility that PEG3 may possess both activator and repressor functions. However, seven specific genes (Dusp1, Grb10, Mapk14, Nt5c1a, Ppargc1b, Setd1a, and Syap1) demonstrated either a statistically significant increase or reduction in their expression when the expression levels of Peg3 were reduced. This data illustrates that PEG3 is able to directly regulate the expression of a subset of its target genes. Overall, these data demonstrate that the genomic binding of PEG3 near gene regions can affect the transcriptional levels of specific genes.
Figure 5. qRT-PCR RNA expression analysis of PEG3 bound genes.
The expression levels of PEG3 bound genes were compared between two sets of total RNA isolated from mouse neonatal brains that were either wild-type (WT) or have reduced expression of Peg3 (KO). Individual gene expression was first normalized to β-actin levels, and then normalized against the WT levels. All data are graphed as the percentage of expression in the KO mice as compared to expression in the WT mice. Bars with asterisks represent values that were statistically different by a p-value less than 0.05.
DISCUSSION
The reported data is the first evidence describing PEG3’s ability to bind DNA and its capability to do so in a sequence specific manner. This study is also one of the few documented cases of an imprinted gene functioning as a DNA-binding protein. Though specific examples of genomic imprinting have been identified in marsupials (but not monotremes), there is currently no published data identifying a homologous Peg3 imprinted region in marsupials. Swaney et al has recently proposed that Peg3 evolved for the purposes of coadaptation between offspring and mother through shared behaviors, suggesting that this imprinted region may have newly evolved for the specific functions of placental mammals. Given that Peg3 is an evolutionarily new gene, it is possible that its target regions are in locations not already occupied by traditional transcription factors and RNA-producing machinery [30-32].
PEG3 binding was also determined to be dependent on the consensus sequence AGTnnCnnnTGGCT (Figures 2-4). While the entire sequence of this motif was necessary for maximal binding of PEG3, the core nucleotide bases TGGCT alone could promote PEG3 binding to DNA. This flexibility in the consensus motif of PEG3 may allow for binding to numerous genomic regions with different protein partners. For other proteins containing multiple adjacent zinc fingers, such as TFIIIA and WT1, it has been demonstrated that only a fraction of these fingers bind DNA tightly with the remaining fingers having a weak DNA association or none at all [13]. Zinc finger regions that are not bound to DNA are frequently determined to interact with other proteins. Given that some of the zinc fingers in PEG3 are more evolutionarily conserved than others, it may be that this highly conserved subset is responsible for DNA binding. Since the nuclear protein partners of PEG3 have yet to be identified, a diverse set of proteins may partner with PEG3 through a subset of the zinc fingers not bound to DNA, ultimately allowing PEG3 to modulate the transcription of different gene sets.
Given that many of the functions of PEG3 are still relatively unexplored, the target genes heavily influenced by PEG3 expression provide several avenues for further research (Figures 1,5). A mutant mouse model targeting Peg3 previously suggested that it has diverse functions that influence physiological traits such as regulating core body temperature, feeding behavior, and obesity in mice [22]. These traits are all linked by changes in energy homeostasis making our discovered connection between PEG3 and a subset of nuclear-encoded mitochondrial genes (Ndufs7, Ndufs8, Ppargc1b, Sdhb) plausible. Furthermore, the same mutant model of Peg3 also exhibited altered maternal behavior resulting in increased offspring death, and this observation was hypothesized to be caused by defective neuronal connectivity [24]. In accord with this observation, knockdown of Peg3 in a zebrafish model impaired tail development thus suggesting Peg3’s ability to inhibit the Wnt signaling pathway in both animal models [17]. Inhibition of the Wnt signaling pathway has been proposed to be necessary for specific tissue types (i.e hypothalamus) to develop properly [33]. Given these observations, our identification of specific genes in tissue development (Dusp1, Mapk14, Wnt9a) being influenced by PEG3 binding and/or expression is intriguing. Future studies to measure the expression of these genes in specific brain regions (namely the hypothalamus) of mutant Peg3 mice may lead to further clarification of the connections between Peg3 and tissue development.
Previous published observations have also generated a large interaction network of co-regulated imprinted genes from which PEG3 was described as a “hub-like node,” suggesting its potential to regulate a large number of genes for a unified specific purpose [7]. The previous analysis concluded that the focus of this network was to regulate embryonic growth and differentiation. Similar to Peg3 mutations, mutation of Zac1, the focus of the previous study, also caused intrauterine growth restriction, but this only occurred in the embryo as Zac1 is not expressed in the placenta. Given that PEG3 is highly expressed in the maternal hypothalamus and placenta, it may function in the imprinted network through DNA-binding activities in the hypothalamus and placenta to regulate embryonic growth. The in vivo expression data suggests that PEG3 may be able to accomplish this task by altering the expression of genes such as Grb10, another imprinted gene. Overall, further experiments in more defined tissues, either specific brain regions or homogeneous tissue types, will provide a detailed characterization of genomic regulation through PEG3’s DNA-binding capabilities.
MATERIALS AND METHODS
Chromatin Immunoprecipitation (ChIP) and qPCR validation
The polyclonal antibody against mouse PEG3 was custom-made by a commercial firm (ProteinTech) using a partial protein that contains all 12 zinc finger domains of mouse PEG3, amino acids 164-1572, which was expressed from the expression vector pGex-4T-1-PEG3. Individual ChIP experiments were performed with mouse brain extracts as detailed in our previous publication with minor changes [34]. For each individual IP, the PEG3 antibody was used with one-tenth of the crosslinked and sonicated brain extract fraction. All immunoprecipitated DNA was precipitated, purified, and resuspended in 50 μl of TE (pH 8.0) for further analysis.
For gel imaging analysis, 1 μl of either input or ChIP DNA was used for PCR with the Maxime PCR Premix kit (iNtRON Biotechnology). The following PCR conditions were used: 95°C for 4 mins, 37 repetitions of the following cycle of 95°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec, and final extension at 72°C for 7 mins. A fraction (2/5) of the PCR product from the input DNA and the entire products from the negative control (n/s IgG) and PEG3 ChIP were separated by agarose gel electrophoresis and visualized with ethidium bromide.
For quantitative PCR (qPCR) analysis, 1 μl of ChIP product was used for PCR with SYBR Green Premix reagents (BioRad) and 100 ng of gene specific primers using the PCR conditions described above. A camera capture setting was added after the 60°C step to monitor the formation of PCR product. A melt curve step ranging from 55-95°C with a hold of 10 sec and a temperature increment of 0.5°C was added at the end of the PCR to monitor the quality of PCR product. Data were analyzed by normalizing to a standard curve (i.e. serial dilution made from input DNA) and graphed as the ratio of the immunoprecipitated DNA to the input DNA. The primer sequences for each gene are available as supplemental data (Supplemental Table 1).
Derivation of a consensus DNA-binding motif of PEG3
The program MEME, a motif exploratory tool, was used to derive a consensus DNA-binding motif for the PEG3 protein [26]. Initial analysis of all ChIP-Seq regions provided a non-specific T-rich motif. This analysis was refined by using only the genes that had been validated to be bound by PEG3 through qPCR. The DNA sequence for each of the validated genes was used for alignment in the MEME software. Finally, four genes were chosen to produce a potential consensus motif given the likelihood of binding and representative nature of the motif produced.
Electromobility shift assay (EMSA)
EMSA was performed according to instructions from the Gel Shift Assay System (Promega). Briefly, various combinations of assay buffer, 2.72 μg mouse brain nuclear extract (Active Motif), PEG3 antibody (3 μL), YY1 antibody (3 μL, sc-1703X, Santa Cruz Biotech.), n/s IgG antibody (3 μL, sc-2338, Santa Cruz Biotech.), and 0.7 pmol unlabeled competitor oligo duplex were incubated at room temperature for 10 minutes. Next, a concentration of 0.07 pmol [γ-32P]ATP end-labeled oligo duplex probe (always containing the Pgm2l1 WT sequence) was added and the complexes were incubated at room temperature for another 20 minutes. The entire reaction was separated on a 5% TBE gel (BioRad) and exposed to film for 1 to 4 hours at −80°C before imaging.
Promoter assay
The promoter assay used the modified version of the β-geo vector, which has been previously described [37]. All DNA fragments were inserted into the NotI site of the β-geo vector. The wild-type (WT) 544 Pgm2l1 promoter was cloned from the BAC clone RP23-344G10 (Invitrogen). The WT 130 construct was made by first digesting WT 544 with FatI and later subcloning the appropriate digested fragment. Mutant 6 and Mutant 12 constructs were made through amplifying WT 130 with primers containing mutations at the desired locations. To generate full-length PEG3, we purchased a cDNA clone (GenBank Accession No. BC072661) for mouse PEG3 which has two deleted regions in the middle of the 9th exon causing a premature stop codon. Thus, a full-length cDNA clone for mouse PEG3 was reconstructed by first amplifying the missing 1055 bp of exon 9 from mouse genomic DNA with primers containing restriction site sequences for either NdeI (5′) or ApaI (3′). These two sites singular sites are inside exon 9, flanking the missing region, and were used to insert the correct portion into the cDNA construct. The final construct was sequenced to confirm the intact full-length ORF, amplified with primers containing NotI restriction sites, and subcloned into the pcDNA 3.1 (-) vector (Invitrogen).
For the β-galactosidase assay, Neuro2a cells (ATCC) were cultured in DMEM plus GlutaMAX medium with 5% fetal bovine serum and 1% antibiotic-antimycotic (GibcoBRL), and plated in 12-well plates (1 × 105 per well) for plasmid transfection. On the following day, the cells were transfected with 4 μL Lipofectamine 2000 (Invitrogen) and 1.5 μg of a reporter construct either alone or with the expression vector containing full-length PEG3 in serum and antibiotic free media. DNA concentrations were kept constant among the experimental groups by the addition of empty pcDNA3.1 plasmid DNA. For the PEG3 concentration gradient, the addition of 10 ng of the PEG3 construct is equivalent to 1X, 50 ng is equivalent to 5X, and so forth. Fresh complete media was added 6 hrs post transfection, and total cell lysates were harvested in 75 μL of cell lysis buffer 48 hrs later according to our previously published protocol [37]. The β-galactosidase assay was performed as previously published with each promoter construct being tested in triplicate. This assay was normalized to the total amount of protein, which was determined by the Coomassie Plus Assay (Thermo Scientific).
Expression level analysis of potential target genes of PEG3 in Peg3 mutant mice
Whole brains were harvested from one-day-old littermates that were either wild-type (WT) or heterozygous (KO) for a mutant allele with paternal transmission lacking the 2.5-kb 1st intron region of Peg3 [29]. The harvested brain tissue was homogenized in Trizol (1 ml per 100 mg tissue, Invitrogen) and total RNA was isolated by phenol-chloroform extraction and alcohol precipitation. The total RNA (500 ng) was reverse-transcribed using random hexamers under the following conditions: 25°C for 10 mins, 37°C for 50 mins, 70°C for 15 mins. The cDNA (200 ng) was used for qRT-PCR analysis under the identical conditions mentioned above for the ChIP experiments. All data were analyzed using the 2-ΔΔCt equation by normalizing first to the expression levels of an internal control, β-actin, then to the expression levels of individual genes in WT tissue. For statistical analysis, Student’s t-test was performed using the 1 tailed-type 2 criterion. The primer sequences for each gene were designed from different exons to exclude genomic DNA contamination (Supplemental Table 1).
Supplementary Material
Supplemental Figure 1. Validation of PEG3 antibody. Antibody produced against mouse PEG3 protein displayed specific binding to both exogenous (A) and endogenous (B) PEG3 protein by immunoblot analysis. For detecting exogenous expression, the full-length PEG3 protein was over-expressed in 293T cells. For detecting endogenous expression, total PEG3 protein was determined before and after siRNA knockdown in Neuro2a cells. For the Neuro2a cell line, but not the 293T, a presumably non-specific band (n/s) was detected at 72 kDa based on size analysis. For all analyses, immunoblots were probed with antibody to actin to demonstrate equivalent protein amounts between samples.
Supplemental Figure 2. Pgm2l1 promoter sequence conservation. The sequence conservation for a portion of the Pgm2l1 promoter containing the PEG3 DNA binding motif was analyzed among 6 placental mammals. The alignment is based on 50 bp from mouse chromosome 7 (bases 107,376,130-107,376,179) near PEG3 sequenced fragments from ChIP-Seq analysis originally depicted in Figure 1C.
Supplemental Figure 3. Electromobility shift assay (EMSA) with the Syap1 region probe.
The sequences for the 32P -labeled probe and all of the unlabeled competitors are provided in the bottom boxes. The red-colored nucleotide bases in the right box indicate the nucleotide bases mutated to A. The labels for the gel picture are as follows: P, 32P -labeled probe; C, complex formed between probe and proteins in the nuclear extract; n/s, non-specific complex formation between probe and antibody; S, supershifted complex composed of probe, PEG3 protein, and PEG3 antibody. The Syap1 probe complexes were competed with either unlabeled self-probe, a mutated version of the Syap1 probe, a wild-type Pgm2l1 probe, or a mutated Pgm2l1 probe.
Highlights.
ChIP analysis was used to identify specific genomic target regions bound by the PEG3 protein.
PEG3 was demonstrated to bind the consensus DNA-binding motif AGTnnCnnnTGGCT.
PEG3 controls the expression of a subset of the target genes in a mutant mouse model with reduced levels of PEG3 protein.
Overall, PEG3 was established to be a DNA-binding protein controlling specific cellular functions.
ACKNOWLEDGEMENTS
We would like to thank Drs. Mark Batzer and Scott Herke for their advice and expertise with the ChIP-Seq experimental process and the Kim laboratory members for their detailed suggestions pertaining to the proofreading of this manuscript.
FUNDING
This work was supported by the National Institute of Health [R01-GM066225 to J.K., R15-ES019118 to J.K.].
Abbreviations
- Peg3
paternally expressed gene 3
- Klf4
Krueppel-like factor 4
- Wt1
Wilm’s tumor protein homolog 1
- Zac1
zinc finger protein 1 regulating apoptosis and cell cycle arrest
- Igf2
insulin-like growth factor 2
- Grb10
growth factor receptor-bound protein 10
- Cdkn1c
cyclin-dependent kinase inhibitor 1C isoform 1
- Traf2
TNF receptor-associated factor 2
- Pgm2l1
glucose 1,6-bisphosphate synthase
- Aebp2
adipocyte enhancer binding protein 2
- Eml2
echinoderm microtubule-associated protein-like 2
- Ndufs7
NADH dehydrogenase [ubiquinone] iron-sulfur 7
- Setd1a
SET domain containing 1a
- Syap1
synapse-associated protein 1
- Wnt9a
protein Wnt-9a precursor
- Dusp1
dual specificity protein phosphatase 1
- Mapk14
mitogen-activated protein kinase 14
- Nt5c1a
cytosolic 5′-nucleotidase 1a
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY DATA
Supplementary Data are available online: Supplementary Tables 1-2, Supplementary Figures 1-3.
REFERENCES
- [1].Barlow DP. Genomic imprinting: a Mammalian epigenetic discovery model. Annual review of genetics. 2011;45:379–403. doi: 10.1146/annurev-genet-110410-132459. [DOI] [PubMed] [Google Scholar]
- [2].Sha K. A mechanistic view of genomic imprinting. Annual review of genomics and human genetics. 2008;9:197–216. doi: 10.1146/annurev.genom.122007.110031. [DOI] [PubMed] [Google Scholar]
- [3].Ivanova E, Kelsey G. Imprinted genes and hypothalamic function. Journal of molecular endocrinology. 2011;47:R67–74. doi: 10.1530/JME-11-0065. [DOI] [PubMed] [Google Scholar]
- [4].Miyoshi N, Barton SC, Kaneda M, Hajkova P, Surani MA. The continuing quest to comprehend genomic imprinting. Cytogenetic and genome research. 2006;113:6–11. doi: 10.1159/000090808. [DOI] [PubMed] [Google Scholar]
- [5].Truty MJ, Lomberk G, Fernandez-Zapico ME, Urrutia R. Silencing of the transforming growth factor-beta (TGFbeta) receptor II by Kruppel-like factor 14 underscores the importance of a negative feedback mechanism in TGFbeta signaling. The Journal of biological chemistry. 2009;284:6291–6300. doi: 10.1074/jbc.M807791200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roberts SG. Transcriptional regulation by WT1 in development. Current opinion in genetics & development. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- [7].Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Developmental cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- [8].Sandhu KS. Systems properties of proteins encoded by imprinted genes. Epigenetics : official journal of the DNA Methylation Society. 2010;5:627–636. doi: 10.4161/epi.5.7.12883. [DOI] [PubMed] [Google Scholar]
- [9].Andrade AC, Lui JC, Nilsson O. Temporal and spatial expression of a growth-regulated network of imprinted genes in growth plate. Pediatr Nephrol. 2010;25:617–623. doi: 10.1007/s00467-009-1339-y. [DOI] [PubMed] [Google Scholar]
- [10].Fauque P, Ripoche MA, Tost J, Journot L, Gabory A, Busato F, Le Digarcher A, Mondon F, Gut I, Jouannet P, Vaiman D, Dandolo L, Jammes H. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Human molecular genetics. 2010;19:1779–1790. doi: 10.1093/hmg/ddq059. [DOI] [PubMed] [Google Scholar]
- [11].Relaix F, Weng X, Marazzi G, Yang E, Copeland N, Jenkins N, Spence SE, Sassoon D. Pw1, a novel zinc finger gene implicated in the myogenic and neuronal lineages. Developmental biology. 1996;177:383–396. doi: 10.1006/dbio.1996.0172. [DOI] [PubMed] [Google Scholar]
- [12].Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li LL, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, Barton SC, Ishino F, Surani MA. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nature genetics. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- [13].Iuchi S. Three classes of C2H2 zinc finger proteins. Cellular and molecular life sciences : CMLS. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deng Y, Wu X. Peg3/Pw1 promotes p53-mediated apoptosis by inducing Bax translocation from cytosol to mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12050–12055. doi: 10.1073/pnas.97.22.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Relaix F, Wei X, Li W, Pan J, Lin Y, Bowtell DD, Sassoon DA, Wu X. Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2105–2110. doi: 10.1073/pnas.040378897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yamaguchi A, Taniguchi M, Hori O, Ogawa S, Tojo N, Matsuoka N, Miyake S, Kasai K, Sugimoto H, Tamatani M, Yamashita T, Tohyama M. Peg3/Pw1 is involved in p53- mediated cell death pathway in brain ischemia/hypoxia. The Journal of biological chemistry. 2002;277:623–629. doi: 10.1074/jbc.M107435200. [DOI] [PubMed] [Google Scholar]
- [17].Jiang X, Yu Y, Yang HW, Agar NY, Frado L, Johnson MD. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. The Journal of biological chemistry. 2010;285:8472–8480. doi: 10.1074/jbc.M109.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Relaix F, Wei XJ, Wu X, Sassoon DA. Peg3/Pw1 is an imprinted gene involved in the TNF-NFkappaB signal transduction pathway. Nature genetics. 1998;18:287–291. doi: 10.1038/ng0398-287. [DOI] [PubMed] [Google Scholar]
- [19].Nicolas N, Marazzi G, Kelley K, Sassoon D. Embryonic deregulation of muscle stress signaling pathways leads to altered postnatal stem cell behavior and a failure in postnatal muscle growth. Developmental biology. 2005;281:171–183. doi: 10.1016/j.ydbio.2005.02.022. [DOI] [PubMed] [Google Scholar]
- [20].Schwarzkopf M, Coletti D, Sassoon D, Marazzi G. Muscle cachexia is regulated by a p53-PW1/Peg3-dependent pathway. Genes & development. 2006;20:3440–3452. doi: 10.1101/gad.412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Swaney WT, Curley JP, Champagne FA, Keverne EB. The paternally expressed gene Peg3 regulates sexual experience-dependent preferences for estrous odors. Behavioral neuroscience. 2008;122:963–973. doi: 10.1037/a0012706. [DOI] [PubMed] [Google Scholar]
- [22].Curley JP, Pinnock SB, Dickson SL, Thresher R, Miyoshi N, Surani MA, Keverne EB. Increased body fat in mice with a targeted mutation of the paternally expressed imprinted gene Peg3. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:1302–1304. doi: 10.1096/fj.04-3216fje. [DOI] [PubMed] [Google Scholar]
- [23].Curley JP, Barton S, Surani A, Keverne EB. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proceedings. Biological sciences / The Royal Society. 2004;271:1303–1309. doi: 10.1098/rspb.2004.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- [25].Kim J, Kim JD. In vivo YY1 knockdown effects on genomic imprinting. Human molecular genetics. 2008;17:391–401. doi: 10.1093/hmg/ddm316. [DOI] [PubMed] [Google Scholar]
- [26].Elkan T.L.B.a.C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; 1994; pp. 28–36. [PubMed] [Google Scholar]
- [27].Kim J, Ashworth L, Branscomb E, Stubbs L. The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4. Genome research. 1997;7:532–540. doi: 10.1101/gr.7.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim J, Bergmann A, Lucas S, Stone R, Stubbs L. Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics. 2004;84:47–58. doi: 10.1016/j.ygeno.2004.02.007. [DOI] [PubMed] [Google Scholar]
- [29].Kim J, Ekram MB, Kim H, Faisal M, Frey W, Huang JM, Tran K, Kim MM, Yu S. Imprinting Control Region (ICR) of the Peg3 domain. Human molecular genetics. 2012 doi: 10.1093/hmg/dds092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Renfree MB, Ager EI, Shaw G, Pask AJ. Genomic imprinting in marsupial placentation. Reproduction. 2008;136:523–531. doi: 10.1530/REP-08-0264. [DOI] [PubMed] [Google Scholar]
- [31].Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. Evolution of genomic imprinting: insights from marsupials and monotremes. Annual review of genomics and human genetics. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- [32].Swaney WT. Genomic imprinting and mammalian reproduction. Hormones and behavior. 2011;59:369–374. doi: 10.1016/j.yhbeh.2010.05.012. [DOI] [PubMed] [Google Scholar]
- [33].Kapsimali M, Caneparo L, Houart C, Wilson SW. Inhibition of Wnt/Axin/beta-catenin pathway activity promotes ventral CNS midline tissue to adopt hypothalamic rather than floorplate identity. Development. 2004;131:5923–5933. doi: 10.1242/dev.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Human molecular genetics. 2003;12:233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- [35].Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Blahnik KR, Dou L, O’Geen H, McPhillips T, Xu X, Cao AR, Iyengar S, Nicolet CM, Ludascher B, Korf I, Farnham PJ. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic acids research. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim JD, Yu S, Kim J. YY1 is autoregulated through its own DNA-binding sites. BMC molecular biology. 2009;10:85. doi: 10.1186/1471-2199-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Validation of PEG3 antibody. Antibody produced against mouse PEG3 protein displayed specific binding to both exogenous (A) and endogenous (B) PEG3 protein by immunoblot analysis. For detecting exogenous expression, the full-length PEG3 protein was over-expressed in 293T cells. For detecting endogenous expression, total PEG3 protein was determined before and after siRNA knockdown in Neuro2a cells. For the Neuro2a cell line, but not the 293T, a presumably non-specific band (n/s) was detected at 72 kDa based on size analysis. For all analyses, immunoblots were probed with antibody to actin to demonstrate equivalent protein amounts between samples.
Supplemental Figure 2. Pgm2l1 promoter sequence conservation. The sequence conservation for a portion of the Pgm2l1 promoter containing the PEG3 DNA binding motif was analyzed among 6 placental mammals. The alignment is based on 50 bp from mouse chromosome 7 (bases 107,376,130-107,376,179) near PEG3 sequenced fragments from ChIP-Seq analysis originally depicted in Figure 1C.
Supplemental Figure 3. Electromobility shift assay (EMSA) with the Syap1 region probe.
The sequences for the 32P -labeled probe and all of the unlabeled competitors are provided in the bottom boxes. The red-colored nucleotide bases in the right box indicate the nucleotide bases mutated to A. The labels for the gel picture are as follows: P, 32P -labeled probe; C, complex formed between probe and proteins in the nuclear extract; n/s, non-specific complex formation between probe and antibody; S, supershifted complex composed of probe, PEG3 protein, and PEG3 antibody. The Syap1 probe complexes were competed with either unlabeled self-probe, a mutated version of the Syap1 probe, a wild-type Pgm2l1 probe, or a mutated Pgm2l1 probe.