Abstract
The Rad52 protein was largely ignored in humans and other mammals when the mouse knockout revealed a largely “no-effect” phenotype. However, using synthetic lethal approaches to investigate context dependent function, new studies have shown that Rad52 plays a key survival role in cells lacking the function of the BRCA1-BRCA2 pathway of homologous recombination. Biochemical studies also showed significant differences between yeast and human Rad52, in which yeast Rad52 can promote strand invasion of RPA-coated single-stranded DNA in the presence of Rad51, but human Rad52 cannot. This results in the paradox of how is human Rad52 providing Rad51 function: presumably there is something missing in the biochemical assays that exists in-vivo, but the nature of this missing factor is currently unknown. Recent studies have suggested that Rad52 provides back-up Rad51 function for all members of the BRCA1-BRCA2 pathway, suggesting that Rad52 may be a target for therapy in BRCA pathway deficient cancers. Screening for ways to inhibit Rad52 would potentially provide a complementary strategy for targeting BRCA-deficient cancers in addition to PARP inhibitors.
Background
Eukaryotic cells are exposed to both endogenous and exogenous insults to their genome. To foster genomic maintenance and protection, an elaborate DNA damage response (DDR) and DNA repair network evolved to encompass multiple repair pathways, each specializing in specific types of DNA lesions. Various sensor, mediator, and effector proteins are required to initiate and complete repair of damaged DNA. Deficiencies in essential components of the DDR or DNA repair pathways results in cell death or accumulation of mutations that can progress to cancer or other disease. For example, ataxia telangiectasia, ataxia-telangectasia-like disorder (AT-LD), Njimegen Breakage syndrome (NBS), and xeroderma pigmentosum are caused by mutations of ATM(1), Mre11(2), NBS1, and XPA respectively. In addition, mutations in the tumor suppressors genes BRCA1 and BRCA2 greatly increase susceptibility to breast, ovarian, and other cancers(3).
One of the most threatening forms of DNA damage is the DNA double-strand break (DSB), as both strands of the DNA duplex are impaired simultaneously. The major repair pathways that cope with DSBs are non-homologous end-joining (NHEJ) and homologous recombination (HR). In many organisms, Rad52 is a key protein involved in the HR pathway. This review will focus on Rad52, its role in HR and the translational opportunities available through understanding this protein and its function.
Homologous recombination pathways
Homologous recombination is the exchange of genetic information between allelic sequences and has an essential role in both meiosis and mitosis. In meiosis, HR allows for exchange of genetic material between paternal and maternal alleles within the gamete, and also coordinates proper segregation of homologous chromosome pairs during the first meiotic division.
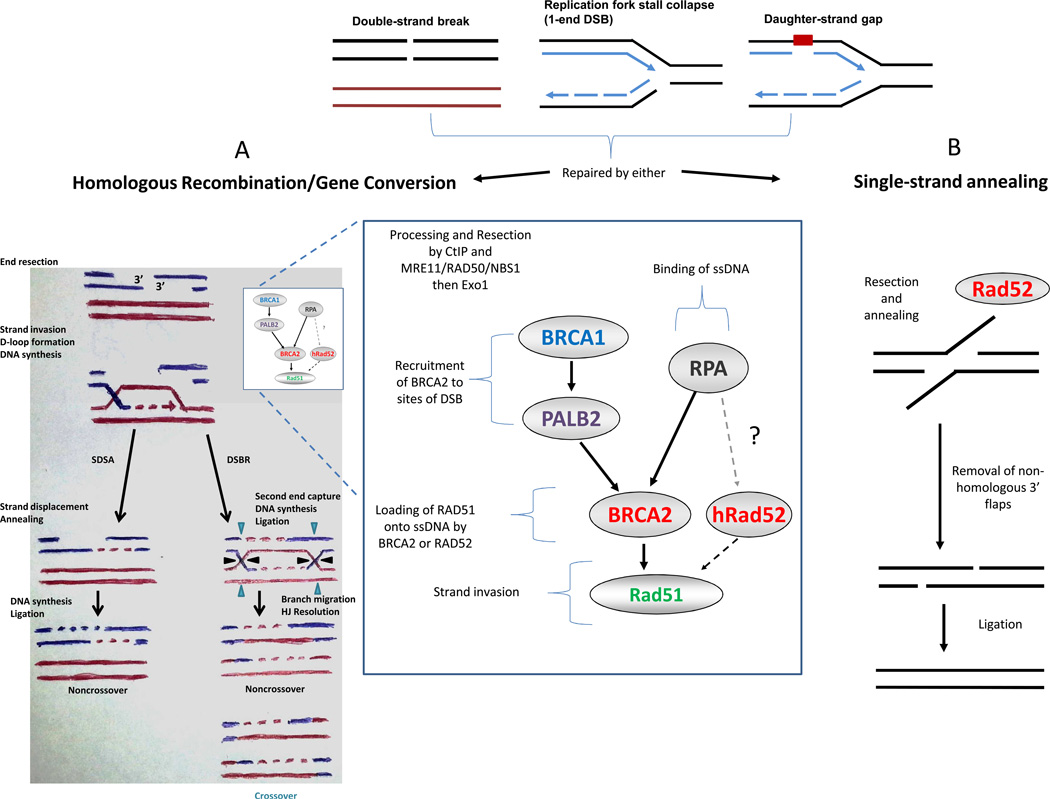
In the somatic cell, HR maintains genomic integrity by promoting accurate repair of DSBs, damaged replication forks, and DNA interstrand crosslinks. HR repair requires a second, homologous DNA sequence to function as a donor template. In brief, the steps of DSB repair (DSBR) by HR are as follows: (a) recognition of a DSB, (b) processing of the DSB by nucleases to generate 3’ single-stranded DNA (ssDNA) tails, (c) formation of a Rad51 recombinase filament on ssDNA ends, (d) strand invasion into the homologous sequence with formation of the D-loop intermediate, (e) DNA polymerase extension from the 3’ end of the invading strand, (f) capture of the second end of the DSB by strand annealing after extension of the D-loop, (g) formation of two Holliday junctions (HJ), and (h) dissolution or resolution of the HJs to form crossover or noncrossover products (Fig 1, left). Of note, the Rad51 recombinase is the essential enzyme that mediates strand invasion of a DNA duplex resulting in an exchange of DNA strands during HR.
Fig 1.
The BRCA and RAD52 pathways of DNA double-strand break (DSB) repair. Various DNA lesions lead to DSBs that can be repaired by homologous recombination (HR) or single-strand annealing (SSA). A) In the HR pathway, after activation of the DNA damage response pathway, DNA ends are resected to expose 3’ single-stranded DNA (ssDNA) which become substrates for binding by RPA. BRCA2 or, in its absence, Rad52 can recruit Rad51 for loading onto ssDNA, displacing RPA, allowing for homology searching and strand invasion by the Rad51 nucleofilament. Subsequently, noncrossover or crossover ligation products are generated in the double-strand break repair (DSBR) pathway or only noncrossover products when synthesis-dependent strand-annealing (SDSA) is utilized. B) SSA requires repetitive sequences around the DSB. After resection, Rad52 mediates annealing of the exposed complementary sequences. After removal of the 3’-flaps, ligation leads to repair, with loss of the intervening sequence. *There is currently no clear evidence that 1-end DSBs or daughter-strand gaps are repaired by single-strand annealing.
The left half of panel A is adapted by permission from Macmillan Publishers Ltd: Nature Reviews Molecular Cell Biology (66), copyright 2006.
The synthesis-dependent strand-annealing (SDSA) pathway of HR differs from the DSBR pathway by diverging following step (e) and is not dependent on the second HJ formation. Rather, SDSA relies upon displacement of the invading strand and annealing it with the second resected DSB end, ultimately, producing a noncrossover product (Fig 1, left).
When a DSB is flanked closely by sequence repeats, repair of the DSB may occur through a process called single-strand annealing (SSA). In SSA the DSB ends are resected, but rather than identifying a homologous DNA template for strand invasion, the resected strands anneal to one another using the repeat sequences for annealing. The result is a deletion of the sequence between the direct repeats (Fig 1, right). Since SSA is independent of strand invasion, the components involved in this process, such as Rad51 filament formation, and the downstream events, including HJ resolution, are not required(4).
Single-ended DSBs can occur at telomeres or at broken replication forks. These can be repaired by HR through a single-ended invasion process known as break-induced replication (BIR)(5). Most BIR events are dependent on Rad51 and other HR factors used in DSBR and SDSA. This process can also occur without Rad51 protein although the Rad51-independent process is thought to be only a minor component of this reaction.
A number of excellent reviews have been published with more in-depth discussion of HR and its sub-pathways (SSA, BIR)(4; 6; 7).
Saccharomyces cerevisiae Rad52 protein and its function in mediating recombination
The S. cerevisiae Rad52 protein (ScRad52) was identified in a genetic screen for mutants that are sensitive to ionizing radiation (IR)(8) and is the most studied recombination mediator. Indeed, many other HR genes are labeled under the heading of RAD52 epistasis group, which includes RAD50, RAD51, RAD54, RAD55, RAD57, RAD59, MRE11, and XRS2. Of all members of the RAD52 epistasis group, the absence of RAD52 confers the most severe defects due to its involvement in all known pathways of HR, both RAD51-dependent (DSBR, SDSA) and the RAD51-independent pathway of SSA(6).
Rad52 forms an oligomeric ring, and the oligomerization is mediated by the N-terminal portion of the protein(9). This N-terminal portion specifically binds ssDNA(10). ScRad52 mediates Rad51 strand invasion by physically associating with the Rad51 protein(11) and allowing for highly efficient reversal of replication protein A (RPA)-imposed inhibition of the ssDNA-dependent ATPase and recombinase activity of Rad51(12–14).
Rad52 also directly associates with RPA(15), a hetero-trimeric ssDNA binding protein that coats the resected ends of the DSB (Fig 1). Both the largest (RPA70) and intermediate (RPA32) subunits demonstrate direct binding with ScRad52(16; 17). The specific interaction of RPA and ScRad52 appears important for the recombination mediator function of ScRad52, as demonstrated when RPA is substituted with the Escherichia coli single-stranded DNA-binding protein (SSB), ScRad52 is not able to overcome SSB-inhibition and subsequently fails to promote Rad51-mediated homology search and strand exchange(13).
Additional functions of S.cerevisiae Rad52
ScRad52 is required for the Rad51-independent SSA and BIR reactions(4; 5). In concordance with its role in SSA, ScRad52 is able to anneal DNA strands that are either bare or coated with RPA(16; 18). There is some evidence to suggest that Rad52-mediated annealing of RPA-coated ssDNA strands is important for second ssDNA end capture in the DSBR pathway of HR(19).
Human recombination mediators: Rad52 and BRCA2
Human Rad52 (hRad52) is similar to ScRad52 structurally and biochemically. hRad52 exists in an oligomeric form, binds ssDNA, promotes ssDNA annealing and, under certain specialized conditions, simulates Rad51-mediated homologous DNA pairing(20). Like ScRad52, hRad52 has conserved the ability to directly interact with RPA(17).
Subsequent studies have examined the N-terminal portion of hRad52 to determine in detail its interaction with ssDNA. Investigation by X-ray crystallography revealed an undecameric (11-subunit) ring structure with a deep groove on the surface that is lined by a vast number of positively charged basic and aromatic residues(21; 22). These hRad52 residues have been shown to be important for ssDNA binding by mutational analysis(22; 23) and deletions of equivalent residues in ScRad52 demonstrate deficiencies in DNA repair and HR(24).
More recently, a second DNA binding domain has been discovered in hRad52, which appears to bind dsDNA(25). Mutations of these amino acid residues are defective in promoting Rad51-mediated D-loop formation in ScRad52(26). Further study of this second DNA binding domain is necessary to understand how it contributes to the known functions of Rad52.
Interesting, although hRad52 did not demonstrate recombination mediator activity in reconstituted biochemical assays(27), it appears to mediate Rad51 function in human cancer cells deficient in BRCA1, PALB2 (28)or BRCA2(29). We postulate that hRad52 may perform its mediator function in combination with partner proteins. It has been suggested that the Rad51 paralogs, Rad51B/C/D-XRCC2, may fulfill this role. However, the Rad51 paralogs appear to function primarily in the BRCA2 pathway and operate independently of hRad52 in mediating the Rad51 recombinase (Chun J and Powell SN, unpublished observations). There remains other Rad51 paralogs that may be candidate co-factors, including hSWS1 (homolog of yeast Shu2) and SWSAP1(30). Alternatively, hRad52 recombination mediator activity may be dependent on specific posttranslational modifications (e.g. phosphorylation, SUMOylation, etc.) that have yet to be examined biochemically.
Besides its role in Rad51 mediation, it has recently been suggested that BRCA2 participates in the stabilization of RAD51 filaments from degradation by MRE11 through interaction of BRCA2’s conserved C-terminal domain with Rad51(31). It is unknown whether Rad52 has any role to play in this proposed mechanism of replication fork protection.
Species spectrum of Rad52 and BRCA2
BRCA2 and its homologs appear to have appropriated subsets of Rad52 function, especially in mediating recombination by Rad51, and are present in a variety of multi-cellular organisms(See Table 1)(32–39).
Table 1.
The spectrum of BRCA2 and RAD52 across various species.
| Biochemical activity | ||||
|---|---|---|---|---|
| Species | Mediator | Annealer | HR Phenotype | |
| S. cerevisiae | No BRCA2 | |||
| Rad52 | Yes | Yes | ++ | |
| U. maydis | Brh2 | Yes | Yes | ++ |
| Rad52 | Low | Yes | − | |
| A. thaliana | BRCA2 | ? | ? | + |
| Rad52 | ? | ? | + | |
| D. melanogaster | BRCA2 | ? | ? | + |
| No Rad52 | ||||
| C. elegans | BRC-2 | Yes | Yes | ++ |
| No Rad52 | ||||
|
H. sapiens M. musculus |
BRCA2 | Yes | No | ++ |
| RAD52 | No | Yes | (+) | |
++ = strong phenotype, + = intermediate phenotype, (+) = weak phenotype; ? = unknown/no published reports identified.
A speculative assumption is that BRCA2 has evolved for the greater level of insults to the genome in multi-cellular organisms in comparison to unicellular organisms. Seen from another perspective, increased -- but regulated -- mutagenesis is evolutionarily preferential in unicellular life; however in multi-cellular organisms more stringent control of DNA repair, especially in stem cells, via BRCA2 is essential for vitality. U. maydis exists as a unique unicellular exception to possessing a BRCA2 homolog, possibly, to enable it to tolerate the greater levels of exogenous DNA damage (e.g. high UV exposure, etc.) it encounters in its native environment budding on crops of corn or in support of its multi-cellular filamentous form.
Rad52 and its synthetically lethal interactions across various species
The dual role of ScRad52 in both mediating Rad51 function and performing the annealing step in second end capture and SDSA explains why ScRad52 mutants display more severe phenotypes than defects in Rad51 protein. Unexpectedly, in organisms containing a BRCA2 homolog, including U. maydis, chicken, and mice, inactivation of Rad52 causes minimal or no HR and DNA repair defects(39–41). However, in human cancer cell-lines deficient in BRCA1, PALB2(28), or BRCA2(29), RAD52 depletion increases damage-induced chromosomal abnormalities, decreases clonogenic survival, and further reduces the rates or frequency of HR(28, 29). Thus, RAD52 appears to exist in a relationship with BRCA1, PALB2, or BRCA2, known as synthetic lethality, where simultaneous inactivation of two genes leads to cell death, whereas inactivation of only one of these genes does not affect viability. This observation is in accordance with other RAD52 synthetically lethal phenotypes seen in chicken DT40 cells, where inactivation of rad52 is lethal with a defect in XRCC3, a RAD51 paralog(42), and, in U. maydis, where a Rad51 paralog mutant, rec2, demonstrates synthetic lethality with loss of rad52(39).
Interestingly, in DT40 cells, rad52 deletion in a BRCA2 mutant background did not enhance cytotoxicity, instead epistasis was observed between BRCA2 and Rad52, as well as other HR proteins(43). Also, in U. maydis, Brh2 mutants in combination with Rad52 defects demonstrate a more subtle synthetically lethal phenotype, which the study’s authors interpret as a compensatory interaction(39). These observed divergences from human and other studied organisms will require further investigation to delineate the hierarchy and functional nuances of these various HR proteins across the evolutionary spectrum.
Dependency of BRCA1-PALB2-BRCA2 deficient human tumor cells on Rad52-Rad51-mediated HR
BRCA1, PALB2 and BRCA2 appear to be linked in a sequential manner. BRCA2 recruitment to foci requires interaction with PALB2, and abolishment of the physical interface between BRCA1 and PALB2 impairs BRCA2 function and subsequently Rad51-mediated HR(44–46). Evidence suggests Rad52 provides an alternative mediator pathway to the BRCA1-PALB2-BRCA2 pathway and allows tumor cells to proliferate in the absence of the BRCA pathway (Fig 1). The loss of Rad52 in a BRCA1, PALB2 (28), or BRCA2(29) mutant leads to cell death.
These observations set the potential foundation for any BRCA1-PALB2-BRCA2 pathway deficient tumor to be therapeutically targeted by Rad52 inactivation. This approach for targeting BRCA pathway deficient cancers is distinct from other strategies that take advantage of synthetic lethality in BRCA mutant tumors, namely poly-(ADP-ribose) polymerase (PARP) inhibition. These translational approaches and discoveries that exploit HR defective cancers will be highlighted in the subsequent section.
Clinical–Translational Advances
Exploiting synthetic lethal interactions in HR deficient tumors
Synthetic lethality can be exploited therapeutically by identifying cells with a cancer-related mutation or loss of a single gene that can then be evaluated for a second gene or protein target that would render the cancer cells non-viable. Since the loss of the second gene alone is not lethal to normal cells, tumor-specific kill can be achieved. This approach has been employed to great promise and effect in HR deficient BRCA1 and BRCA2 mutant cancers, and is being expanded to many cancers with tumor-specific alterations that are potentially exploitable, such as oncogene addiction.
PARP inhibition in BRCA1-BRCA2 deficient tumors
PARP inhibitors (PARPi) are synthetically lethal with BRCA defective tumor cells(47; 48). The presumed mechanistic model for this lethal interaction is the inhibition of PARP prevents repair of single-strand DNA breaks which accumulate and are then converted into DSBs during replication. The DSB repair HR pathway is impaired in BRCA1-BRCA2 mutant tumors, leading to a lethal amount of DSBs after PARP inhibition. However, new evidence has suggested other models that build upon or modify this assumed mechanism. One model proposes that PARPi may cause PARP-1 to be trapped onto DNA repair intermediates that then stall replication forks, or another suggests that PARP is directly involved in catalyzing the restart of stalled replication forks. There are excellent reviews for further discussion of this topic(49; 50).
Regardless of the mechanism, the use of PARPi to target BRCA mutant cancers has moved beyond the laboratory and into clinical trials with promising results(51–54). However, even in these studies, some BRCA mutant carriers respond poorly(51) and resistance to PARPi therapy via BRCA2 reversion mutations has been documented(55). In addition, expression of the multi-drug resistance transporter also results in resistance to PARPi. These observations highlight the need to identify biomarkers that will predict patient response to PARP inhibition and the development of additional strategies for exploiting BRCA pathway deficiency.
Targeting Rad52 in BRCA1-BRCA2 deficient tumors
The rationale for targeting Rad52 in BRCA-deficient tumors was established by the pre-clinical evidence covered in the preceding sections. In addressing the inactivation of Rad52 in the clinical setting, a variety of approaches can be examined. One approach would be to target Rad52 directly. Currently, there are no known enzymatic or kinase functions of Rad52, which preclude the more well-studied pharmacologic approaches. However, as advances are made by molecular pharmacologists and medicinal chemists, creating or identifying a compound that disrupts the oligomer ring structure or binds in the vicinity of the DNA binding groove of Rad52 to prevent access by the DNA substrate may be potential molecular mechanisms. These approaches will require high-throughput screening with libraries of compounds as a top-down approach coupled with other investigations from a bottom-up mechanistically driven strategy.
Targeting post-translational modifications of Rad52 that are essential for function is another promising avenue of approach. Rad52 is phosphorylated by the c-Abl kinase which affects its ability to form sub-nuclear foci(56) and enhances its annealing functions(57). Disrupting the c-Abl kinase through genetic and pharmacological approaches in preliminary studies appeared to disrupt Rad52 function (Lok BH and Powell SN, unpublished observations). Another post-translational modification that contributes to Rad52 function is SUMOylation, which may be another potential target for impeding Rad52 function(58–62). The interaction of Rad52 with RPA may also be a potentially targetable site, but developing drugs to inhibit protein-protein interactions has always been a challenging problem. Whether these strategies will result in clinically applicable therapeutics remains to be seen.
Identification of BRCA pathway deficient cancers susceptible to synthetically lethal therapeutic approaches
Even with the mechanistic understanding of these realized and potential therapeutic approaches, proper identification of tumors susceptible to these interventions is equally important. A potent intervention applied to an improper scenario/disease-state will result in disappointingly ineffectual or even harmful outcomes.
Indeed, one of the primary target proteins of PARPi, PARP-1, displays a spectrum of protein expression levels even within genetically-similar tumor types. In this regard, not all BRCA1-associated breast cancers are created equal, with up to 18% of these tumors expressing none or low levels of nuclear PARP-1 protein(63). This is suggestive that investigating target protein expression levels in tumors may be one approach to predict patient response to PARPi therapy.
The HR proficiency of a tumor can be measured by observing sub-nuclear focus formation of HR proteins induced by ex-vivo irradiation, including BRCA1 and Rad51 amongst others(64; 65), and these HR foci formation assays correlate with the PARPi sensitivity of these tumors(65). This functionally-driven approach allows for more precise patient identification, in addition to expanding the potential pool of targetable tumors, through establishing a predictive biomarker for HR-deficient tumors that are not identified by the traditional genetic test for the BRCA1 or BRCA2 mutation.
Conclusions
We are entering an exciting era of precision-targeted and personalized cancer therapy. With the greater understanding and continual investigation of the complex genetic interactions of oncogenesis and tumor proliferation, rational design of and molecular target identification by various therapeutic methods can be accomplished. Success will also depend on the development of biomarkers of DNA repair function to identify appropriate patients for targeted therapy. Continual and further elucidation of the mechanisms of DNA repair will be paramount to devising therapeutics, identifying appropriate patients, evaluating responsiveness, and preventing resistance to targeted therapeutic strategies.
Acknowledgements
We apologize to those investigators whose work could not be cited owing to space limitations. Work on homologous recombination in S.N.P.’s laboratory is supported by grants from the National Cancer Institute (CA58985, CA86140) and the Susan G. Komen for the Cure (KG081528). B.H.L. was supported in part by a medical research training fellowship from the Howard Hughes Medical Institute (57007004) and a medical student research grant from the Radiological Society of North America (RMS1009).
Footnotes
Conflicts of interest.
The authors have no potential conflict of interest to report.
References
- 1.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. [Internet] [cited 2012 Apr 23];Science (New York, N.Y.) 1995 Jun;268(5218):1749–1753. doi: 10.1126/science.7792600. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7792600. [DOI] [PubMed] [Google Scholar]
- 2.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, et al. [Internet] [cited 2012 Apr 23];Cell. 1999 Dec;99(6):577–587. doi: 10.1016/s0092-8674(00)81547-0. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10612394. [DOI] [PubMed] [Google Scholar]
- 3.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. [Internet] Nature reviews. Cancer. 2012 Jan;12(1):68–78. doi: 10.1038/nrc3181. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22193408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symington LS. Role of RAD52 Epistasis Group Genes in Homologous Recombination and Double-Strand Break Repair [Internet] Microbiology and Molecular Biology Reviews. 2002;66(4):630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents.Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=134659&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. [Internet] [cited 2012 Mar 16];Annual review of biochemistry. 2006 Jan;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16756487. [DOI] [PubMed] [Google Scholar]
- 6.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. [Internet] [cited 2012 Mar 20];Annual review of biochemistry. 2008 Jan;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18275380. [DOI] [PubMed] [Google Scholar]
- 7.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis [Internet] Nature Publishing Group. 2010;11 doi: 10.1038/nrm2851. (mARCH)Available from: http://dx.doi.org/10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Game JC, Mortimer RK. A genetic study of X-ray sensitive mutants in yeast [Internet] [cited 2012 Apr 24];Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1974 Sep;24(3):281–292. doi: 10.1016/0027-5107(74)90176-6. Available from: http://dx.doi.org/10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 9.Ranatunga W, Jackson D, Lloyd JA, Forget AL, Knight KL, Borgstahl GE. Human RAD52 exhibits two modes of self-association. [Internet] [cited 2012 Mar 17];The Journal of biological chemistry. 2001 May;276(19):15876–15880. doi: 10.1074/jbc.M011747200. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11278978. [DOI] [PubMed] [Google Scholar]
- 10.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. [Internet] Proceedings of the National Academy of Sciences of the United States of America. 1996 Oct;93(20):10729–10734. doi: 10.1073/pnas.93.20.10729. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=38223&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne GT, Weaver DT. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. [Internet] [cited 2012 Apr 27];Genes & Development. 1993 Sep;7(9):1755–1765. doi: 10.1101/gad.7.9.1755. Available from: http://www.genesdev.org/cgi/doi/10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara a, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. [Internet] Nature. 1998 Jan;391(6665):404–407. doi: 10.1038/34943. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9450759. [DOI] [PubMed] [Google Scholar]
- 13.New J, Sugiyama T, Zaitseva E. Rad52 protein stimulates DNA strand exchange by Rad51andreplicationproteinA [Internet] [cited 2012 Apr 27];Nature. 1998 391(January):407–410. doi: 10.1038/34950. Available from: http://microbiology.ucdavis.edu/sklab/PDF_files/New et al (1998) Nature 391, 407-410.pdf. [DOI] [PubMed] [Google Scholar]
- 14.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase [Internet] [cited 2012 Apr 29];Journal of Biological Chemistry. 1997 :28194–28197. doi: 10.1074/jbc.272.45.28194. Available from: http://www.jbc.org/content/272/45/28194.short. [DOI] [PubMed]
- 15.Hays SL, Firmenich AA, Massey P, Berg P. Studies of the Interaction between Rad52 Protein and the Yeast Single-Stranded DNA Binding Protein RPA Studies of the Interaction between Rad52 Protein and the Yeast Single-Stranded DNA Binding Protein RPA. 1998 doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. [Internet] Genes to cells?: devoted to molecular & cellular mechanisms. 1998 Mar;3(3):145–156. doi: 10.1046/j.1365-2443.1998.00176.x. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9619627. [DOI] [PubMed] [Google Scholar]
- 17.Jackson D. Analysis of the Human Replication Protein A:Rad52 Complex: Evidence for Crosstalk Between RPA32, RPA70, Rad52 and DNA [Internet] [cited 2012 Apr 16];Journal of Molecular Biology. 2002 Aug;321(1):133–148. doi: 10.1016/s0022-2836(02)00541-7. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0022283602005417. [DOI] [PubMed] [Google Scholar]
- 18.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. [Internet] Proceedings of the National Academy of Sciences of the United States of America. 1998 May;95(11):6049–6054. doi: 10.1073/pnas.95.11.6049. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=27583&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. [Internet] [cited 2012 Mar 28];The EMBO journal. 2006 Nov;25(23):5539–5548. doi: 10.1038/sj.emboj.7601412. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1679760&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann P, West SC. Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. [Internet] Journal of molecular biology. 1999 Aug;291(2):363–374. doi: 10.1006/jmbi.1999.2954. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10438626. [DOI] [PubMed] [Google Scholar]
- 21.Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. [Internet] Proceedings of the National Academy of Sciences of the United States of America. 2002 Oct;99(21):13492–13497. doi: 10.1073/pnas.212449899. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=129701&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, et al. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. [Internet] Molecular cell. 2002 Aug;10(2):359–371. doi: 10.1016/s1097-2765(02)00587-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12191481. [DOI] [PubMed] [Google Scholar]
- 23.Lloyd JA, McGrew DA, Knight KL. Identification of residues important for DNA binding in the full-length human Rad52 protein. [Internet] [cited 2012 Mar 17];Journal of molecular biology. 2005 Jan;345(2):239–249. doi: 10.1016/j.jmb.2004.10.065. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15571718. [DOI] [PubMed] [Google Scholar]
- 24.Mortensen UH, Erdeniz N, Feng Q, Rothstein R. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. [Internet] Genetics. 2002 Jun;161(2):549–562. doi: 10.1093/genetics/161.2.549. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1462154&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagawa W, Kagawa A, Saito K, Ikawa S, Shibata T, Kurumizaka H, et al. Identification of a second DNA binding site in the human Rad52 protein. [Internet] [cited 2012 Apr 16];The Journal of biological chemistry. 2008 Aug;283(35):24264–24273. doi: 10.1074/jbc.M802204200. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3259773&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arai N, Kagawa W, Saito K, Shingu Y, Mikawa T, Kurumizaka H, et al. Vital roles of the second DNA-binding site of Rad52 protein in yeast homologous recombination. [Internet] [cited 2012 Apr 27];The Journal of biological chemistry. 2011 May;286(20):17607–17617. doi: 10.1074/jbc.M110.216739. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21454474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. [Internet] [cited 2012 Mar 9];Nature. 2010 Oct;467(7316):678–683. doi: 10.1038/nature09399. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2952063&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. [Internet] [cited 2012 Sep 15];Oncogene. 2012 Sep;(July):1–7. doi: 10.1038/onc.2012.391. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22964643. [DOI] [PMC free article] [PubMed]
- 29.Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. [Internet] [cited 2011 Jun 19];Proceedings of the National Academy of Sciences of the United States of America. 2011 Jan;108(2):686–691. doi: 10.1073/pnas.1010959107. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3021033&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Wan L, Wu Y, Chen J, Huang J. hSWS1·SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. [Internet] [cited 2012 May 9];The Journal of biological chemistry. 2011 Dec;286(48):41758–41766. doi: 10.1074/jbc.M111.271080. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21965664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. [Internet] [cited 2012 Jul 31];Cell. 2011 May;145(4):529–542. doi: 10.1016/j.cell.2011.03.041. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3261725&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer K, Schüller C, Wambutt R, Murphy G, Volckaert G, Pohl T, et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. [Internet] Nature. 1999 Dec;402(6763):769–777. doi: 10.1038/47134. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10617198. [DOI] [PubMed] [Google Scholar]
- 33.Abe K, Osakabe K, Ishikawa Y, Tagiri A, Yamanouchi H, Takyuu T, et al. Inefficient double-strand DNA break repair is associated with increased fasciation in Arabidopsis BRCA2 mutants. [Internet] [cited 2012 Apr 12];Journal of experimental botany. 2009 Jan;60(9):2751–2761. doi: 10.1093/jxb/erp135. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2692019&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brough R, Wei D, Leulier S, Lord CJ, Rong YS, Ashworth A. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. [Internet] [cited 2012 May 2];DNA repair. 2008 Jan;7(1):10–19. doi: 10.1016/j.dnarep.2007.07.013. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17822964. [DOI] [PubMed] [Google Scholar]
- 35.Klovstad M, Abdu U, Schüpbach T. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. [Internet] [cited 2012 Apr 16];PLoS genetics. 2008 Feb;4(2):e31. doi: 10.1371/journal.pgen.0040031. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2233675&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petalcorin MIR, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. [Internet] [cited 2012 Apr 16];Journal of molecular biology. 2006 Aug;361(2):231–242. doi: 10.1016/j.jmb.2006.06.020. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16843491. [DOI] [PubMed] [Google Scholar]
- 37.Petalcorin MIR, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. [Internet] Proceedings of the National Academy of Sciences of the United States of America. 2007 May;104(20):8299–8304. doi: 10.1073/pnas.0702805104. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1895944&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. [Internet] Molecular cell. 2002 Sep;10(3):683–691. doi: 10.1016/s1097-2765(02)00632-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12408834. [DOI] [PubMed] [Google Scholar]
- 39.Kojic M, Mao N, Zhou Q, Lisby M, Holloman WK. Compensatory role for Rad52 during recombinational repair in Ustilago maydis. [Internet] [cited 2012 Feb 15];Molecular microbiology. 2008 Mar;67(5):1156–1168. doi: 10.1111/j.1365-2958.2008.06116.x. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18208529. [DOI] [PubMed] [Google Scholar]
- 40.Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. [Internet] Molecular and cellular biology. 1998 Nov;18(11):6423–6429. doi: 10.1128/mcb.18.11.6423. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=109228&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi-iwai Y, Sonoda E, Bezzubova O, Morrison C, Buerstedde J-marie Homologous Recombination, but Not DNA Repair, Is Reduced in Vertebrate Cells Deficient in RAD52 Homologous Recombination, but Not DNA Repair, Is Reduced in Vertebrate Cells Deficient in RAD52. 1998 doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimori a, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, et al. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. [Internet] The EMBO journal. 2001 Oct;20(19):5513–5520. doi: 10.1093/emboj/20.19.5513. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=125654&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, et al. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. [Internet] [cited 2012 Apr 3];PLoS genetics. 2011 Jul;7(7)::e1002148. doi: 10.1371/journal.pgen.1002148. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3136442&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7(7):1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sy SMH, Huen MSY, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. [Internet] Proceedings of the National Academy of Sciences of the United States of America. 2009 Apr;106(17):7155–7160. doi: 10.1073/pnas.0811159106. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2678481&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, et al. PALB2 Links BRCA1 and BRCA2 in the DNA-Damage Response [Internet] Current Biology. 2009;19(6):524–529. doi: 10.1016/j.cub.2009.02.018. Available from: http://dx.doi.org/10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryant H, Schultz N, Thomas H, Parker K. Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase [Internet] [cited 2012 May 3];Nature. 2005 7(d):913–918. doi: 10.1038/nature03443. Available from: http://www.nature.com/nature/journal/v434/n7035/full/nature03443.html?free=2. [DOI] [PubMed] [Google Scholar]
- 48.Farmer H, Mccabe N, Lord CJ. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;239(1991):236–239. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 49.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. [Internet] [cited 2012 Apr 3];Molecular oncology. 2011 Aug;5(4):387–393. doi: 10.1016/j.molonc.2011.07.001. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21821475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. [Internet] [cited 2012 Mar 19];Nature reviews. Cancer. 2010 Apr;10(4):293–301. doi: 10.1038/nrc2812. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2910902&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers [Internet] [cited 2012 Feb 22];New England Journal of Medicine. 2009 361(2):123–134. doi: 10.1056/NEJMoa0900212. Available from: http://www.nejm.org/doi/full/10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 52.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. [Internet] [cited 2012 Apr 3];Lancet. 2010 Jul;376(9737):245–251. doi: 10.1016/S0140-6736(10)60893-8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20609468. [DOI] [PubMed] [Google Scholar]
- 53.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. [Internet] [cited 2012 Mar 10];Lancet. 2010 Jul;376(9737):235–244. doi: 10.1016/S0140-6736(10)60892-6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20609467. [DOI] [PubMed] [Google Scholar]
- 54.O’Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer [Internet] [cited 2012 Feb 22];New England Journal of Medicine. 2011 364(3):205–214. doi: 10.1056/NEJMoa1011418. Available from: http://www.nejm.org/doi/full/10.1056/nejmoa1011418. [DOI] [PubMed] [Google Scholar]
- 55.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. [Internet] [cited 2011 Sep 12];Nature. 2008 Feb;451(7182):1111–1115. doi: 10.1038/nature06548. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18264088. [DOI] [PubMed] [Google Scholar]
- 56.Kitao H, Yuan Z-M. Regulation of ionizing radiation-induced Rad52 nuclear foci formation by c-Abl-mediated phosphorylation. [Internet] [cited 2012 Mar 17];The Journal of biological chemistry. 2002 Dec;277(50):48944–48948. doi: 10.1074/jbc.M208151200. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12379650. [DOI] [PubMed] [Google Scholar]
- 57.Honda M, Okuno Y, Yoo J, Ha T, Spies M. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. [Internet] [cited 2011 Dec 24];The EMBO journal. 2011 Aug;30(16):3368–3382. doi: 10.1038/emboj.2011.238. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3160658&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito K, Kagawa W, Suzuki T, Suzuki H, Yokoyama S, Saitoh H, et al. The putative nuclear localization signal of the human RAD52 protein is a potential sumoylation site. [Internet] [cited 2012 Feb 22];Journal of biochemistry. 2010 Jun;147(6):833–842. doi: 10.1093/jb/mvq020. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20190268. [DOI] [PubMed] [Google Scholar]
- 59.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. [Internet] [cited 2011 Jul 16];Nature cell biology. 2007 Aug;9(8):923–931. doi: 10.1038/ncb1619. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17643116. [DOI] [PubMed] [Google Scholar]
- 60.Ohuchi T, Seki M, Branzei D, Maeda D, Ui A, Ogiwara H, et al. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. [Internet] [cited 2012 Feb 15];DNA repair. 2008 Jun;7(6):879–889. doi: 10.1016/j.dnarep.2008.02.005. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18396468. [DOI] [PubMed] [Google Scholar]
- 61.Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. [Internet] [cited 2011 Jul 21];Nature cell biology. 2006 Nov;8(11):1284–1290. doi: 10.1038/ncb1488. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17013376. [DOI] [PubMed] [Google Scholar]
- 62.Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, Damborsky J, et al. Rad52 SUMOylation affects the efficiency of the DNA repair. [Internet] [cited 2011 Jul 21];Nucleic acids research. 2010 Aug;38(14):4708–4721. doi: 10.1093/nar/gkq195. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2919706&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W. PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy [Internet] [cited 2012 May 3];Breast Cancer Research and Treatment. 2011 Mar;127(3):861–869. doi: 10.1007/s10549-011-1441-2. Available from: http://www.springerlink.com/index/10.1007/s10549-011-1441-2. [DOI] [PubMed] [Google Scholar]
- 64.Willers H, Taghian AG, Luo C-M, Treszezamsky A, Sgroi DC, Powell SN. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. [Internet] [cited 2012 May 1];Molecular cancer research?: MCR. 2009 Aug;7(8):1304–1309. doi: 10.1158/1541-7786.MCR-09-0149. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19671671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukhopadhyay A, Elattar A, Cerbinskaite A, Wilkinson SJ, Drew Y, Kyle S, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. [Internet] [cited 2012 Apr 16];Clinical cancer research?: an official journal of the American Association for Cancer Research. 2010 Apr;16(8):2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20371688. [DOI] [PubMed] [Google Scholar]
- 66.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nature Reviews Molecular Cell Biology. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]