Abstract
The present study investigated the association between variants in the vitamin D receptor gene (VDR) and protein tyrosine phosphatase, non-receptor type 2 gene (PTPN2), as well as an interaction between VDR and PTPN2 and the risk of islet autoimmunity (IA) and progression to type 1 diabetes (T1D). The Diabetes Autoimmunity Study in the Young (DAISY) has followed children at increased risk of T1D since 1993. Of the 1692 DAISY children genotyped for VDR rs1544410, VDR rs2228570, VDR rs11568820, PTPN2 rs1893217, and PTPN2 rs478582, 111 developed IA, defined as positivity for GAD, insulin or IA-2 autoantibodies on 2 or more consecutive visits, and 38 IA positive children progressed to T1D. Proportional hazards regression analyses were conducted.
There was no association between IA development and any of the gene variants, nor was there evidence of a VDR*PTPN2 interaction. Progression to T1D in IA positive children was associated with the VDR rs2228570 GG genotype (HR: 0.49, 95% CI: 0.26–0.92) and there was an interaction between VDR rs1544410 and PTPN2 rs1893217 (pinteraction = 0.02). In children with the PTPN2 rs1893217 AA genotype, the VDR rs1544410 AA/AG genotype was associated with a decreased risk of T1D (HR: 0.24, 95% CI: 0.11–0.53, p = 0.0004), while in children with the PTPN2 rs1893217 GG/GA genotype, the VDR rs1544410 AA/AG genotype was not associated with T1D (HR: 1.32, 95% CI: 0.43–4.06, p = 0.62). These findings should be replicated in larger cohorts for confirmation. The interaction between VDR and PTPN2 polymorphisms in the risk of progression to T1D offers insight concerning the role of vitamin D in the etiology of T1D.
Keywords: Vitamin D, Genetics, Epidemiology, Islet autoimmunity, Type 1 diabetes, Vitamin D receptor
1. Introduction
Type 1 diabetes (T1D) is an autoimmune disease in which the insulin-producing beta cells of the pancreas are destroyed. There is typically a preclinical phase of circulating autoantibodies, called islet autoimmunity (IA) that precedes the clinical diagnosis of T1D.
Vitamin D deficiency has been associated with a number of diseases, including multiple sclerosis [1], rheumatoid arthritis, and T1D [2], although not consistently. The mechanism by which vitamin D may exert its effects on these diseases is still not understood completely, particularly with regard to underlying genetic risk. The gene for the vitamin D receptor (VDR), through which vitamin D acts, has long been a candidate gene for T1D. Initial small studies found VDR polymorphisms to be associated with T1D [3–9]. However, Nejentsev et al. analyzed association of the 98 VDR single nucleotide polymorphisms (SNPs) in up to 3763 type 1 diabetic families and found no evidence of association with T1D in the populations tested [10]. Moreover, in a meta-analysis, Guo et al. found no evidence of an association between VDR gene polymorphisms (FokI, BsmI, ApaI, and TaqI) and T1D risk [11]. Finally, in a recent analysis of 19 genes for association with T1D in the Type 1 Diabetes Genetics Consortium families, none of the forty SNPs genotyped in the VDR region were associated with T1D [12]. A recent study [13] showed that the vitamin D receptor binds to a number of genomic positions across the genome, including a novel intronic binding site in the protein tyrosine phosphatase, non-receptor type 2 gene (PTPN2), which has also been associated with T1D through a genome-wide association scan in 2007 [14,15]. This suggests the possibility of a more complex relationship in which variation in both VDR and PTPN2 is necessary to have an effect on diabetes risk, which may explain why previous findings regarding VDR have been inconsistent.
The Diabetes Autoimmunity Study in the Young (DAISY) has been prospectively following children at increased T1D risk for the development of IA and progression to T1D since 1993. The purpose of this study was to examine the associations between 5 particular VDR and PTPN2 SNPs and the development of IA and progression to T1D in the prospective DAISY cohort. We also aimed to explore a potential gene–gene interaction between VDR and PTPN2 polymorphisms and the risk of IA and progression to T1D.
2. Materials and methods
2.1. Subjects
DAISY is a prospective study composed of two groups of children at increased risk for T1D who were recruited between 1993 and 2004 and are being followed prospectively for the development of IA and T1D. One group is made up of first degree relatives of patients with type 1 diabetes mellitus, recruited between birth and eight years of age. The second group consists of infants born at St. Joseph’s Hospital in Denver, Colorado, whose umbilical cord blood was screened for diabetes-susceptibility genotypes in the HLA region. The St. Joseph’s Hospital newborn population is representative of the general population of the Denver metropolitan area. Details of the newborn screening, sibling and offspring recruitment, and follow-up of both cohorts have been published previously [16,17]. Cord blood or the first available blood sample (depending on enrolment group) was sent to Roche Molecular Systems, Inc., Alameda, CA, for PCR-based HLA class II typing. All study protocols were approved by the Colorado Multiple Institutional Review Board, and informed consent was given by parents of all participating children.
2.2. Measurement of autoantibodies
Autoantibodies were tested at 9, 15, and 24 months, and annually thereafter, or at their first visit and annually thereafter if the child enrolled after birth. Radio-immunoassays were used to measure serum autoantibodies to insulin, GAD-65, and IA-2 (BDC512), as previously described [18–21], with rigorous confirmation of all positive and a subset of negative results. The cut-off for positivity was established as the 99th percentile of healthy controls. Children who tested autoantibody positive were put on an accelerated testing schedule of every 3–6 months.
Cases of persistent IA were defined as those children positive for at least one islet autoantibody (IAA, GAD-65, IA-2) on two or more consecutive visits. Type 1 diabetes was diagnosed by a physician and defined as random blood glucose >200 mg/dL and/or HbA1c (A1C) >6.2% with clinical symptoms of diabetes.
2.3. VDR and PTPN2 genotyping
DAISY children were genotyped for VDR rs1544410 (BsmI), VDR rs2228570 (FokI), VDR rs11568820 (Cdx2), PTPN2 rs1893217, and PTPN2 rs478582. The three VDR SNPs were chosen based on previous associations with T1D [3,4,8,9,22] as well as function [10,23–25]. The VDR rs1544410 SNP is located at the 3′ end of VDR and is not known to alter the structure or function of VDR [22,26,27]. The VDR rs2228570 polymorphism is a T-C transition at the translation initiation codon of VDR that results in a shorter protein with increased biological activity [23–25]. VDR rs11568820 is in the 5′ promoter region of VDR, results in reduced transcriptional activity of the promoter, and affects calcium absorption in the intestine [27–30]. The PTPN2 SNPs, both of which are intronic, were chosen for their association with both T1D and celiac disease [15]. Linkage disequilibrium (LD) between the three VDR SNPs and between the two PTPN2 SNPs was tested in our population using Haploview version 4.2 and none of the SNPs were found to be in LD as measured by r2, with r2 ≤ 0.005 for the VDR SNPs, and an r2 =0.124 for the PTPN2 SNPs.
VDR rs1544410 was genotyped using a linear array method at Roche Molecular Systems, Inc., as described in Mirel et al. [31]. PTPN2 rs1893217 was genotyped as part of a second project using Illumina 48-plex VeraCode technology following the manufacturer’s protocol. Genotyping data analysis and clustering was performed in Illumina’s GenomeStudio. Clustering clouds were manually investigated and adjusted if necessary. All plates included one duplicate sample and one positive control. VDR rs2228570, VDR rs11568820, and VDR rs478582 were typed utilizing the Taqman SNP genotype-based OpenArray platform [Applied Biosystems, CA, USA]. Custom designed arrays were loaded using the OpenArray AccuFill system and cycling was performed on a GeneAmp 9700 PCR system [Applied Biosystems, CA, USA], all according to manufacturer’s protocol. Alleles were analyzed using the OpenArray SNP genotyping analysis software v.1.0.3 and Taqman Genotyper Software 2.0 [Applied Biosystems, CA, USA].
All five SNPs had a 95% call rate or higher and were in Hardy–Weinberg equilibrium. Each SNP was tested for consistency with Hardy–Weinberg proportions using a 1-degree of freedom χ2 goodness-of-fit test with a p-value of 0.001 considered as evidence of a departure from Hardy–Weinberg equilibrium.
2.4. Analysis population
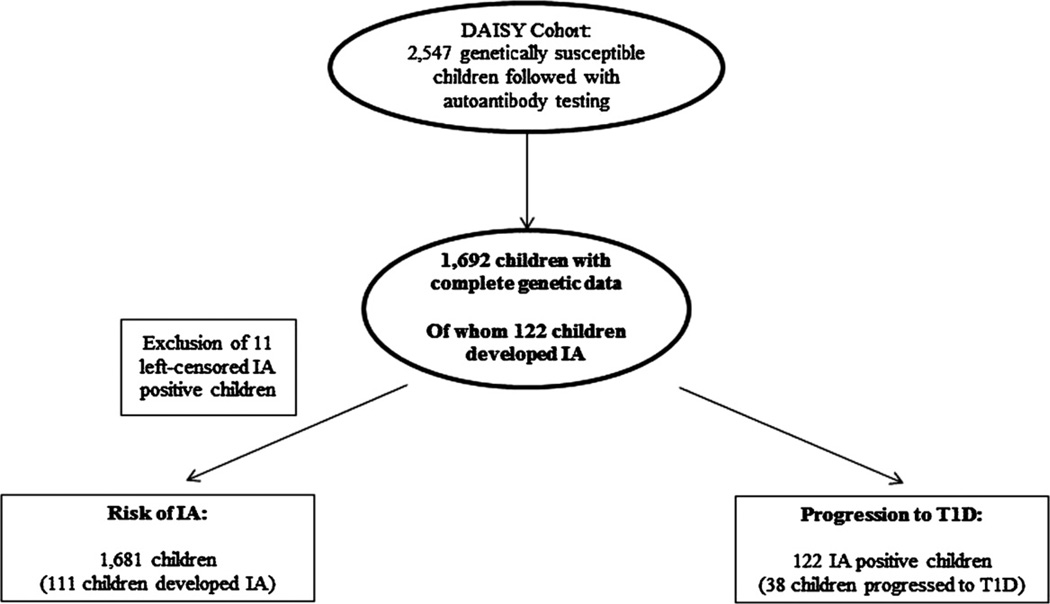
We obtained genetic data for all five SNPs on 1692 children in the DAISY cohort. A flow-chart showing selection of subjects for the analyses is shown in Fig. 1. Comparisons of the children who have complete genetic data with those who do not are presented in Supplemental Table 1 for the cohort examining risk of IA, and Supplemental Table 2 for the cohort examining progression to T1D in IA positive children. There were small differences in HLA, first-degree relative status and sex between children with and without complete genetic data in the cohort examining risk of IA, and no differences in the cohort examining progression to T1D in IA positive children. The 1692 children with complete genetic data included 122 children who developed persistent IA, of whom 38 went on to develop T1D. However, 11 IA cases were positive on their first clinic visit, thus considered left censored, and were removed from the development of IA analysis cohort.
Fig. 1.
Flow chart illustrating formation of analysis cohorts from the DAISY study population. IA, islet autoimmunity; T1D, type1 diabetes.
2.5. Statistical analyses
SAS version 9.3 (SAS Institute Inc., Cary, NC, USA) statistical software package was used for all statistical analyses. SNPs were analyzed for their association both with development of IA and with progression from IA to T1D. For each model described below, hazard ratios (HR) and 95% confidence intervals (CI) were estimated using Cox proportional hazards regression. A clustered time to event analysis was performed treating siblings from the same family as clusters, and robust sandwich variance estimates [32] were used for statistical inference. Analyses of time to development of IA were adjusted for the HLA-DR genotype (HLA-DR3/4, DQB1*0302 vs. other genotypes), presence of a first degree relative with T1D, and self-reported ethnicity (non-Hispanic white vs. other). Analyses of time to progression to T1D were adjusted, in addition, for age at first positive autoantibody. The significance threshold was defined a priori as α < 0.05. Because our analyses were based on an a priori hypothesis, p-values were not corrected for multiple testing.
Genotype frequencies of the five SNPs are presented in Supplemental Table 3 for the cohort examining risk of IA, and in Supplemental Table 4 for the cohort examining progression to T1D in IA positive children. To avoid sparse cell counts, the genotypes of each SNP were dichotomized in the following manner: VDR rs1544410 genotypes were dichotomized as AA/AG vs. GG, VDR rs2228570 genotypes as AA/AG vs. GG, VDR rs11568820 genotypes as TT/TC vs. CC, PTPN2 rs1893217 genotypes as GG/GA vs. AA, and the PTPN2 rs478582 genotypes as CC/CT vs. TT for all analyses.
We analyzed VDR and PTPN2 in separate models before testing the interaction. For the VDR gene analysis, all three VDR SNPs were included in the same model, adjusting for HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, and non-Hispanic white ethnicity. Similarly, for the PTPN2 gene analysis, the two PTPN2 SNPs were included in the same model, adjusting for HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, and non-Hispanic white ethnicity. These analyses were performed for both the development of IA as well as for progression to T1D in IA positive children. Then, based on the finding by Ramagopalan et al., all possible VDR*PTPN2 SNP interactions were examined for association with development of IA and progression to T1D [13], with separate models for each interaction tested. For significant interaction terms, we computed HRs and 95% CIs from the coefficient and standard error estimates of the main effect and interaction terms to describe the interaction.
3. Results
3.1. Development of persistent islet autoimmunity
We first examined whether variants in VDR were associated with development of persistent islet autoimmunity, and whether these interacted with variants in PTPN2. Table 1 describes the children by whether or not they developed persistent IA. Of a total of 122 IA positive children in DAISY, 11 had to be excluded from the subsequent proportional hazards analyses of IA because they tested autoantibody positive on their first study visits (i.e., they were left-censored). The mean age at first IA positive visit was 5.5 years, and the mean age at last follow-up visit in children who did not develop IA was 9.1 years. IA positive children, were more likely to have the HLA-DR3/4, DQB1 *0302 genotype or a first degree relative with T1D compared to children who did not develop IA.
Table 1.
Characteristics of the analysis population by islet autoimmunity (IA) status.
| Characteristic | Children positive for IA n = 111 |
Children negative for IA n = 1570 |
Univariate HR and 95% CI | p-Value |
|---|---|---|---|---|
| Mean agea (years) | 5.5 (3.8) | 9.1 (5.6) | N/A | N/A |
| HLA-DR3/4, DQB1*0302 | 34 (30.6%) | 274 (17.5%) | 2.18 (1.45, 3.26) | 0.0002 |
| First degree relative with type 1 diabetes | 67 (60.4%) | 652 (41.5%) | 1.66 (1.13, 2.43) | 0.01 |
| Ethnicity (non-Hispanic white) | 90 (81.1%) | 1120 (71.3%) | 1.38 (0.85, 2.25) | 0.20 |
| Sex (female) | 53 (47.8%) | 730 (46.5%) | 1.07 (0.75, 1.54) | 0.71 |
| VDR rs1544410 (AA/AG) | 76 (68.5%) | 959 (61.1%) | 1.32 (0.90, 1.95) | 0.16 |
| VDR rs2228570 (AA/AG) | 69 (62.2%) | 955 (60.8%) | 0.99 (0.68, 1.45) | 0.98 |
| VDR rs11568820 (TT/TC) | 36 (32.4%) | 630 (40.1%) | 0.72 (0.49, 1.07) | 0.11 |
| PTPN2 rs1893217 (GG/GA) | 39 (35.1%) | 438 (27.9%) | 1.34 (0.90, 1.99) | 0.15 |
| PTPN2 rs478582 (CC/CT) | 78 (70.3%) | 1077 (68.6%) | 1.08 (0.72, 1.62) | 0.72 |
Age at first IA positive visit in autoantibody positive children or age at last follow-up in autoantibody negative children.
Table 2 shows the association between the SNPs and the development of IA for the model containing all three VDR SNPs and the model containing both of the PTPN2 SNPs, adjusted for HLA DR3/4 status, having a first degree relative with T1D, and ethnicity. There was no evidence of significant association between the three VDR SNPs together and development of IA. The two PTPN2 variants (rs1893217 and rs478582) were also not associated with risk of IA.
Table 2.
Association between variants in VDR and PTPN2 and risk of islet autoimmunity (n = 111 affecteds and 1570 unaffecteds).
| Variable | HR | 95% CI | p-Value |
|---|---|---|---|
| Model of VDR Variants | |||
| VDR rs1544410 (AA/AG vs. GG) | 1.39 | 0.94, 2.07 | 0.10 |
| VDR rs2228570 (AA/AG vs. GG) | 0.99 | 0.68, 1.45 | 0.96 |
| VDR rs11568820 (TT/TC vs. CC) | 0.73 | 0.49, 1.10 | 0.13 |
| Model of PTPN2 Variants | |||
| PTPN2 rs1893217 (GG/GA) | 1.36 | 0.90, 2.04 | 0.15 |
| PTPN2 rs478582 (CC/CT) | 1.23 | 0.80, 1.87 | 0.35 |
In the Model of VDR variants, the three VDR SNPs were included in a single model, adjusting for HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, and non-Hispanic white ethnicity. Likewise, in the Model of PTPN2 variants, the two PTPN2 SNPs were included in a single model, adjusting for HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, and non-Hispanic white ethnicity.
We then explored whether variants in VDR and PTPN2 interacted to affect risk of IA. With three VDR SNPs and two PTPN2 SNPs, we tested six potential VDR*PTPN2 interactions for their associations with risk of IA and found no evidence of VDR*PTPN2 interactions, adjusting for HLA DR3/4 status, having a first degree relative with T1D, and ethnicity (data not shown).
3.2. Progression to T1D in children with IA
We examined whether variants in VDR were associated with progression to T1D in IA positive children, and whether these variants interacted with variants in PTPN2. The study population of IA positive children is described in Table 3. Of the 122 IA positive children in DAISY, 38 developed T1D; the mean age at T1D diagnosis was 8.2 years. The mean age at last follow-up visit in non-diabetic children with IA was 12.8 years. Children who develop T1D were younger when they first tested positive for an autoantibody than IA positive children who have not progressed to T1D, 3.6 and 6.1 years, respectively. They were also more likely to have the HLA-DR3/4, DQB1*0302 genotype compared to children who did not progress to T1D.
Table 3.
Characteristics of islet autoantibody (IA) positive subjects by type 1 diabetes (T1D) status.
| Characteristic | IA positive children who progressed to T1D n = 38 |
IA positive children who have not progressed to T1D n = 84 |
Univariate HR and 95% CI | p-Value |
|---|---|---|---|---|
| Mean agea (years) | 8.2 (4.0) | 12.8 (4.6) | N/A | N/A |
| Mean age at first IA positive visit (years) | 3.6 (2.9) | 6.1 (3.8) | 0.88 (0.78, 0.98) | 0.03 |
| HLA-DR3/4, DQB1*0302 | 23 (60.5%) | 14 (16.7%) | 3.98 (2.13, 7.41) | <.0001 |
| First degree relative with T1D | 26 (68.4%) | 52 (61.9%) | 1.11 (0.57, 2.17) | 0.76 |
| Ethnicity (non-Hispanic white) | 35 (92.1%) | 66 (78.6%) | 1.95 (0.57, 6.62) | 0.29 |
| Sex (female) | 20 (52.6%) | 37 (44.1%) | 1.47 (0.76, 2.85) | 0.26 |
| VDR rs1544410 (AA/AG) | 24 (63.2%) | 57 (67.9%) | 0.67 (0.34, 1.29) | 0.23 |
| VDR rs2228570 (AA/AG) | 18 (47.4%) | 56 (66.7%) | 0.63 (0.34, 1.17) | 0.14 |
| VDR rs11568820 (TT/TC) | 14 (36.8%) | 30 (35.7%) | 1.37 (0.70, 2.71) | 0.36 |
| PTPN2 rs1893217 (GG/GA) | 12 (31.6%) | 31 (36.9%) | 0.77 (0.40, 1.48) | 0.43 |
| PTPN2 rs478582 (CC/CT) | 28 (73.7%) | 57 (67.9%) | 1.23 (0.61, 2.45) | 0.57 |
Age at T1D diagnosis in diabetic children or age at last follow-up in non-diabetic children.
Table 4 displays the three VDR SNPs modeled together in one model and their association with the progression to T1D in IA positive children, adjusting for HLA DR3/4 status, having a first degree relative with T1D, ethnicity, and age at first IA positive visit. One of the two functional VDR SNPs, VDR rs2228570, was significantly associated with development of T1D when the three VDR SNPs were modeled together. The two PTPN2 SNPs were analyzed together similarly in a separate model to more completely describe the variation in the PTPN2 gene, but there was no evidence of significant associations with progression to T1D in IA positive children. We then explored whether variants in VDR and PTPN2 interacted to affect progression to T1D in IA positive children. We found a significant interaction between VDR 1544410 and PTPN2 rs1893217 on progression to T1D, adjusting for HLA DR3/4 status, having a first degree relative with T1D, ethnicity and age at first IA positive visit (Table 4 and Fig. 2).
Table 4.
Association between variants in VDR and PTPN2 and risk of progression to type 1 diabetes in IA positive children (n = 38 affecteds and 84 unaffecteds).
| Variable | HR | 95% CI | p-Value |
|---|---|---|---|
| Model of VDR variants | |||
| VDR rs1544410 (AA/AG) | 0.61 | 0.31, 1.21 | 0.16 |
| VDR rs2228570 (AA/AG) | 0.50 | 0.26, 0.95 | 0.03 |
| VDR rs11568820 (TT/TC) | 1.87 | 0.93, 3.74 | 0.08 |
| Model of PTPN2 variants | |||
| PTPN2 rs1893217 (GG/GA) | 0.65 | 0.27, 1.60 | 0.35 |
| PTPN2 rs478582 (CC/CT) | 1.07 | 0.43, 2.67 | 0.89 |
| VDR model with interaction term | |||
| VDR rs1544410*PTPN2 rs1893217 interaction | * | * | 0.02 |
| PTPN2 rs1893217 | * | * | 0.007 |
| VDR rs1544410 (AA/AG) | * | * | 0.0004 |
| VDR rs2228570 (AA/AG) | 0.49 | 0.26, 0.92 | 0.03 |
| VDR rs11568820 (TT/TC) | 1.82 | 0.90, 3.66 | 0.10 |
In the model of VDR variants, the three VDR SNPs were included in a single model, adjusting for HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, non-Hispanic white ethnicity, and age at first IA positive visit. Likewise, in the Model of PTPN2 variants, the two PTPN2 SNPs were included in a single model, adjusting for HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, non-Hispanic white ethnicity, and age at first IA positive visit. In the VDR model with interaction term, the variables included were VDR rs1544410, VDR rs2228570, VDR rs11568820, PTPN2 rs1893217, the VDR rs1544410*PTPN2 rs1893217 interaction term, HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, non-Hispanic white ethnicity, and age at first IA positive visit.
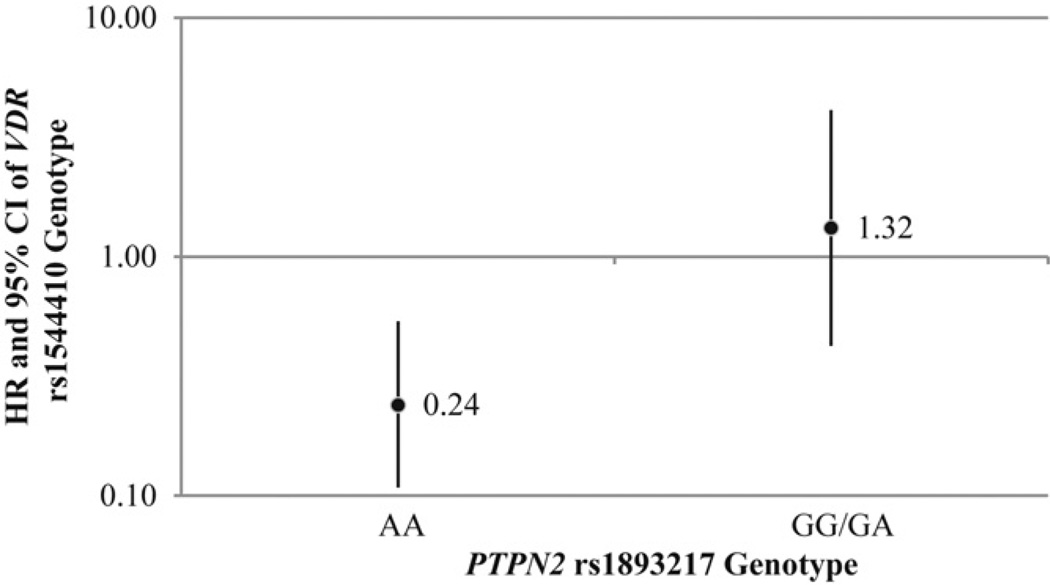
Fig. 2.
The association between VDR rs1544410 and the risk of progression to T1D in IA positive children differs by PTPN2 rs1893217 genotype. Black dots represent the hazard ratio for the VDR rs1544410 AA/AG genotype (with GG genotype as the referent). The solid lines represent the 95% confidence intervals for the hazard ratios. In children with the PTPN2 rs1893217 AA genotype (n = 79), the VDR rs1544410 AA/AG genotype (n = 54) was associated with a significantly lower risk of progressing to T1D compared to children with the VDR rs1544410 GG genotype (n = 25) (HR: 0.24, 95% CI: 0.11–0.53, p-value = 0.0004). In IA positive children with the PTPN2 rs1893217 GG/GA genotype (n = 43), the VDR rs1544410 AA/AG genotype (n = 27) was not associated with progression to T1D (HR: 1.32, 95% CI: 0.43, 4.06, p-value = 0.62).
The association between VDR rs1544410 and the progression to T1D in IA positive children differs by PTPN2 rs1893217 genotype, as shown in Fig. 2. In children with the PTPN2 rs1893217 AA genotype, the VDR rs1544410 AA/AG genotype is associated with a significantly lower risk of progressing to T1D compared to children with the VDR rs1544410 GG genotype (HR: 0.24, 95% CI: 0.11–0.53, p-value = 0.0004). In IA positive children with the PTPN2 rs1893217 GG/GA genotype, the VDR rs1544410 AA/AG genotype was not associated with progression to T1D. A subanalysis on non-Hispanic whites only produced similar results to those reported herein for development of IA and for progression to T1D in IA positive children (data not shown).
4. Discussion
In the DAISY population, the three VDR SNPs, VDR rs1544410, VDR rs2228570, and VDR rs11568820 we tested were not significantly associated with the appearance of IA. However, VDR rs2228570 was found to be associated with progression to T1D in IA positive children, and VDR rs1544410 significantly interacted with PTPN2 rs1893217 on risk of progression to T1D, suggesting that the role of VDR in risk of T1D is complex. Besides the aforementioned interaction between PTPN2 rs1893217 and VDR rs1544410, the two PTPN2 SNPs we tested were not significantly associated with the appearance of IA or progression to T1D as main effects. This is in slight contrast to our recent report in which we found PTPN2 rs1893217 to be weakly associated with development of islet autoimmunity [33]. This discrepancy can be attributed to the use of different IA case definitions. DAISY uses two definitions of IA, one that defines IA as the presence of at least one islet autoantibody on two consecutive visits (which is the definition used in the present study); and the other that further requires that the children still be autoantibody positive or diabetic on their most recent visits (which is the definition used in the previous study).
The studies that have examined VDR variants and the outcome of T1D have been inconsistent, and it is possible that the effects of VDR are only important for a faster development of T1D in the presence of IA, but not for the overall risk of T1D among genetically (HLA-DR,DQ) susceptible children. We found the functional VDR SNP, VDR rs2228570, to be a significant predictor in the progression to development of T1D. It also appears that VDR rs1544410, presumed to be non-functional because of its location in an intron, becomes mechanistically important for the progression to T1D in IA positive children when in combination with PTPN2 rs1893217. This, coupled with the finding that the vitamin D receptor has an intronic binding site in the PTPN2 gene [13], may provide insight into one of the ways in which the vitamin D receptor exerts its effects on T1D, which currently are still widely unknown. VDR rs1544410 has been shown to be in LD with a poly(A) microsatellite located in the 3′-untranslated region of the VDR gene, which has been discussed to influence VDR mRNA stability [34,35]. Ramagopalan et al. identified two VDR binding intervals in the PTPN2 gene; and the SNP involved in the interaction (PTPN2 rs1893217) is 5 kb and 58 kb away from these two intervals [13]. Thus, it is possible that this SNP is in LD with a variant in one of the two identified VDR binding intervals. This unique finding should be confirmed in other populations.
Ramagopalan et al. found increased VDR binding in intronic and intergenic regions compared with the basal state upon stimulation with calcitriol, the hormonally active form of vitamin D [13]. We were unable to examine what influence calcitriol had on the observed associations between VDR and progression to T1D because we do not have measures of 1,25-dihydroxyvitamin D on our children. Circulating 1,25-dihydroxyvitamin D has a short half-life of 15 h, is closely regulated by parathyroid hormone, calcium, and phosphate [36], and measurement requires a large amount of plasma for our pediatric subjects. Therefore, DAISY measures annually only plasma 25[OH]D as an estimate of vitamin D status. Only in situations of severe vitamin D deficiency do levels of 1,25-dihydroxyvitamin D decrease [2,37]. Simpson et al. recently reported no association between 25(OH)D levels and the risk of IA, nor progression to diabetes in IA positive children in DAISY [38].
The observation that VDR rs2228570 is associated with progression to T1D but not development of IA suggests that the role of VDR may be in the acceleration or deceleration of progression to T1D in autoimmune children. VDR 2228570 has been inconsistently associated with T1D [10,22,26,39–45]. VDR rs2228570 is a coding non-synonymous SNP located in the translational initiation codon that determines the formation of two protein variants: a longer version of the VDR protein (427-amino acids) that corresponds to the A allele and a form shortened by three amino acids corresponding to the G allele (424-amino acids) [23,46,47]. Functional studies have shown that the shorter version of the protein has greater transcriptional activity and is more effective in trans-activation of the vitamin D signal [23,46–49]. Therefore, the VDR rs2228570 minor allele is associated with the production of a longer VDR protein that is less transcriptionally active [46,50,51]. In this particular study, we found the VDR rs2228570 minor allele (A) to be significantly protective for the development of T1D in IA positive children, which may imply that once autoimmune, less active transcription is beneficial. It is also interesting that VDR rs2228570 only becomes significant when adjusting for VDR 1544410 and VDR rs11568820, which may be an indication of the complexity of the way in which these polymorphisms act together.
The major strength of this study is the prospective long-term follow-up from birth of children at an increased risk for T1D, which allowed us to differentiate between genetic risk factors for the appearance of autoimmunity and the subsequent progression to T1D. However, the cost of assembling and following such a unique cohort has limited the number of IA positive children and children who progress to T1D that we could include in our analysis. While our study had adequate power to detect the novel interaction between variants in VDR and PTPN2 that we present herein, it is possible that we have missed other interactions of smaller magnitude due to limited power. This is the first study to observe this gene–gene interaction in the progression phase to T1D, and it is important that this be investigated in other populations, to rule out type 1 error and lend further evidence to this association.
In conclusion, a functional variant in VDR, VDR rs2228570 was found to be significantly associated with progression to T1D in IA positive children of the DAISY population. A significant interaction between a non-coding VDR variant, VDR rs1544410, and a variant in PTPN2, PTPN2 rs1893217, was also found to be associated with progression to T1D in IA positive children. Interestingly, these two VDR variants, VDR 2228570 and VDR 1544410, were not found to be associated with the appearance of IA. These findings should be explored in other prospective cohorts following children with IA for progression to T1D, as they may offer insight concerning the complex role of vitamin D in the etiology of T1D, as well as other autoimmune diseases associated with PTPN2, e.g., Crohn’s disease and celiac disease.
Supplementary Material
Acknowledgements
This research was supported by NIH grants R01-DK49654, R01-DK32493, DERC Molecular Biology Core NIH P30 DK57516, R21 DK84568, and JDRF Grant 11-2010-206. We acknowledge Cisca Wijmenga for providing us with the PTPN2 genotypes.
Abbreviations
- IA
islet autoimmunity
- PTPN2
protein tyrosine phosphates, non-receptor type 2 gene
- SNP
single nucleotide polymorphism
- T1D
type 1 diabetes
- VDR
vitamin D receptor gene
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsbmb.2012.08.012.
References
- 1.George CE. Environmental factors and multiple sclerosis. The Lancet Neurology. 2008;7:268–277. doi: 10.1016/S1474-4422(08)70042-5. [DOI] [PubMed] [Google Scholar]
- 2.Holick MF. Vitamin D deficiency. New England Journal of Medicine. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 3.McDermott MF, Ramachandran A, Ogunkolade BW, Aganna E, Curtis D, Boucher BJ, et al. Allelic variation in the vitamin D receptor influences susceptibility to IDDM in Indian Asians. Diabetologia. 1997;40:971–975. doi: 10.1007/s001250050776. [DOI] [PubMed] [Google Scholar]
- 4.Pani MA, Knapp M, Donner H, Braun J, Baur MP, Usadel KH, et al. Vitamin D receptor allele combinations influence genetic susceptibility to type 1 diabetes in Germans. Diabetes. 2000;49:504–507. doi: 10.2337/diabetes.49.3.504. [DOI] [PubMed] [Google Scholar]
- 5.Chang TJ, Lei HH, Yeh JI, Chiu KC, Lee KC, Chen MC, et al. Vitamin D receptor gene polymorphisms influence susceptibility to type 1 diabetes mellitus in the Taiwanese population. Clinical Endocrinology (Oxford) 2000;52:575–580. doi: 10.1046/j.1365-2265.2000.00985.x. [DOI] [PubMed] [Google Scholar]
- 6.Gyorffy B, Vasarhelyi B, Krikovszky D, Madacsy L, Tordai A, Tulassay T. Gender-specific association of vitamin D receptor polymorphism combinations with type 1 diabetes mellitus. European Journal of Endocrinology. 2002;147:803–808. doi: 10.1530/eje.0.1470803. [DOI] [PubMed] [Google Scholar]
- 7.Skrabić V, Zemunik T, Situm M, Terzić J. Vitamin, D receptor polymorphism and susceptibility to type 1 diabetes in the Dalmatian population. Diabetes Research and Clinical Practice. 2003;59:31–35. doi: 10.1016/s0168-8227(02)00195-x. [DOI] [PubMed] [Google Scholar]
- 8.Ban Y, Taniyama M, Yanagawa T, Yamada S, Maruyama T, Kasuga A. Vitamin D receptor initiation codon polymorphism influences genetic susceptibility to type 1 diabetes mellitus in the Japanese population. BMC Medical Genetics. 2001;2:7. doi: 10.1186/1471-2350-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motohashi Y, Yamada S, Yanagawa T, Maruyama T, Suzuki R, Niino M, et al. Vitamin D receptor gene polymorphism affects onset pattern of type 1 diabetes. Journal of Clinical Endocrinology and Metabolism. 2003;88:3137–3140. doi: 10.1210/jc.2002-021881. [DOI] [PubMed] [Google Scholar]
- 10.Nejentsev S, Cooper JD, Godfrey L, Howson JMM, Rance H, Nutland S, et al. Analysis of the vitamin D receptor gene sequence variants in type 1 diabetes. Diabetes. 2004;53:2709–2712. doi: 10.2337/diabetes.53.10.2709. [DOI] [PubMed] [Google Scholar]
- 11.Guo S-W, Magnuson VL, Schiller JJ, Wang X, Wu Y, Ghosh S, et al. Meta-analysis of vitamin D receptor polymorphisms and type 1 diabetes: a HuGE review of genetic association studies. American Journal of Epidemiology. 2006;164:711–724. doi: 10.1093/aje/kwj278. [DOI] [PubMed] [Google Scholar]
- 12.Howson JMM, Walker NM, Smyth DJ, Todd JA. Analysis of 19 genes for association with type I diabetes in the Type I Diabetes Genetics Consortium families. Genes & Immunity. 2009;1(Suppl. 10):S74–S84. doi: 10.1038/gene.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Research. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nature Genetics. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JHM. Shared and distinct genetic variants in Type 1 diabetes and celiac disease. New England Journal of Medicine. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, et al. Newborn screening for HLA markers associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY) Diabetologia. 1996;39:807–812. doi: 10.1007/s001250050514. [DOI] [PubMed] [Google Scholar]
- 17.Norris JM, Yin X, Lamb MM, Barriga K, Seifert J, Hoffman M, et al. Omega-3 polyunsaturated fatty acid intake and islet autoimmunity in children at increased risk for type 1 diabetes. Journal of American Medical Association. 2007;298:1420–1428. doi: 10.1001/jama.298.12.1420. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, Rewers M, Gianani R, Kawasaki E, Zhang Y, Verge C, et al. Antiislet autoantibodies usually develop sequentially rather than simultaneously. Journal of Clinical Endocrinology & Metabolism. 1996;81:4264–4267. doi: 10.1210/jcem.81.12.8954025. [DOI] [PubMed] [Google Scholar]
- 19.Yu L, Robles DT, Abiru N, Kaur P, Rewers M, Kelemen K, et al. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proceedings of the National Academy of Sciences. 2000;97:1701–1706. doi: 10.1073/pnas.040556697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grubin C, Daniels T, Toivola B, Landin-Olsson M, Hagopian W, Karlsen A, et al. A novel radioligand binding assay to determine diagnostic accuracy of isoform-specific glutamic acid decarboxylase antibodies in childhood IDDM. Diabetologia. 1994;37:344–350. doi: 10.1007/BF00408469. [DOI] [PubMed] [Google Scholar]
- 21.Gianani R, Rabin DU, Verge CF, Yu L, Babu SR, Pietropaolo M. ICA512 autoantibody radioassay. Diabetes. 1995;44:1340–1344. doi: 10.2337/diab.44.11.1340. [DOI] [PubMed] [Google Scholar]
- 22.Panierakis C, Goulielmos G, Mamoulakis D, Petraki E, Papavasiliou E, Galanakis E. Vitamin, D receptor gene polymorphisms and susceptibility to type 1 diabetes in Crete, Greece. Clinical Immunology. 2009;133:276–281. doi: 10.1016/j.clim.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Cox MB, Ban M, Bowden NA, Baker A, Scott RJ, Lechner-Scott J. Potential association of vitamin D receptor polymorphism Taq1 with multiple sclerosis. Multiple Sclerosis Journal. 2012;18:16–22. doi: 10.1177/1352458511415562. [DOI] [PubMed] [Google Scholar]
- 24.Jurutka PW, Remus LS, Whitfield GK, Thompson PD, Hsieh J-C, Zitzer H. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Molecular Endocrinology. 2000;14:401–420. doi: 10.1210/mend.14.3.0435. [DOI] [PubMed] [Google Scholar]
- 25.Uitterlinden AG, Fang Y, van Meurs JBJ, Pols HAP, van Leeuwen JPTM, et al. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Lemos MC, Fagulha A, Coutinho E, Gomes L, Bastos M, Barros L, et al. Lack of association of vitamin D receptor gene polymorphisms with susceptibility to type 1 diabetes mellitus in the Portuguese population. Human Immunology. 2008;69:134–138. doi: 10.1016/j.humimm.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Fang Y, Van Meurs JB, Bergink AP, Hofman A, Van Duijn CM, Van Leeuwen JP, et al. Cdx-2 polymorphism in the promoter region of the human vitamin D receptor gene determines susceptibility to fracture in the elderly. Journal of Bone and Mineral Research. 2003;18:1632–1641. doi: 10.1359/jbmr.2003.18.9.1632. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto H, Miyamoto K-I, Li B, Taketani Y, Kitano M, Inoue Y, et al. The Caudal-related homeodomain protein Cdx-2 regulates vitamin D receptor gene expression in the small intestine. Journal of Bone and Mineral Research. 1999;14:240–247. doi: 10.1359/jbmr.1999.14.2.240. [DOI] [PubMed] [Google Scholar]
- 29.Slattery ML, Herrick J, Wolff RK, Caan BJ, Potter JD, Sweeney C, et al. CDX2 VDR polymorphism and colorectal cancer. Cancer Epidemiology, Biomarkers and Prevention. 2007;16:2752–2755. doi: 10.1158/1055-9965.EPI-07-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arai H, Miyamoto K-I, Yoshida M, Yamamoto H, Taketani Y, Morita K, et al. The polymorphism in the Caudal-related homeodomain protein Cdx-2 binding element in the human vitamin D receptor gene. Journal of Bone and Mineral Research. 2001;16:1256–1264. doi: 10.1359/jbmr.2001.16.7.1256. [DOI] [PubMed] [Google Scholar]
- 31.Mirel DB, Valdes AM, Lazzeroni LC, Reynolds RL, Erlich HA, Noble JA. Association of IL4R haplotypes with type 1 diabetes. Diabetes. 2002;51:3336–3341. doi: 10.2337/diabetes.51.11.3336. [DOI] [PubMed] [Google Scholar]
- 32.Wei LJ, Lin DY, Weissfeld L. Regression analysis of multivariate incomplete failure time data by modeling marginal distributions. Journal of the American Statistical Association. 1989;84:1065–1073. [Google Scholar]
- 33.Steck AK, Wong R, Wagner B, Johnson K, Liu E, Romanos J, et al. Effects of Non-HLA gene polymorphisms on development of islet autoimmunity and type 1 diabetes in a population with high-risk HLA-DR,DQ genotypes. Diabetes. 2012;61:753–758. doi: 10.2337/db11-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingles SA, Haile RW, Henderson BE, Kolonel LN, Nakaichi G, Shi CY, et al. Strength of linkage disequilibrium between two vitamin D receptor markers in five ethnic groups: implications for association studies, Cancer Epidemiology. Biomarkers and Prevention. 1997;6:93–98. [PubMed] [Google Scholar]
- 35.Whitfield GK, Remus LS, Jurutka PW, Zitzer H, Oza AK, Dang HT, et al. Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Molecular and Cellular Endocrinology. 2001;177:145–159. doi: 10.1016/s0303-7207(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 36.Jones G, et al. Pharmacokinetics of vitamin D toxicity. American Journal of Clinical Nutrition. 2008;88:582S–586S. doi: 10.1093/ajcn/88.2.582S. [DOI] [PubMed] [Google Scholar]
- 37.Cranney A, Horsley T, O’Donnell S, Weiler H, Puil L, Ooi D, et al. Effectiveness and safety of vitamin D in relation to bone health. Evidence Report/Technology Assessment (Full Report) 2007:1–235. [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson M, Brady H, Yin X, Seifert J, Barriga K, Hoffman M, et al. No association of vitamin D intake or 25-hydroxyvitamin D levels in childhood with risk of islet autoimmunity and type 1 diabetes: the Diabetes Autoimmunity Study in the Young (DAISY) Diabetologia. 2011;54:2779–2788. doi: 10.1007/s00125-011-2278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadnejad Z, Ghanbari M, Ganjali R, Afshari JT, Heydarpour M, Taghavi SM, et al. Association between vitamin D receptor gene polymorphisms and type 1 diabetes mellitus in Iranian population. Molecular Biology Reports. 2012;39:831–837. doi: 10.1007/s11033-011-0805-3. [DOI] [PubMed] [Google Scholar]
- 40.Mory DB, Rocco ER, Miranda WL, Kasamatsu T, Crispim F, Dib SA, et al. Prevalence of vitamin D receptor gene polymorphisms FokI and BsmI in Brazilian individuals with type 1 diabetes and their relation to β-cell autoimmunity and to remaining β-cell function. Human Immunology. 2009;70:447–451. doi: 10.1016/j.humimm.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Capoluongo E, Pitocco D, Concolino P, Santonocito C, Di Stasio E, d’ Onofrio G. Slight association between type 1 diabetes and ff VDR FokI genotype in patients from the Italian Lazio Region. Lack of association with diabetes complications. Clinical Biochemistry. 2006;39:888–892. doi: 10.1016/j.clinbiochem.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Eerligh P, Koeleman BPC, Dudbridge F, Jan Bruining G, Roep BO, Giphart MJ, et al. Functional genetic polymorphisms in cytokines and metabolic genes as additional genetic markers for susceptibility to develop type 1 diabetes. Genes & Immunity. 2004;5:36–40. doi: 10.1038/sj.gene.6364036. [DOI] [PubMed] [Google Scholar]
- 43.Guja C, Marshall S, Welsh K, Merriman M, Smith A, Todd JA, et al. The study of CTLA-4 and vitamin D receptor polymorphisms in the Romanian type 1 diabetes population. Journal of Cellular and Molecular Medicine. 2002;6:75–81. doi: 10.1111/j.1582-4934.2002.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mimbacas A, Trujillo J, Gascue C, Javiel G, Cardoso H, et al. Prevalence of vitamin D receptor gene polymorphism in a Uruguayan population and its relation to type 1 diabetes mellitus. Genetics and Molecular Research. 2007;6:534–542. [PubMed] [Google Scholar]
- 45.Angel B, Santos JL, Carrasco E, Albala C, Pérez-Bravo F. Vitamin, D receptor polymorphism and susceptibility to type 1 diabetes in Chilean subjects: a case-parent study. European Journal of Epidemiology. 2004;19:1085–1087. doi: 10.1007/s10654-004-1026-z. [DOI] [PubMed] [Google Scholar]
- 46.Anderson LN, Cotterchio M, Cole DEC, Knight JA, et al. Vitamin D-related genetic variants, interactions with vitamin D exposure, and breast cancer risk among caucasian women in Ontario. Cancer Epidemiology Biomarkers & Prevention. 2011;20:1708–1717. doi: 10.1158/1055-9965.EPI-11-0300. [DOI] [PubMed] [Google Scholar]
- 47.Orton S-M, Ramagopalan SV, Para AE, Lincoln MR, Handunnetthi L, Chao MJ, et al. Vitamin D metabolic pathway genes and risk of multiple sclerosis in Canadians. Journal of the Neurological Sciences. 2011;305:116–120. doi: 10.1016/j.jns.2011.02.032. [DOI] [PubMed] [Google Scholar]
- 48.Lurie G, Wilkens LR, Thompson PJ, Carney ME, Palmieri RT, Pharoah PDP, et al. Vitamin D receptor rs2228570 polymorphism and invasive ovarian carcinoma risk: pooled analysis in five studies within the Ovarian Cancer Association Consortium. International Journal of Cancer. 2011;128:936–943. doi: 10.1002/ijc.25403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, Yang MH, Zeng CA, Wu WL, Huang XF, Ji Y, et al. Association of vitamin D receptor gene polymorphisms in Chinese patients with generalized aggressive periodontitis. Journal of Periodontal Research. 2008;43:360–363. doi: 10.1111/j.1600-0765.2007.01044.x. [DOI] [PubMed] [Google Scholar]
- 50.McKay JD, McCullough ML, Ziegler RG, Kraft P, Saltzman BS, Riboli E, et al. Vitamin D receptor polymorphisms and breast cancer risk: results from the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Cancer Epidemiology Biomarkers & Prevention. 2009;18:297–305. doi: 10.1158/1055-9965.EPI-08-0539. [DOI] [PubMed] [Google Scholar]
- 51.Barroso E, Fernandez LP, Milne RL, Pita G, Sendagorta E, Floristan U. Genetic analysis of the vitamin D receptor gene in two epithelial cancers: melanoma and breast cancer case–control studies. BMC Cancer. 2008;8:385. doi: 10.1186/1471-2407-8-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.