Abstract
A panel of clinically used tyrosine kinase inhibitors was compared and nilotinib was found to most potently sensitize specific anticancer agents by blocking the functions of ABCB1/P-glycoprotein, ABCG2/BCRP and ABCC10/MRP7 transporters involved in multi-drug resistance. Nilotinib appreciably enhanced the antitumor response of 1) paclitaxel in the ABCB1- and novel ABCC10-xenograft models, and 2) doxorubicin in a novel ABCG2-xenograft model. With no apparent toxicity observed in the above models, nilotinib attenuated tumor growth synergistically and increased paclitaxel concentrations in ABCB1-overexpressing tumors. The beneficial actions of nilotinib warrant consideration as viable combinations in the clinic with agents that suffer from MDR-mediated insensitivity.
Keywords: ABC transporters, ABCB1/P-gp, ABCC10/MRP7, ABCG2/BCRP, Nilotinib, multidrug resistance
1. Introduction
Multidrug resistance (MDR) is characterized by energy-dependent efflux of drugs from cancer cells by ATP-binding cassette (ABC) transporters that may cause tumors to be insensitive to chemotherapy [1; 2]. The ABC transporters are prevalent throughout the body as well, and serve important functions related to drug and metabolite disposition, and in fact protect vital organs by hastening drug elimination and preventing tissue access [1; 3; 4]. The overexpression of certain ABC transporters such as ABCB1 (Multiple Drug Resistance 1, P-glycoprotein), ABCC/MRP subfamily (Multidrug Resistance Protein) and ABCG2 (Breast Cancer Resistance Protein, MitoXantrone Resistance protein, ATP-Binding Cassette of Placenta) are significantly correlated with MDR in tumor cells, where they actively efflux certain antineoplastic drugs, thereby reducing the intracellular concentration of drugs below an effective cytotoxic threshold [1; 3; 5; 6]. Numerous in vitro studies have shown that overexpression of ABCB1 in tumor cells significantly reduces the accumulation of a broad range of neutral and cationic hydrophobic chemotherapeutic substrates, including taxanes, epipodophyllotoxins, vinca alkaloids, anthracyclines and more recently certain tyrosine kinase inhibitors (TKIs) [4; 6; 7; 8; 9; 10; 11; 12]. ABCC1 has a high affinity for negatively charged lipophilic compounds, yet its substrate profiles overlap with that of ABCB1 and its overexpression confers resistance to anthracyclines, camptothecin analogs, vinca alkaloids, epipodophyllotoxins and anthracenediones (with a few exceptions, such as taxanes and bisantrene) [6; 12; 13; 14]. The ABCG2 transporter confers resistance to a wide spectrum of chemotherapeutic agents, ranging from organic anion conjugates, nucleoside analogs, organic dyes, and TKIs to anthracyclines [15; 16]. Recent reports have shown that ABCC10, a lipophilic anionic transporter conferred resistance to taxanes, vinca alkaloids, daunorubicin, etoposide, nucleoside-based drugs including cytarabine (Ara-C) and gemcitabine, and to microtubule stabilizing agent, epothilone B, which was originally designed to surmount ABCB1 mediated MDR [17; 18]. Therefore, each of the transporters examined in this investigation have been associated with cancer cell resistance.
Significant efforts have been made to devise new strategies to overcome MDR to cancer chemotherapy, and the addition of ABC transport inhibitors to combination regimens have gained traction. In this regard, in vitro investigations have demonstrated that TKIs are able to modulate ABC transporter function and their use in combination regimens may serve dual purposes of anticancer activity as well as inhibition of MDR [12; 19; 20; 21]. Specifically, we and others have shown that the human epidermal growth factor receptor type (HER/EGFR) TKIs such as erlotinib, lapatinib, AG1478, BCR-ABL TKIs such as nilotinib, imatinib and vascular endothelial growth factor inhibitor (VEGF) TKI such as sunitinib are likely to play a key role in modulation of ABC transporters-ATPase activity and inhibit active drug efflux at clinically achievable concentrations, and hence can overcome drug resistance in cells with the MDR phenotype [7; 8; 9; 10; 22; 23; 24]. There have been in vitro data to support the use of these inhibitors as potential MDR-modulating agents for clinical use [10; 25; 26]. Translational significance of these modulators lies in developing a targeted combination strategy with pharmacokinetic and Pharmacodynamic [PK/PD] parameters in mind, which lays the foundation for a clinical trial to further optimize pharmacotherapy. Therefore, we assessed and compared the potency of most of the above mentioned TKIs to modulate and sensitize anticancer activity of doxorubicin and paclitaxel in in vitro models first and then chose a potent TKI to ascertain its effect further in an in vivo model of ABCB1 and newly generated ABCG2 and ABCC10-xenograft models. These findings should help optimize the design of present and future clinical trials elucidating potential pharmacokinetic interactions of anticancer drugs with MDR modulators.
2. Materials and methods
2.1 Materials
Monoclonal antibodies BXP-34 (against ABCG2), C-219 (against ABCB1), and MRPm5 (against ABCC1) were acquired from Signet Laboratories, Inc. (Dedham, MA). A polyclonal goat antibody against human ABCC10 (D-19), and anti-actin monoclonal antibody (sc-8432) was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Nilotinib was obtained as a gift from Novartis Pharmaceuticals (Basel, Switzerland). Fumitremorgin C (FTC) was synthesized by Thomas McCloud Developmental Therapeutics Program, Natural Products Extraction Laboratory, NCI, NIH (Bethesda, MD). Paclitaxel was obtained from LC labs (Woburn, MA). Mitoxantrone (MX), vincristine, colchicine, doxorubicin, cisplatin, verapamil, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma Chemical Co. (St. Louis, MO). All other reagents and solvents were purchased from VWR (West Chester, PA).
2.2 Cell lines and cell culture
HEK293/pcDNA3.1, HEK293/ABCB1, HEK293/ABCC1 and HEK293/MRP7 cells were established by transfecting HEK293 with either the empty pcDNA3.1 vector or pcDNA3.1 vector containing the full length ABCB1 (HEK293/ABCB1) or ABCC1 (HEK293/ABCC1) or ABCC10 (HEK293/MRP7), and were cultured in a medium with 2 mg/ml of G418 [18; 26]. The parental human epidermoid carcinoma cell line KB-3-1 was selected in a stepwise manner using increasing concentrations of colchicine to establish the ABCB1/P-gp-overexpressing drug-resistant cell line, KB-C2, and was cultured in medium with 2 μg/ml of colchicine [27]. Another ABCB1-overexpressing cell line KB-V1 was cultured in medium containing 1 μg/ml vinblastine. An ABCC1-overexpressing MDR cell line, KB-CV60, was also cloned from KB-3-1 cells and was maintained in medium with 1 μg/ml of cepharanthine and 60 ng/ml of vincristine [28]. In addition, non-small cell lung cancer cells H460 was cultured with mitoxantrone upto 20 μmol/L to produce ABCG2 overexpressing H460/MX-20 cells. All of the cell lines were grown as adherent monolayers in flasks with DMEM culture medium (Hyclone Co., UT) supplemented with 10% fetal bovine serum in a humidified incubator containing of 5% CO2 at 37°C.
2.3 Cell cytotoxicity by MTT assay
The MTT colorimetric assay with slight modifications from that previously described [29], was used to detect the sensitivity of cells against anticancer drugs. Cells were harvested with trypsin and resuspended in a final concentration of 4 × 104 cells/ml for KB-3-1, 7.5 × 104 cells/ml for KB-C2, 5 × 104 cells/ml for H460 and 8 × 104 for all the other cell lines. Cells were seeded evenly into (180 μl/well) 96-well multiplates. For the reversal experiments, nilotinib, verapamil, or FTC (10 μl/well) were added, followed by different concentrations of chemotherapeutic drugs (10 μl/well) into designated wells. After 72 h of incubation, 20 μl of MTT solution (4 mg/ml) was added to each well, and the plate was further incubated for 4 h, allowing viable cells to convert the yellow-colored MTT into dark-blue formazan crystals. Subsequently, the medium was discarded, and 100 μl of dimethylsulfoxide (DMSO) was added into each well to dissolve the formazan crystals. The absorbance was determined at 570 nm by an OPSYS microplate Reader from DYNEX Technologies, Inc. (Chantilly, VA). The degree of resistance was calculated by dividing the IC50 (concentrations required to inhibit growth by 50%) for the MDR cells by that of the parental sensitive cells. The degree of the reversal of MDR was calculated by dividing the IC50 for cells with the anticancer drug in the absence of nilotinib or other reversal agents by that obtained in the presence of nilotinib/reversal agent. The IC50 were calculated from survival curves using the Bliss method [30].
2.4 Western blot analysis
Equal amounts of total cell lysates (80 μg protein) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto polyvinylidene fluoride (PVDF) membranes. After incubation in a blocking solution in TBST buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween 20) for 1 h at room temperature, the membranes were immunoblotted overnight with primary monoclonal antibodies against either ABCB1 or actin at 1:200 dilution or ABCG2 at 1:500 dilution at 4°C, and were then incubated for 3 h at room temperature with horseradish peroxide (HRP)-conjugated secondary antibody (1:1000 dilution). The protein-antibody complex was detected by enhanced chemiluminescence detection system (Amersham, NJ). The protein expression was quantified by Scion Image software (Scion Co, MD) [9].
2.5 Molecular Modeling - ABCB1 and ABCG2
2.5.1 Ligand structure preparation
Nilotinib structure was built using the fragment dictionary of Maestro v9.0 and energy minimized by Macromodel program v9.7 (Schrödinger, Inc., New York, NY, 2009) using the OPLSAA force field with the steepest descent followed by truncated Newton conjugate gradient protocol. The low-energy 3D structures of nilotinib were generated by LigPrep v2.3 and the parameters were defined based on different protonation states at physiological pH±2, and all possible tautomers and ring conformations. Ligand structures obtained from the LigPrep v2.3 run were further used for generating 100 ligand conformations for each protonated structure using the default parameters of mixed torsional/low-mode sampling function. The conformations were filtered with a maximum relative energy difference of 5 kcal/mol to exclude redundant conformers. The output conformational search (Csearch) file containing at most 100 unique conformers of nilotinib were used as input for docking simulations into each binding site of human ABCB1 and ABCG2.
2.5.2 Protein structure preparation
The X-ray crystal structure of mouse ABCB1 in apoprotein state (PDB ID: 3G5U), in complex with inhibitors QZ59-RRR (PDB ID: 3G6O), QZ59-SSS (PDB ID: 3G61) [31] and bacterial co-crystal structure of LmrA ATP-binding domain (PDB ID: 1MV5) as the template obtained from the RCSB Protein Data Bank were used to generate the homology model of human ABCB1, based on our homology modeling protocol [32]. The refined human ABCB1 homology model was further used to generate different receptor grids for different sites (site 1-4) by selecting QZ59-RRR (site-1) and QZ59-SSS (site-2) bound ligands, all amino acid residues known to contribute to verapamil binding (site-3), two residues (Phe728 and Val982) known to be common to three previous sites (site-4) as previously reported by us [32], and additionally to evaluate any possibility of nilotinib interaction at the ATP-binding site, bound ATP ligand was selected for grid generation and ensuing docking simulation of nilotinib at ATP-binding site. Previously, Shukla et al. suggested that nilotinib displaces [125I]-IAAP in a dose dependent manner through a photoaffinity-labeling assay; hence share the same binding site [26]. We also docked IAAP to these sites for comparison and found that IAAP have best binding score at site-1 as nilotinib on the ABCB1. Homology model of ABCG2 was built based on the mouse apoprotein (PDB ID: 3G5U) [31] as template and has been generated previously by Rosenberg et al. [33]. The homology model of ABCG2 was energy minimized before initiating grid preparation. To identify the druggable sites on ABCG2 homology model, we have generated various grids based on the following residues as centroids, for example, Arg482 (grid 1), Asn629 (grid 2), Arg383 (grid 3) and Leu241 along with Gly83 (grid 4). The choice of these residues was based on their involvement in ABCG2 function as determined through mutational experiments [34; 35]. The grid 1 generated using Arg482 as the centroid was found to have the best docking score; hence, docking discussion was based on binding mode of nilotinib at this site. Glide v5.0 docking protocol was followed with the default functions (Schrödinger, Inc., New York, NY, 2009). The top scoring nilotinib conformation at Arg482 site of ABCG2 and site 1 of ABCB1 was used for graphical analysis. All computations were carried out on a Dell Precision 470n dual processor with the Linux OS (Red Hat Enterprise WS 4.0).
2.6 Animals
Male athymic NCR (nu/nu) nude mice (18 – 25 g, age 10 – 15 wk), were purchased from the Taconic Farms (NCRNU-M, Homozygous, Albino color) and were used for ABCB1 ABCG2 and ABCC10 xenograft model. All the animals were maintained on an alternating 12 h light/dark cycle with free access to water and rodent chow ad libitum. The mice were maintained at the St. John's University Animal Facility and were monitored closely for tumor growth by palpation and visual examination. Institutional Animal Care & Use Committee (IACUC) of St. John's University approved this project, and the research was conducted in compliance with the Animal Welfare Act and other federal statutes.
2.7 Nude mice MDR xenograft models
The ABCB1 overexpressing KB-C2 model was designed for the first time with a slight modification of KBv200 cell xenograft model previously established by Chen and colleagues [36]. The ABCG2 overexpressing non-small cell lung cancer H460/MX-20 model and ABCC10 overexpressing HEK/MRP7 model were designed for the first time after series of pilot experiment to decide on the proper inoculums of cells, tumor growth rate and maximum tolerated dose of doxorubicin and paclitaxel, respectively in the nude mice.. Briefly, KB-3-1 (1.2 × 106), KB-C2 (1 × 107), H460 (4 × 106), H460/MX20 (6 × 106), HEK293 and HEK/MRP7 (1.2 × 107) cells were injected s.c. under the armpits as shown in the Supplementary Fig. 1. Tumors that fail to reach a volume of 20 mm3 at the start of treatment were not used in this study. When the tumors reached a mean diameter of 0.5 cm (day 0), the mice were randomized into four groups (n=10) and treated with one of the following regimens: (a) vehicle (10% N-methyl-pyrrolidinone, 90% polyethylene glycol 300) (q3d × 6), (b) paclitaxel (18 mg/kg, i.p., q3d × 6) in ABCB1 or ABCC10 arm or doxorubicin (1.8 mg/kg, i.p., q3d × 6) in ABCG2 arm, (c) nilotinib diluted in 10% N-methyl-pyrrolidinone, 90% polyethylene glycol 300 (75 mg/kg, p.o., q3d × 6), and (d) paclitaxel (18 mg/kg, i.p., q3d × 6) in ABCB1 or ABCC10 arm or doxorubicin (1.8 mg/kg, i.p., q3d × 6) in ABCG2 arm + nilotinib (75 mg/kg, p.o., q3d × 6, given 1 h before giving paclitaxel/doxorubicin) see Supplementary Fig. 1. Paclitaxel for injection was prepared by dissolving 6mg paclitaxel, 527 mg purified polyoxyethylated castor oil and 49.7% of dehydrated alcohol in each ml of distilled water. Doxorubicin for injection was prepared by dissolving it in sterile water. Tumor volume was measured using calipers and body weights were recorded [7]. The body weight of the animals was monitored every 3rd day to adjust the drug dosage and to access treatment related toxicities as well as disease progression. The two perpendicular diameters of tumors (termed A and B) were recorded every 3rd day and tumor volume (V) was estimated according to the following formula published previously [7; 36]:
The increase in tumor volume from the start of treatment (Vo) until the value at any given time (Vt) was calculated for each tumor and expressed as relative tumor volume (Vt:Vo) on the day of the measurement. The mean of these values was then used to determine the ratio between treated (T) and control (C) tumors (T:C × 100%). Growth inhibition or the rate of inhibition (IR) was calculated according to the formula given below [7; 36].
When the mean of tumor weights went over 1 g in the control group, all the animals were killed by terminal bleeding through cardiac puncture under isofluorane anesthesia, plasma, different organs and tumor tissue were excised and stored at -80°C.
In a separate group of experiments mice bearing KB-C2 tumor were divided into two groups, where paclitaxel was administered via tail vein with or without nilotinib (given orally 1 hour before the paclitaxel treatment) (n=3-6) at 7.5, 15, 30, 60, 120 or 240 min after paclitaxel administration. At the end of treatment, through cardiac puncture blood was taken from anaesthetized animals into heparinized tubes and plasma was harvested. In addition, the tumors were removed, weighed, snap frozen in liquid nitrogen, and stored at –80°C until analysis. Paclitaxel was quantified using HPLC analysis, as described below.
2.8 HPLC analysis of paclitaxel in plasma and tissues
2.8.1 Chromatographic conditions
Quantification of paclitaxel was conducted using isocratic Shimadzu LC-20AB liquid chromatograph equipped with the Shimadzu SIL-20A HT autosampler and LC-20AB pump connected to a Dgu-20A3 degasser (Columbia, MD) according to the method described by Gill et al. [37]. The column used was a reversed-phase, Phenomenex Luna C18 column (250 × 4.6 mm i.d., 5 μm; Phenomenex, Torrance, CA) with an ODS guard column (4 mm × 3 mm; Phenomenex). The injection volume was 20 μl, and the mobile phase used for the separation of paclitaxel in plasma and tissue homogenate samples consisted of acetonitrile and water (53:47, v/v) delivered at 1.0 ml/min flow rate. For paclitaxel detection, Shimadzu UV SPD-20A detector set at 227 nm was used. Data acquisition and analysis was achieved using LC Solution software version 1.22 SP1 (Shimadzu). All samples were analyzed in duplicate. Under these chromatographic conditions, the total run time was 15 min with a retention time of 12 min for paclitaxel. Standard curves for paclitaxel in plasma and tissue homogenates were prepared in the ranges of 25–5000 ng/ml. The analytical method described in this work has been already established and validated previously [37].
2.8.2 Extraction of paclitaxel from plasma and tissue homogenate samples
Simple one-step protein precipitation with acetonitrile was used for sample preparation. Tissues were homogenized in saline in the ratio of 1:2 (v/v). Paclitaxel was extracted from plasma and tissue homogenate samples by precipitation with acetonitrile in 1:1 and 1:2 ratios (v/v), respectively. Samples were then vortexed for 1.0 min followed by centrifugation for 10 min at 10,000 rpm. The supernatant was transferred to insert vials from which 20 μl was injected onto the HPLC column. Samples with concentrations higher than the calibration range limit were appropriately diluted to fit within the working calibration curve.
2.9 Statistical analysis
All experiments were repeated at least 3 times and the differences were determined using the Student's t-test using GraphPad Prism version 5.04. The area under the curve for tumor and plasma concentrations was calculated by non-compartmental analysis using WinNonlin Phoenix 6.2. The statistical significance was determined at P < 0.05.
3. Results
3.1 Nilotinib synergistic activity towards anticancer drugs compared to a panel of TKIs
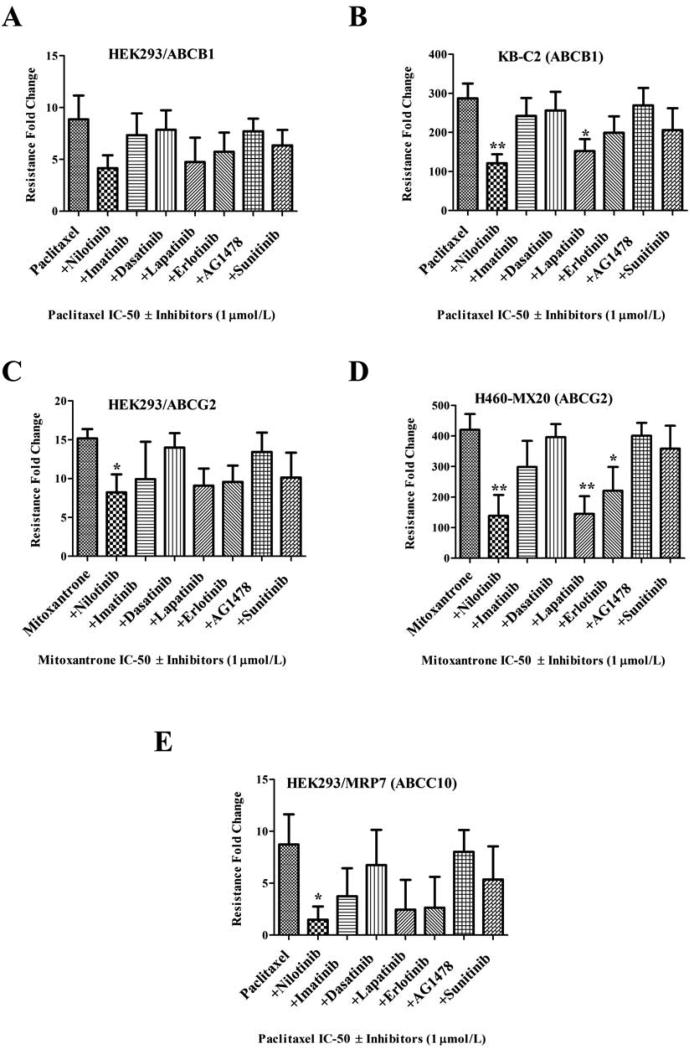
Pre-clinical and clinical development of MDR modulator to overcome anticancer drug-resistance would require a safe and potent compound. Specific TKIs have shown promising interactions with certain ABC transporters in in vitro studies [7; 8; 9; 10; 11; 12; 19; 24; 25; 38]. Therefore we compared a panel of clinically approved or under development TKIs belonging to BCR-ABLTKIs (nilotinib, imatinib and dasatinib), EGFR TKIs (lapatinib, erlotinib, AG1478) and a multikinase VEGF TKI (sunitinib) for its ability to enhance anticancer sensitivity at a low 1 μmol/L concentrations in ABCB1 (HEK293/ABCB1 and KB-C2), ABCG2 (HEK293/ABCG2 and H460-MX20) and ABCC10 (HEK293/MRP7) overexpressing cells. In Fig. 1A, although statistical significance was not reached in transfected HEK293/ABCB1 cells at 1 μmol/L, the increased sensitivity of paclitaxel in nilotinib combination group can be easily perceived. The effect of nilotinib towards paclitaxel sensitivity was significant in KB-C2 cells and even more profound than lapatinib at 1 μmol/L concentrations (Fig. 1B). When compared with other TKIs at 1 μmol/L, the nilotinib produced most significant sensitization towards mitoxantrone resistance in both transfected HEK293/ABCG2 and drug selected H460-MX20 cells (Figs. 1C and 1D). Similar results were also observed in ABCC10 overexpressing cells, where nilotinib was most potent in sensitizing paclitaxel cytotoxicity in HEK293/MRP7 cells (Fig. 1E). No significant effect were observed in the parental HEK293, KB-3-1 or H460 cells in any of the treatment group at 1 μmol/L (data not shown), suggesting the effects produced by nilotinib or other TKIs were solely due to their ability to modulate ABC transporters.
Figure 1. Resistance reversal of ABCB1, ABCG2 and ABCC10 anticancer substrates with or without TKI's.
The results of the MTT cytotoxicity assays were used to measure cell survival and the bar graph represents the change in resistance-fold (RF) in the presence or absence of TKI's in resistant cells. Resistance-fold was calculated by dividing the IC50 value for paclitaxel or mitoxantrone with or without the TKIs in the resistant cells shown in A-E with IC50 value for paclitaxel or mitoxantrone without inhibitors in the respective parental cells (not shown) as described in Materials and Methods. The MDR reversal effect of TKIs at 1 μmol/L in combination with paclitaxel is shown in ABCB1 overexpressing A. HEK293/ABCB1 B. KB-C2 cells; in combination with mitoxantrone in C. HEK293/R2 D. H460/MX20 cells; and in combination with paclitaxel in E. HEK293/MRP7 (ABCC10) cells is shown. The data points represent the mean ± SD of at least three independent experiments performed in triplicate. Statistically, *, P < 0.05; **,P < 0.01, versus the control group (Student t-test).
Previously, we reported that nilotinib could modulate drug resistance in drug selected ABCB1 (KB-C2 and KB-V1) cells as well in ABCG2-transfected (HEK293/ABCG2-482-R2, HEK293/ABCG2-482-G2 and HEK293/ABCG2-482-T7) cells [10]. In the current study, we extended this work to investigate the effect of nilotinib on ABCB1-tranfected HEK293 cells as well as mitoxantrone (MX) selected H460 cells overexpressing ABCG2 transporters (Supplementary Fig. 2). Based on our previous experience [10], we used well-tolerated concentrations of nilotinib up to 5 μmol/L for MDR reversal studies, a concentration that caused < 10% inhibition of growth in all the cell lines used in this study (Supplementary Fig. 3) and is clinically attainable. Nilotinib in a concentration-dependent manner significantly decreased the IC50 values for vincristine and paclitaxel, which are substrates of ABCB1, in the transfected HEK293/ABCB1 cells (Supplementary Table 1A), and at 5 μmol/L the inhibitory effect of nilotinib on ABCB1 was comparable to verapamil, a well-established inhibitor of ABCB1. However, nilotinib neither produced any significant alterations in the cytotoxicity of vincristine or paclitaxel on parental HEK293/pcDNA3.1 cells nor on cisplatin (a non-ABCB1 substrate) toxicity in either HEK293/pcDNA3.1 or HEK293/ABCB1 cells that supports nilotinib as being a specific modulator of ABCB1 transporters in these cells (Supplementary Table 1A).
As shown in Supplementary Table 1B, nilotinib in a concentration dependent manner at 2.5 and 5 μmol/L significantly decreased the IC50 values of ABCG2 substrates mitoxantrone (MX) and doxorubicin in the MX-selected H460/MX20 cells. Furthermore, 5 μmol/L of nilotinib produced a proportionate and significantly greater degree of sensitization to ABCG2-overexpressing cells with both MX and doxorubicin when compared to nilotinib at 2.5 μmol/L. The magnitude of the sensitization produced by 5 μmol/L nilotinib was comparable to that induced by the known specific ABCG2 inhibitor FTC at 2.5 μmol/L. Again, in the parental HEK293/pcDNA3.1 cells, nilotinib did not alter the growth inhibitory effects of either doxorubicin or MX, and had no effect on cisplatin, a non-substrate of ABCG2, in all the cell lines, similar to FTC (Supplementary Table 1B).
Finally, the IC50 values of vincristine were determined in HEK293/ABCC1 cells in the absence and presence of both nilotinib and ONO-1078, a known ABCC1 inhibitor, wherein it was found that only ONO-1078 reduced the IC50 values of vincristine (Supplementary Table 2). Collectively, the above findings indicate that nilotinib is one of the most potent TKI among the screened panel of TKIs and can reverse MDR mediated by ABCB1- or ABCG2- or ABCC10-overexpressing cells but not ABCC1-mediated MDR. Nonetheless, critical in vivo data on the ability of nilotinib to modulate ABC transport function in MDR-xenograft models is lacking.
3.2 Nilotinib potentiates the anticancer activity in in vivo models of ABCB1-, ABCG2- and ABCC10-mediated MDR
We successfully developed new ABCB1-, ABCG2 and ABCC10- xenograft MDR models in athymic mice after a series of optimizations and pilot experiments (see Supplementary Fig. 1). The intra-peritoneal doses of paclitaxel (18 mg/kg) and doxorubicin (3 mg/kg) were determined on the basis that they produced significant resistance in MDR-xenograft models compared to parental-xenograft models (data not shown) It was found that nilotinib alone up to 150 mg/kg oral doses had no visible toxicity or phenotypic changes in the male athymic NCR nude mice, and attained weight gains similar to that of vehicle group (data not shown). Oral doses of 75 mg/kg were chosen as this dose in nude mice produced plasma concentrations that ranged from 25 - 30 μmol/L [39; 40], which should be sufficient to inhibit ABC transporter function.
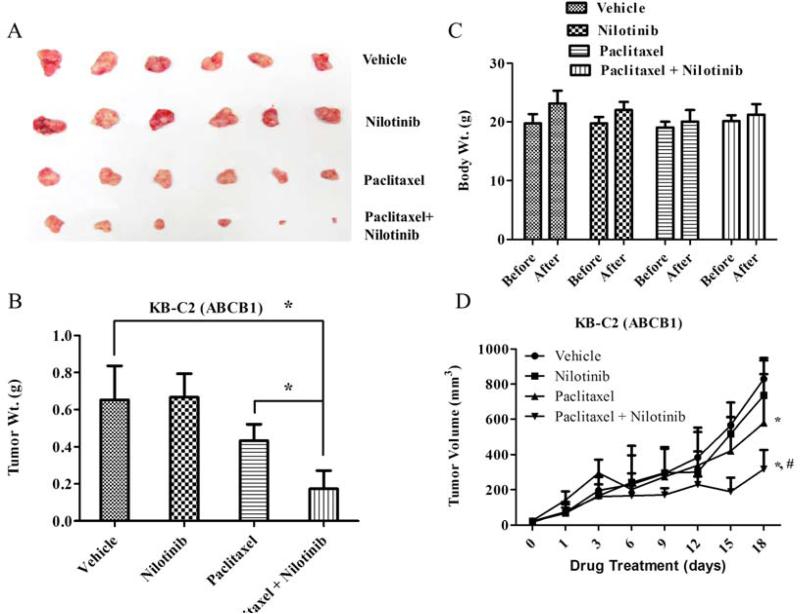
In the first series of studies, the ability of nilotinib to augment the activity of paclitaxel was tested in the parental KB-3-1 and ABCB1 overexpressing KB-C2 xenograft models (Fig. 2 and Supplementary Fig. 4). At the end of an 18-day treatment period, nilotinib (75 mg/kg, p.o.) in combination with paclitaxel (18 mg/kg, i.p.) produced a significantly greater inhibitory effect on KB-C2 tumor growth (size and weight) compared to vehicle, nilotinib or paclitaxel treated groups (P < 0.05; Figs. 2A and 2B). No weight loss or phenotypic changes were observed in any of the treatment groups (Fig. 2C). While nilotinib alone had no effect on tumor volume in KB-C2 xenograft models (Figs. 2A and 2B), its combination with paclitaxel produced significant reduction of tumor growth curves when plotted against treatment duration after tumor implantation (Fig. 2D). The nilotinib-pacliataxel combination significantly reduced the tumor volume when compared to vehicle and the paclitaxel alone group in the ABCB1 xenograft model (Fig. 2D, P < 0.05).
Figure 2. The effect of nilotinib on ABCB1 xenograft models.
Potentiation of antitumor effects of paclitaxel by nilotinib in ABCB1 overexpressing (KB-C2) oral epidermoid carcinoma xenograft model is shown. Atleast two independent experiments were carried out using athymic NCR nude mice implanted s.c. with KB-C2 cells. A. A representative picture of the excised KB-C2 tumor sizes from different mice is shown on the 18th day after implantation. B. The bar graph represents the mean tumor weight (n=6-10) of the excised KB-C2 tumor from different mice. The treatments were as follows: (a) vehicle (q3d × 6), (b) paclitaxel (18 mg/kg, i.p., q3d × 6) (c) nilotinib (75 mg/kg, p.o., q3d × 6) (d) paclitaxel (18 mg/kg, i.p., q3d × 6) + nilotinib (75 mg/kg, p.o., q3d × 6, given 1 h before giving paclitaxel). Each column represent the mean determinations and the bars represent SD. *, P < 0.05; **, P < 0.001 versus the control group. C. Changes in mean body weight before and after treatment for ABCB1-xenograft model are shown in the bar graph. D. Changes in tumor volume with time in ABCB1-xenograft model are shown. Points represent mean tumor volume for each group (n=6) after implantation. Each point on line graph represent the mean tumor volume (mm3) at a particular day after implantation and the bars represent SD. *, P < 0.05 versus the vehicle group; *,#, P < 0.05 versus paclitaxel alone group.
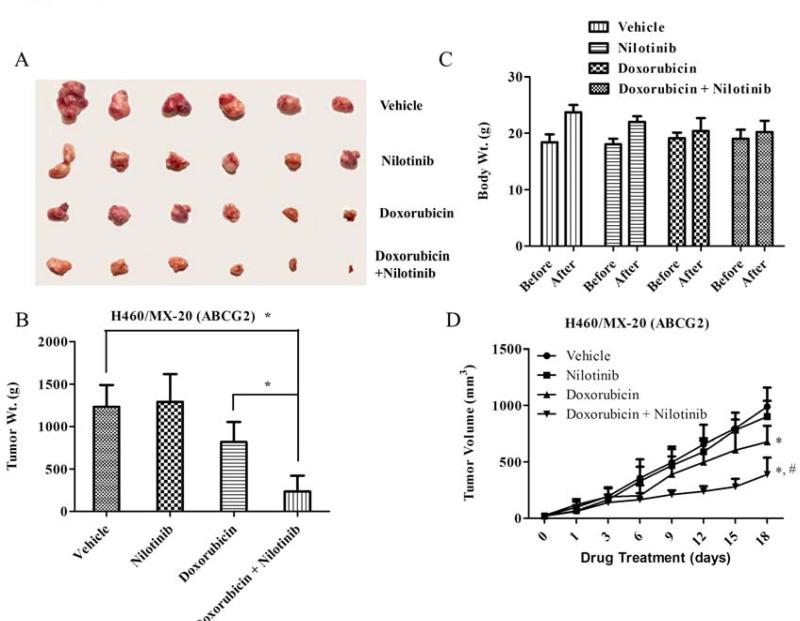
Next, the parental large cell lung cancer H460 and mitoxantrone-selected ABCG2 overexpressing H460/MX-20 xenograft models were tested for mitoxantrone toxicity, with or without nilotinib. Mitoxantrone alone was lethal to mice at doses (0.8 – 1.5 mg/kg) required to inhibit the parental H460 xenografts (Data not shown), hence, we switched to another well-known ABCG2 substrate doxorubicin, which has previously been shown to be significantly active against H460 bearing tumor mice at both 5 and 10 mg/kg dose [41]. We found that doxorubicin even at a 3 mg/kg i.p dose caused appreciable growth retardation in parental H460 xenografts (Supplementary Fig. 4B), while analogous doses in H460/MX-20 xenografts were unaffected (P < 0.05). Nilotinib in combination with doxorubicin produced a significant reduction in tumor size and weight (Figs. 3A and 3B). In addition, the tumor volume recorded over a period of 18 days showed a significant decrease in the nilotinib-doxorubicin combination group compared to vehicle, nilotinib or doxorubicin alone groups (P < 0.05; Fig. 3D). It should be noted that doxorubicin at 3 mg/kg with or without nilotinib produced no apparent toxicity or weight loss (Fig. 3C).
Figure 3. The effect of nilotinib on ABCG2 xenograft model.
Potentiation of antitumor effects of doxorubicin by nilotinib in ABCG2 overexpressing NSCLC H460/MX-20-xenograft model is shown on the 18th day after implantation. At least two independent experiments were carried out using athymic NCR nude mice implanted s.c. with H460/MX-20 cells. A. A representative picture of the excised H460/MX-20 tumor sizes from different mice is shown on the 18th day after implantation. B. The bar graph represents the mean tumor weight (n=6-10) of the excised H460/MX-20 tumor from different mice. The treatment were as follows: (a) vehicle (q3d × 6), (b) doxorubicin (1.8 mg/kg, i.p., q3d × 6) (c) nilotinib (75 mg/kg, p.o., q3d × 6) (d) doxorubicin (1.8 mg/kg, i.p., q3d × 6) + nilotinib (75 mg/kg, p.o., q3d × 6, given 1 h before giving doxorubicin). Each column represent the mean determinations and the bars represent SD. *, P < 0.05; **, P < 0.001 versus the control group. C. Changes in mean body weight before and after treatment for ABCG2-xenograft model are shown in the bar graph. D. Changes in tumor volume with time in ABCG2-xenograft model are shown. Points represent mean tumor volume for each group (n=6) after implantation. Each point on line graph represent the mean tumor volume (mm3) at a particular day after implantation and the bars represent SD. *, P < 0.05 versus the vehicle group; *,#, P < 0.05 versus doxorubicin alone group.
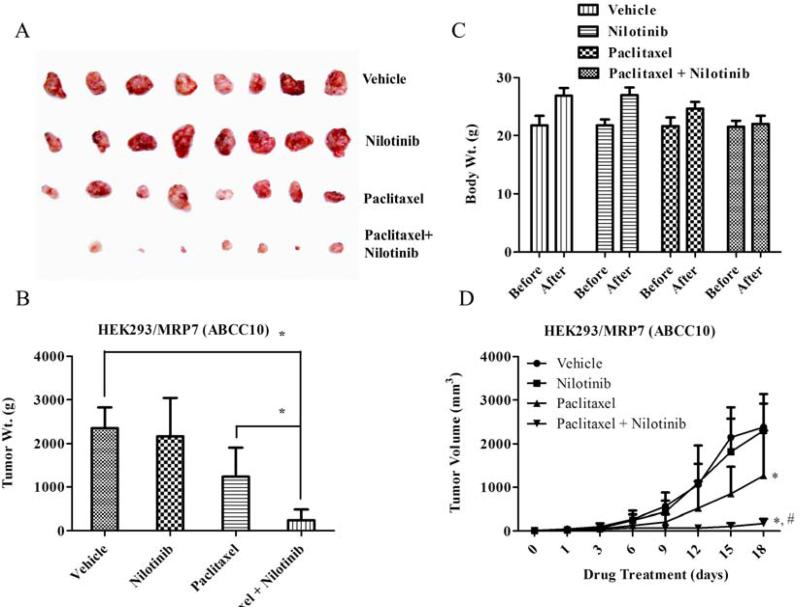
A similar in vivo model was designed to study the nilotinib effects on paclitaxel, a ABCC10/MRP7 substrate, in a ABCC10 transfected HEK293/MRP7 MDR-xenograft model. At a dose of 18 mg/kg i.p., equal to that used in the ABCB1 xenograft model, HEK293/MRP7 produced significant resistance to paclitaxel (Figs. 4A, 4B and 4D), in contrast to parental HEK293 tumors that were almost completely eliminated at this dose (Supplementary Fig. 4C) and no apparent weight loss was observed among treatment groups (Fig. 4C). Nilotinib (75 mg/kg, p.o.) in combination with paclitaxel significantly decreased the HEK293/MRP7 tumor size, weight and its growth curve over a period of 18 days when compared to vehicle, nilotinib or paclitaxel alone groups (P < 0.05; Figs. 4A, 4B and 4D, respectively).
Figure 4. The effect of nilotinib on ABCC10/MRP7 xenograft model.
Potentiation of antitumor effects of paclitaxel by nilotinib in human embryonic kidney (HEK293) cells transfected with ABCC10/MRP7-xenograft model is shown on the 18th day after implantation. Atleast two independent experiments were carried out using athymic NCR nude mice implanted s.c. with HEK293/MRP7. A. A representative picture of the excised HEK293/MRP7 tumor sizes from different mice is shown on the 18th day after implantation. B. The bar graph represents the mean tumor weight (n=6-10) of the excised HEK293/MRP7 tumor from different mice. The treatments were as follows: (a) vehicle (q3d × 6), (b) paclitaxel (18 mg/kg, i.p., q3d × 6) (c) nilotinib (75 mg/kg, p.o., q3d × 6) (d) paclitaxel (18 mg/kg, i.p., q3d × 6) + nilotinib (75 mg/kg, p.o., q3d × 6, given 1 h before giving paclitaxel). Each column represent the mean determinations and the bars represent SD. *, P < 0.05; **, P < 0.001 versus the control group. C. Changes in mean body weight before and after treatment for ABCC10-xenograft model are shown in the bar graph. D. Changes in tumor volume with time in ABCC10-xenograft model are shown. Points represent mean tumor volume for each group (n=6) after implantation. Each point on line graph represent the mean tumor volume (mm3) at a particular day after implantation and the bars represent SD. *, P < 0.05 versus the vehicle group; *,#, P < 0.05 versus paclitaxel alone group.
It should be noted that nilotinib by itself had no significant effect on the inhibition of ABCB1, ABCG2 and ABCC10 overexpressing MDR-xenografts (Figs. 1, 2 and 3) or parental-xenograft models (Supplementary Fig. 4). In addition, the nilotinib combination regimens did not result in increased toxicity; instead it uniformly improved the efficacy of paclitaxel and doxorubicin in the ABCB1, ABCG2 and ABCC10 MDR-xenograft models compared to paclitaxel or doxorubicin treatment alone (Figs. 2, 3 and 4).
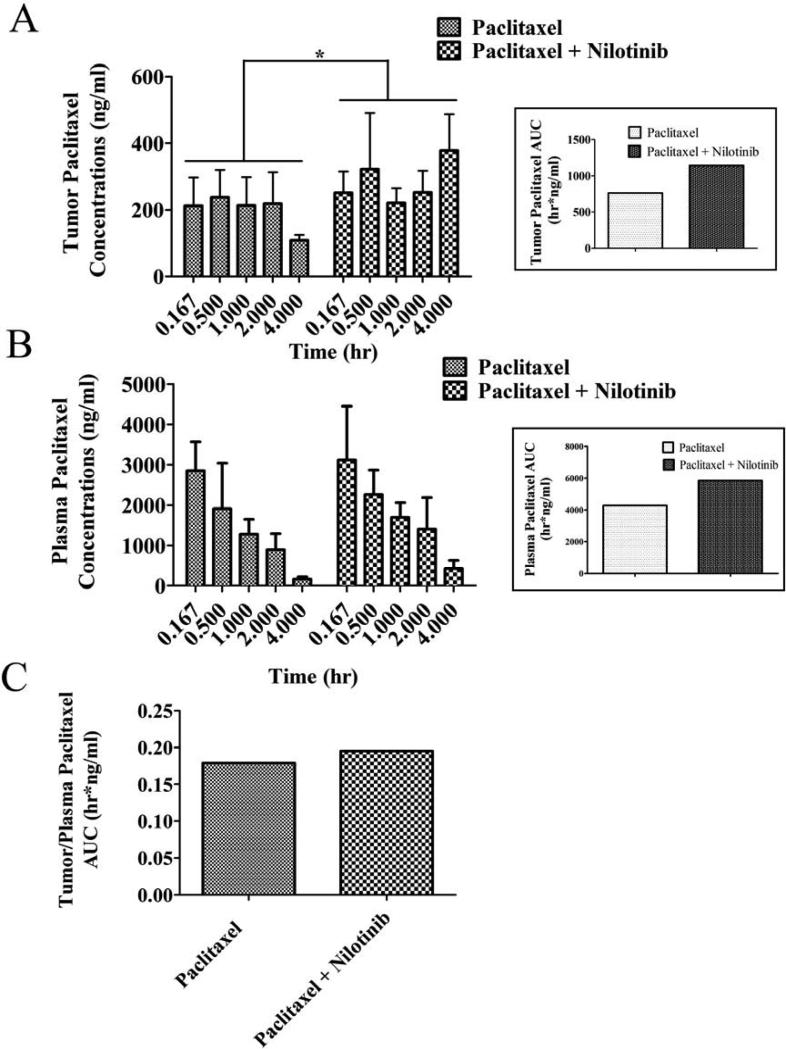
In a separate study to understand the pharmacokinetic implications of nilotinib on tumor distribution of paclitaxel, paclitaxel was given intravenously with or without oral nilotinib for 18-days. Pairwise comparisons of paclitaxel tumor concentrations between paclitaxel alone and in combination with nilotinib revealed a significant elevation at 4 hours in the combination group. The total tumor exposure to paclitaxel as indicated by the area under the drug concentration-time curve or AUC indicated an approximate 33% increase, 1141.6 hr*ng/ml vs 764.5 hr*ng/ml in the paclitaxel alone group (Fig. 5A). Although, there were increases in paclitaxel plasma concentration in the nilotinib combination group compared to the paclitaxel alone group, none of the pairwise comparisons reached statistical significance (Fig. 5B). The plasma AUC values were 5838.2 hr*ng/ml in the nilotinib-pacliataxel group compared to 4271.4 hr*ng/ml in the paclitaxel alone group or a 27% elevation in the nilotinib group. The corresponding tumor/plasma AUC's indicated a slight increase (~ 10%) in nilotinib combination group over paclitaxel alone group (Fig. 5C). These data suggest that nilotinib can influence the PK behavior of paclitaxel most likely due to its inhibition of ABCB1 transport at the level of biliary excretion and possibly at the tumor cell.
Figure 5. The effect of nilotinib on plasma and tumor concentration of paclitaxel in ABCB1-xenograft model.
Paclitaxel concentration in KB-C2 tumor (box – paclitaxel tumor AUC) A. plasma (box – paclitaxel plasma AUC) B. KB-C2 tumor/plasma AUC ratio C. with or without nilotinib treatment is shown. In the combination group, 75 mg/kg nilotinib was given orally, 1 h before giving 18 mg/kg paclitaxel via tail vein injection. Mice (n=5-6) were euthanized via cardiac puncture at different intervals and tumor and plasma was harvested and stored in -80°. Paclitaxel was quantified as described in materials and methods. Columns, means (n = 3-6); bars, SD. * P < 0.05, compared with only paclitaxel receiving group (unpaired student t-test).
3.3 Nilotinib docking analysis with human ABCB1 and ABCG2 homology models
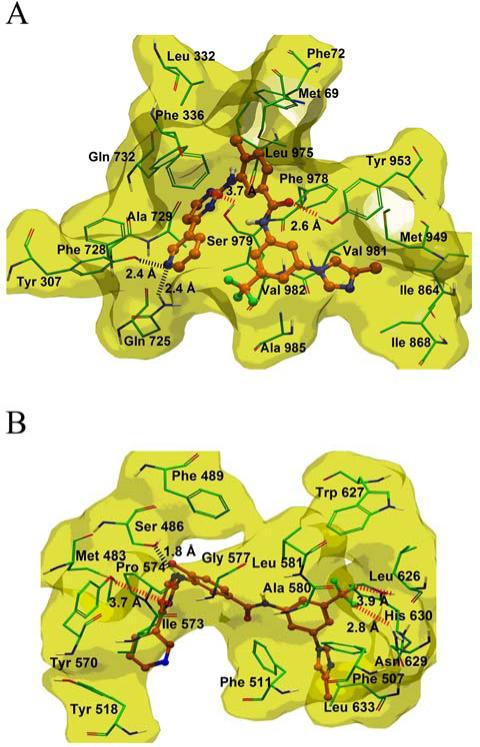
In the absence of the crystal structure of human ABCB1, we developed an ABCB1 homology model based on the crystal structure of ABCB1 from mice [31]. To understand the binding mechanism of nilotinib to the homology model of human ABCB1 at a molecular level, docking studies were performed on all of the possible binding sites [32]. The best docking score for the docked conformation of nilotinib was found at site-1, which was also supported by photoaffinity labeling of [125I]- iodoarylazidoprazosin (IAAP) . Therefore, the binding interaction of nilotinib within the site 1 of ABCB1 is shown in Fig. 6A. The pyridine ring N atom of nilotinib forms hydrogen bonding interactions with the side chains of Tyr307 (-N---HO-Tyr307, 2.4 Å) and Gln725 (-N---H2NOC-Gln725, 2.4 Å). The N-5 atom of the pyrimidine ring of nilotinib forms electrostatic interaction with the side chain hydroxyl group of Ser979 (-N---HO-Ser979, 3.7 Å). Moreover the pyridine and pyrimidine rings of nilotinib are located within a hydrophobic pocket lined by the side chains of Phe336, Phe728, Ala729, Phe732 and Val982. The methyl substituted phenyl ring of nilotinib lies within a hydrophobic pocket bordered by the side chains of Met69, Phe72, leu332, Phe336, Phe732, Leu975, Phe978 and Val982. The carbonyl oxygen atom of the amide linker is involved in electrostatic interaction with the side chain hydroxyl group of Tyr953 (-CO---HO-Tyr953, 2.6 Å). The trifluoromethylphenyl ring of nilotinib is stabilized by hydrophobic contacts with the side chains of Val982 and Ala985. The 4 methyl imidazole group of nilotinib forms hydrophobic contacts with the side chain of Ile864, Ile868, Met949, Tyr953, Val981 and Ala985. The culmination of these numerous interactions suggests that nilotinib binds to the ABCB1 drug-binding site (site-1) with high affinity.
Figure 6. XP-Glide predicted binding mode of nilotinib with homology modeled ABCB1 and ABCG2.
The docked conformations of nilotinib as ball and stick model are shown within the large hydrophobic cavity of A. ABCB1 and B. ABCG2. Important amino acids are depicted as sticks with the atoms colored as carbon – green, hydrogen – white, nitrogen – blue, oxygen – red, sulfur – yellow, whereas nilotinib is shown with the same color scheme as above except carbon atoms are represented in orange and fluorine in light green. Dotted black line indicates hydrogen-bonding interactions, whereas dotted red line indicates electrostatic interactions. ABCB1 and ABCG2 are represented as macromodel surfaces based on residue charge (hydrophobic-yellow).
The Glide predicted docked model of nilotinib at the Arg482 centroid-based grid of ABCG2 exhibited the binding pose as shown in Fig. 6B. The 4-methylimidazole and trifluoromethylphenyl substituents of nilotinib are stabilized through hydrophobic contacts with the side chains of Phe507, Phe511, Ala580, Leu581, Leu626, Trp627 and Leu633. One of the fluorine atoms of the trifluoromethyl substituent may participate in electrostatic interaction with the side chain of Asn629 (-CF3---H2NOC-Asn629, 2.8 Å), whereas another fluorine atom may interact with the side chain of His630 (-CF3---HN-His630, 3.9 Å). As shown in Fig. 6B, the 4-methylphenyl, pyridine and the pyrimidine rings are extensively stabilized through the hydrophobic side chains of Met483, Phe489, Tyr518, Ile573, Pro574 and Gly577. The -NH linker present between the 4-methylphenyl ring and the pyrimidine ring forms a hydrogen bond with the side chain hydroxyl function of Ser486 (-NH---OH-Ser486, 1.8 Å). The N-2 atom of the pyrimidine ring may be involved in an electrostatic interaction with the side chain hydroxyl function of Tyr570 (-N---HO-Tyr570, 3.7 Å) (Fig. 6B). Favorable binding affinity of nilotinib towards ABCG2, suggests that it may significantly modulate the ABCG2 transporter and its substrates.
4. Discussion
Nilotinib seems to be one of the more potent modulators of ABCB1, ABCG2, and ABCC10 when compared to other clinically available TKIs such as imatinib, dasatinib, AG1478, erlotinib, lapatinib and sunitinib in in vitro studies (Fig. 1). Nilotinib is an orally bioavailable TKI administered on a daily basis to BCR-ABL positive CML patients. It was designed with insight from the crystal structure of the imatinib-ABL complex and is a relatively selective inhibitor of the tyrosine kinase activities of BCR-ABL, discoidin domain receptors (DDR), platelet derived growth factors (PDGFR) and mast/stem-cell growth factor (c-KIT) [39; 42]. Deguchi et al. reported that nilotinib may be useful for the treatment of ABCB1-overexpressing leukemic cells as these cells were still quite sensitive to nilotinib yet conferred resistance to imatinib [43]. At the same time, K562 leukemic cells that overexpress ABCB1 and/or ABCG2 were resistant to nilotinib indicating the importance of drug concentration and the need to establish concentration-response relationships [44; 45]. To address this, we demonstrated relatively higher, yet clinically achievable, nilotinib concentrations could competitively block the drug efflux function of ABCB1, ABCG2 and ABCC10 transporters, and thus it could be used to potentiate the anticancer sensitivity in MDR cancer cells [10; 22]. We have shown that a non-toxic concentration of nilotinib in combination with vincristine or paclitaxel and MX or doxorubicin could inhibit ABCB1-, and ABCG2-mediated drug resistance in both transfected and drug resistant cells, respectively. Nonetheless, critical in vivo data on the ability of nilotinib to modulate ABC transport function in MDR-xenograft models is lacking. Novel ABCB1-, ABCG2- and ABCC10 MDR-xenograft mouse models were established to evaluate the combinatorial effect of nilotinib with anticancer drugs paclitaxel and doxorubicin that are substrates of ABCB1, ABCG2 and ABCC10 transporters. Nilotinib significantly potentiated the antitumor effect of paclitaxel in ABCB1 overexpressing (KB-C2) oral epidermoid carcinoma and ABCC10 overexpressing HEK293/MRP7 xenografts in athymic nude mice. In addition, doxorubicin, an anthracycline used in the treatment of lung cancer, when combined with nilotinib, significantly reduced tumor burden in the ABCG2-mediated MDR large cell lung cancer H460/MX-20-xenograft in mice model. Over 40 clinical trials are being pursued currently either with nilotinib alone or in combination (http://www.cancer.gov/clinicaltrials/search/results?protocolsearchid=6392375) for different indications, such as CML, acute lymphoblastic and myeloid leukemias, gastrointestinal stromal tumors, and certain primary and metastatic melanomas. The MDR modulatory effects of nilotinib demonstrated in the preclinical setting here might be an important aspect to further develop and assess in the clinic. Collection of this key information was the focus of the current investigation that was complemented by homology modeling and docking analysis to determine nilotinib's affinity to bind at the drug-binding sites of ABCB1 and ABCG2 transporters.
Until now, attempts to obtain the co-crystal structures of human ABCB1 and ABCG2 have been unsuccessful due to their membrane bound nature and ensuing difficulties in isolation and purification. Currently high-resolution crystal structures of human ABCB1 and ABCG2 are unavailable; hence, we used ABCB1 and ABCG2 homology models. To identify the most appropriate binding site for nilotinib, docking studies were performed on all possible binding sites of ABCB1 as described by Aller et al. [31], and reported critical residues shown by site-directed mutagenesis experiments for ABCG2 [34; 35]. After exploring docking simulations of nilotinib at all of these binding sites, the most favorable binding site for nilotinib against human ABCB1 and ABCG2 was identified and discussed. The ability of nilotinib to inhibit ABCB1 and ABCG2 could be considered as a phenyl-pyrimidine derived BCR-ABL TKIs class effect based on two criterias: a) in general BCR-ABL TKIs are hydrophobic (logP ~ 3 to 6) as is the transmembrane domain (substrate binding sites) of human ABCB1 and ABCG2 [46], b) the BCR-ABL TKIs have been previously shown to interact significantly with ABCB1 and ABCG2 [10; 47]. Moreover, nilotinib appears to exhibit all of the pharmacophoric features, such as hydrophobic groups and/or aromatic ring centers, hydrogen-bond acceptors and hydrogen-bond donor groups that have been described critical for binding at ABCB1 and ABCG2 transporters [48]. By understanding the nature of the molecular interaction gained through molecular modeling studies structural clues to obtain potent inhibitors of these targets may be revealed.
Pharmacokinetic analysis of patients at the recommended 400 mg b.i.d dose of nilotinib showed mean peak-trough plasma concentrations ranged from 3.6 μmol/L to 1.7 μmol/L, with an apparent half-life of 15 h [49]. Our in vitro studies showed that nilotinib at concentrations of 1 μmol/L can effectively block ABCB1, ABCG2 and ABCC10-mediated MDR, and thus, these effects can be attained in patients [10; 22]. A recent in vitro study in soft tissue sarcoma also confirms our results that nilotinib could potentiate doxorubicin cytotoxicity by inhibiting ABCB1 mediated MDR [25]. In fact, the transport inhibiting function of nilotinib in conjunction with its TKI activity has dual implications when combined with drugs whose PK properties and cancer cell toxicity are mediated by ABCB1, ABCG2 or ABCC10. Blockade of these drug efflux pumps could increase oral bioavailability or decrease biliary excretion leading to higher systemic drug exposures that could lead to greater cell toxicity that may be further enhanced by a local action of nilotinib on these transporters. TKIs such as gefitinib and lapainib have shown the ability to enhance the bioavailability of certain anticancer drugs [7; 50]. The elevated paclitaxel plasma and tumor concentrations in the presence of nilotinib support its inhibitory action on ABCB1; however additional studies would be useful to fully define these actions.
Targeting ABC transporters in cancer cells is only one of the many ways to overcome MDR. Other drug-resistance determinants like changes in metabolizing and detoxifying systems, such as DNA repair and the cytochrome P450 oxidases, should also be targeted to overcome MDR to obtain optimal pharmacokinetic properties, tumor penetration, and intracellular concentration of chemotherapeutic drugs in malignant cancerous cells. Although we did not observe significant increases in paclitaxel concentration in the plasma when co-administered with nilotinib (Fig. 5B) there was a trend towards elevated paclitaxel plasma concentrations with a ~36% increase in the AUC. This action could be modulated by the reported ability of nilotinib to both induce and inhibit CYP2C8, the hepatic enzyme primarily responsible for paclitaxel metabolism. The conditions for such dual actions of nilotinib on CYP2C8, and correspondingly, on paclitaxel's metabolism, and whether there is a net negative or positive impact on nilotinib's inhibitory action on the drug efflux pumps will require further studies.
In vivo studies using nude mouse xenograft models indicate that nilotinib potentiates the anticancer effect of paclitaxel in ABCB1- and ABCC10-, and doxorubicin in ABCG2-xenograft models at concentrations that are achieved clinically. These positive findings suggest that nilotinib can be combined with conventional chemotherapeutic drugs as well as other TKIs that are substrates of ABCB1 and ABCG2 in patients with MDR mediated by ABC transporters to attain improved anticancer responses.
Supplementary Material
Acknowledgements
We are thankful to Novartis for providing us nilotinib. We thank Drs. Shin-ichi Akiyama (Kagoshima University, Japan) for ONO-1078, KB-3-1, KB-C2 and KB-CV60 cell lines, Susan E. Bates and Robert W. Robey (NIH, USA) for FTC and ABCG2-overexpressing cell lines including H460/MX-20 cells; Dr. Suresh V. Ambudkar (NIH, USA) for ABCB1-transfected HEK/ABCB1 and ABCC1-transfected HEK/ABCC1 cell line; Xiaoyan Zhang and Shu-Pei Wu (Mount Sinai School of Medicine, NY), Dr. Dong-Hua Yang (Fox Chase Cancer Center, PA) for editorial assistance. We thank Drs. Mark F. Rosenberg and Zsolt Bikadi (University of Manchester, Manchester, UK) for providing coordinates of ABCG2 homology model.
Grant Support: This work was supported by funds from National Institutes of Health (No.1R15CA143701 to Z.S.C.) and St. John's University Seed Grant No. 579-1110, (Z.S. Chen).
Abbreviations
- MDR
multidrug resistance
- ABC
ATP-binding cassette
- ABCB1
also called P-gp (P-glycoprotein)
- ABCG2
also called BCRP (breast cancer resistance protein)/MXR (mitoxantrone resistance protein)
- ABCC1
also called MRP1 (multidrug resistance protein 1)
- EGFR
epidermal growth factor receptor
- HER
human epidermal receptor
- TKI
tyrosine kinase inhibitor
- PBS
phosphate-buffered saline
- FTC
fumitremorgin C
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no potential conflict of interest.
PhD Thesis: This work was a part of Doctoral Thesis work done at Department of Pharmaceutical Sciences, St. John's University, Jamaica, New York, 11439; under the guidance of Dr. ZS Chen
REFERENCES
- 1.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 2.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman MM, Ambudkar SV. Overview: ABC transporters and human disease. J Bioenerg Biomembr. 2001;33:453–458. doi: 10.1023/a:1012866803188. [DOI] [PubMed] [Google Scholar]
- 4.Szakacs G, Varadi A, Ozvegy-Laczka C, Sarkadi B. The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME-Tox). Drug Discov Today. 2008;13:379–393. doi: 10.1016/j.drudis.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 6.Gillet JP, Efferth T, Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim Biophys Acta. 2007;1775:237–262. doi: 10.1016/j.bbcan.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Dai CL, Tiwari AK, Wu CP, Su XD, Wang SR, Liu DG, Ashby CR, Jr., Huang Y, Robey RW, Liang YJ, Chen LM, Shi CJ, Ambudkar SV, Chen ZS, Fu LW. Lapatinib (Tykerb, GW572016) reverses multidrug resistance in cancer cells by inhibiting the activity of ATP-binding cassette subfamily B member 1 and G member 2. Cancer Res. 2008;68:7905–7914. doi: 10.1158/0008-5472.CAN-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai CL, Liang YJ, Wang YS, Tiwari AK, Yan YY, Wang F, Chen ZS, Tong XZ, Fu LW. Sensitization of ABCG2-overexpressing cells to conventional chemotherapeutic agent by sunitinib was associated with inhibiting the function of ABCG2. Cancer Lett. 2009;279:74–83. doi: 10.1016/j.canlet.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 9.Shi Z, Peng XX, Kim IW, Shukla S, Si QS, Robey RW, Bates SE, Shen T, Ashby CR, Jr., Fu LW, Ambudkar SV, Chen ZS. Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 2007;67:11012–11020. doi: 10.1158/0008-5472.CAN-07-2686. [DOI] [PubMed] [Google Scholar]
- 10.Tiwari AK, Sodani K, Wang SR, Kuang YH, Ashby CR, Jr., Chen X, Chen ZS. Nilotinib (AMN107, Tasigna) reverses multidrug resistance by inhibiting the activity of the ABCB1/Pgp and ABCG2/BCRP/MXR transporters. Biochem Pharmacol. 2009;78:153–161. doi: 10.1016/j.bcp.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Shukla S, Robey RW, Bates SE, Ambudkar SV. Sunitinib (Sutent(R), SU11248), a small-molecule receptor tyrosine kinase inhibitor, blocks function of the ABC transporters, P-glycoprotein (ABCB1) and ABCG2. Drug Metab Dispos. 2008 doi: 10.1124/dmd.108.024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari AK, Sodani K, Dai CL, Ashby CR, Jr., Chen ZS. Revisiting the ABCs of multidrug resistance in cancer chemotherapy. Curr Pharm Biotechnol. 2011;12:570–594. doi: 10.2174/138920111795164048. [DOI] [PubMed] [Google Scholar]
- 13.Breuninger LM, Paul S, Gaughan K, Miki T, Chan A, Aaronson SA, Kruh GD. Expression of multidrug resistance-associated protein in NIH/3T3 cells confers multidrug resistance associated with increased drug efflux and altered intracellular drug distribution. Cancer Res. 1995;55:5342–5347. [PubMed] [Google Scholar]
- 14.Kruh GD, Belinsky MG. The MRP family of drug efflux pumps. Oncogene. 2003;22:7537–7552. doi: 10.1038/sj.onc.1206953. [DOI] [PubMed] [Google Scholar]
- 15.Mao Q, Unadkat JD. Role of the breast cancer resistance protein (ABCG2) in drug transport. AAPS J. 2005;7:E118–133. doi: 10.1208/aapsj070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Hopper-Borge E, Chen ZS, Shchaveleva I, Belinsky MG, Kruh GD. Analysis of the drug resistance profile of multidrug resistance protein 7 (ABCC10): resistance to docetaxel. Cancer Res. 2004;64:4927–4930. doi: 10.1158/0008-5472.CAN-03-3111. [DOI] [PubMed] [Google Scholar]
- 18.Hopper-Borge E, Xu X, Shen T, Shi Z, Chen ZS, Kruh GD. Human multidrug resistance protein 7 (ABCC10) is a resistance factor for nucleoside analogues and epothilone B. Cancer Res. 2009;69:178–184. doi: 10.1158/0008-5472.CAN-08-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla S, Chen ZS, Ambudkar SV. Tyrosine kinase inhibitors as modulators of ABC transporter-mediated drug resistance. Drug Resist Updat. 2012 doi: 10.1016/j.drup.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozvegy-Laczka C, Cserepes J, Elkind NB, Sarkadi B. Tyrosine kinase inhibitor resistance in cancer: role of ABC multidrug transporters. Drug Resist Updat. 2005;8:15–26. doi: 10.1016/j.drup.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Hegedus C, Ozvegy-Laczka C, Szakacs G, Sarkadi B. Interaction of ABC multidrug transporters with anticancer protein kinase inhibitors: substrates and/or inhibitors? Curr Cancer Drug Targets. 2009;9:252–272. doi: 10.2174/156800909788166565. [DOI] [PubMed] [Google Scholar]
- 22.Shen T, Kuang YH, Ashby CR, Lei Y, Chen A, Zhou Y, Chen X, Tiwari AK, Hopper-Borge E, Ouyang J, Chen ZS. Imatinib and nilotinib reverse multidrug resistance in cancer cells by inhibiting the efflux activity of the MRP7 (ABCC10). PLoS One. 2009;4:e7520. doi: 10.1371/journal.pone.0007520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuang YH, Shen T, Chen X, Sodani K, Hopper-Borge E, Tiwari AK, Lee JW, Fu LW, Chen ZS. Lapatinib and erlotinib are potent reversal agents for MRP7 (ABCC10)-mediated multidrug resistance. Biochem Pharmacol. 2010;79:154–161. doi: 10.1016/j.bcp.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Z, Tiwari AK, Shukla S, Robey RW, Kim IW, Parmar S, Bates SE, Si QS, Goldblatt CS, Abraham I, Fu LW, Ambudkar SV, Chen ZS. Inhibiting the function of ABCB1 and ABCG2 by the EGFR tyrosine kinase inhibitor AG1478. Biochem Pharmacol. 2009;77:781–793. doi: 10.1016/j.bcp.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villar VH, Vogler O, Martinez-Serra J, Ramos R, Calabuig-Farinas S, Gutierrez A, Barcelo F, Martin-Broto J, Alemany R. Nilotinib counteracts p-glycoprotein-mediated multidrug resistance and synergizes the antitumoral effect of Doxorubicin in soft tissue sarcomas. PloS one. 2012;7:e37735. doi: 10.1371/journal.pone.0037735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S, Skoumbourdis AP, Walsh MJ, Hartz AM, Fung KL, Wu CP, Gottesman MM, Bauer B, Thomas CJ, Ambudkar SV. Synthesis and characterization of a BODIPY conjugate of the BCR-ABL kinase inhibitor Tasigna (nilotinib): evidence for transport of Tasigna and its fluorescent derivative by ABC drug transporters. Mol Pharm. 2011;8:1292–1302. doi: 10.1021/mp2001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akiyama S, Fojo A, Hanover JA, Pastan I, Gottesman MM. Isolation and genetic characterization of human KB cell lines resistant to multiple drugs. Somat Cell Mol Genet. 1985;11:117–126. doi: 10.1007/BF01534700. [DOI] [PubMed] [Google Scholar]
- 28.Taguchi Y, Yoshida A, Takada Y, Komano T, Ueda K. Anti-cancer drugs and glutathione stimulate vanadate-induced trapping of nucleotide in multidrug resistance-associated protein (MRP). FEBS Lett. 1997;401:11–14. doi: 10.1016/s0014-5793(96)01421-4. [DOI] [PubMed] [Google Scholar]
- 29.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 30.Shi Z, Liang YJ, Chen ZS, Wang XW, Wang XH, Ding Y, Chen LM, Yang XP, Fu LW. Reversal of MDR1/P-glycoprotein-mediated multidrug resistance by vector-based RNA interference in vitro and in vivo. Cancer Biol Ther. 2006;5:39–47. doi: 10.4161/cbt.5.1.2236. [DOI] [PubMed] [Google Scholar]
- 31.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Z, Tiwari AK, Shukla S, Robey RW, Singh S, Kim IW, Bates SE, Peng X, Abraham I, Ambudkar SV, Talele TT, Fu LW, Chen ZS. Sildenafil reverses ABCB1- and ABCG2-mediated chemotherapeutic drug resistance. Cancer Res. 2011;71:3029–3041. doi: 10.1158/0008-5472.CAN-10-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg MF, Bikadi Z, Chan J, Liu X, Ni Z, Cai X, Ford RC, Mao Q. The human breast cancer resistance protein (BCRP/ABCG2) shows conformational changes with mitoxantrone. Structure. 2010;18:482–493. doi: 10.1016/j.str.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robey RW, Honjo Y, Morisaki K, Nadjem TA, Runge S, Risbood M, Poruchynsky MS, Bates SE. Mutations at amino-acid 482 in the ABCG2 gene affect substrate and antagonist specificity. Br J Cancer. 2003;89:1971–1978. doi: 10.1038/sj.bjc.6601370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alqawi O, Bates S, Georges E. Arginine482 to threonine mutation in the breast cancer resistance protein ABCG2 inhibits rhodamine 123 transport while increasing binding. Biochem J. 2004;382:711–716. doi: 10.1042/BJ20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen LM, Liang YJ, Ruan JW, Ding Y, Wang XW, Shi Z, Gu LQ, Yang XP, Fu LW. Reversal of P-gp mediated multidrug resistance in-vitro and in-vivo by FG020318. J Pharm Pharmacol. 2004;56:1061–1066. doi: 10.1211/0022357043879. [DOI] [PubMed] [Google Scholar]
- 37.Gill KK, Nazzal S, Kaddoumi A. Paclitaxel loaded PEG(5000)-DSPE micelles as pulmonary delivery platform: formulation characterization, tissue distribution, plasma pharmacokinetics, and toxicological evaluation. Eur J Pharm Biopharm. 2011;79:276–284. doi: 10.1016/j.ejpb.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Hegedus T, Orfi L, Seprodi A, Varadi A, Sarkadi B, Keri G. Interaction of tyrosine kinase inhibitors with the human multidrug transporter proteins, MDR1 and MRP1. Biochim Biophys Acta. 2002;1587:318–325. doi: 10.1016/s0925-4439(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 39.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, Kung AL, Mestan J, Daley GQ, Callahan L, Catley L, Cavazza C, Azam M, Neuberg D, Wright RD, Gilliland DG, Griffin JD. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Kaur P, Feldhahn N, Zhang B, Trageser D, Muschen M, Pertz V, Groffen J, Heisterkamp N. Nilotinib treatment in mouse models of P190 Bcr/Abl lymphoblastic leukemia. Mol Cancer. 2007;6:67. doi: 10.1186/1476-4598-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraus-Berthier L, Jan M, Guilbaud N, Naze M, Pierre A, Atassi G. Histology and sensitivity to anticancer drugs of two human non-small cell lung carcinomas implanted in the pleural cavity of nude mice. Clin Cancer Res. 2000;6:297–304. [PubMed] [Google Scholar]
- 42.Manley PW, Drueckes P, Fendrich G, Furet P, Liebetanz J, Martiny-Baron G, Mestan J, Trappe J, Wartmann M, Fabbro D. Extended kinase profile and properties of the protein kinase inhibitor nilotinib. Biochim Biophys Acta. 2010;1804:445–453. doi: 10.1016/j.bbapap.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 43.Deguchi Y, Kimura S, Ashihara E, Niwa T, Hodohara K, Fujiyama Y, Maekawa T. Comparison of imatinib, dasatinib, nilotinib and INNO-406 in imatinib-resistant cell lines. Leuk Res. 2008;32:980–983. doi: 10.1016/j.leukres.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Brendel C, Scharenberg C, Dohse M, Robey RW, Bates SE, Shukla S, Ambudkar SV, Wang Y, Wennemuth G, Burchert A, Boudriot U, Neubauer A. Imatinib mesylate and nilotinib (AMN107) exhibit high-affinity interaction with ABCG2 on primitive hematopoietic stem cells. Leukemia. 2007;21:1267–1275. doi: 10.1038/sj.leu.2404638. [DOI] [PubMed] [Google Scholar]
- 45.Mahon FX, Hayette S, Lagarde V, Belloc F, Turcq B, Nicolini F, Belanger C, Manley PW, Leroy C, Etienne G, Roche S, Pasquet JM. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res. 2008;68:9809–9816. doi: 10.1158/0008-5472.CAN-08-1008. [DOI] [PubMed] [Google Scholar]
- 46.Nicolle E, Boccard J, Guilet D, Dijoux-Franca MG, Zelefac F, Macalou S, Grosselin J, Schmidt J, Carrupt PA, Di Pietro A, Boumendjel A. Breast cancer resistance protein (BCRP/ABCG2): new inhibitors a nd QSAR studies by a 3D linear solvation energy approach. Eur J Pharm Sci. 2009;38:39–46. doi: 10.1016/j.ejps.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Dohse M, Scharenberg C, Shukla S, Robey RW, Volkmann T, Deeken JF, Brendel C, Ambudkar SV, Neubauer A, Bates SE. Comparison of ATP-binding cassette transporter interactions with the tyrosine kinase inhibitors imatinib, nilotinib, and dasatinib. Drug Metab Dispos. 2010;38:1371–1380. doi: 10.1124/dmd.109.031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klepsch F, Jabeen I, Chiba P, Ecker GF. Pharmacoinformatic approaches to design natural product type ligands of ABC-transporters. Curr Pharm Des. 2010;16:1742–1752. doi: 10.2174/138161210791163992. [DOI] [PubMed] [Google Scholar]
- 49.Kantarjian H, Giles F, Wunderle L, Bhalla K, O'Brien S, Wassmann B, Tanaka C, Manley P, Rae P, Mietlowski W, Bochinski K, Hochhaus A, Griffin JD, Hoelzer D, Albitar M, Dugan M, Cortes J, Alland L, Ottmann OG. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 50.Stewart CF, Leggas M, Schuetz JD, Panetta JC, Cheshire PJ, Peterson J, Daw N, Jenkins JJ, 3rd, Gilbertson R, Germain GS, Harwood FC, Houghton PJ. Gefitinib enhances the antitumor activity and oral bioavailability of irinotecan in mice. Cancer Res. 2004;64:7491–7499. doi: 10.1158/0008-5472.CAN-04-0096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.