Abstract
Understanding the organization of the human brain requires identification of its functional subdivisions. Clustering schemes based on resting-state functional magnetic resonance imaging (fMRI) data are rapidly emerging as non-invasive alternatives to cytoarchitectonic mapping in postmortem brains. Here, we propose a novel spatio-temporal probabilistic parcellation scheme that overcomes major weaknesses of existing approaches by (i) modeling the fMRI time series of a voxel as a von Mises-Fisher distribution, which is widely used for clustering high dimensional data; (ii) modeling the latent cluster labels as a Markov Random Field, which provides spatial regularization on the cluster labels by penalizing neighboring voxels having different cluster labels; and (iii) introducing a prior on the number of labels, which helps in uncovering the number of clusters automatically from the data. Cluster labels and model parameters are estimated by an iterative expectation maximization procedure wherein, given the data and current estimates of model parameters, the latent cluster labels, are computed using α-expansion, a state of the art graph cut, method. In turn, given the current estimates of cluster labels, model parameters are estimated by maximizing the pseudo log-likelihood. The performance of the proposed method is validated using extensive computer simulations. Using novel stability analysis we examine the sensitivity of our methods to parameter initialization and demonstrate that the method is robust to a wide range of initial parameter values. We demonstrate the application of our methods by parcellating spatially contiguous as well as non-contiguous brain regions at both the individual-participant and group levels. Notably, our analyses yield new data on the posterior boundaries of the supplementary motor area and provide new insights into functional organization of the insular cortex. Taken together, our findings suggest that our method is a powerful tool for investigating functional subdivisions in the human brain.
Keywords: Parcellation, Segmentation, Data clustering, Markov Random Fields, Graph Cuts, α-expansion, resting-state fMRI
Introduction
Understanding how the human brain produces cognition depends critically on knowledge of its functional organization; yet demarcating its functional subdivisions remains a significant challenge. Brain regions can be demarcated using different types of anatomical features such as cellular organization, gyral folding patterns, or neurotransmitter profiles (Brodmann, 1994; Zilles and Amunts, 2010). Anatomical parcellations in these atlases are based on post-mortem brains of the elderly, and are less useful for examining individual differences in the functional organization of the human brain. Increasingly, non-invasive imaging techniques such as resting-state fMRI (rs-fMRI) are being used to uncover the functional boundaries between brain regions in normal healthy adults (Barnes et al., 2010; Barnes et al., 2012; Cohen et al., 2008; Deen et al., 2011; Kelly et al., 2012; Kelly et al., 2010; Kim et al., 2010; Nelson et al., 2010; Shen et al., 2010; Zhang et al., 2010), as well as pediatric groups (Barnes et al., 2012), both at the individual participant and group levels.
Several data clustering schemes have been proposed for segmenting brain regions into homogeneous units based on rs-fMRI data. Currently used methods include hierarchical clustering (Achard et al., 2006; Salvador et al., 2005), Gaussian mixture modeling (Golland et al., 2008), graph theory based spectral clustering (Craddock et al., 2011; Shen et al., 2010), computer vision based edge detection (Cohen et al., 2008) and the commonly used K-means based clustering methods (Kim et al., 2010). With the exception of edge detection methods all of these methods require the specification of number of clusters. This is a problem because the choice of number of clusters can significantly influence functional modules uncovered by each method (Delong, 2011). A second major limitation of current methods used to parcellate the brain is that they do not exploit spatial correlations that are inherent in rs-fMRI data. In this study we develop a novel parcellation scheme that identifies the number of clusters automatically from the data while at the same time modeling spatial correlations in rs-fMRI.
Our generative probabilistic model is based on the von Mises-Fisher (VMF) distribution (Banerjee et al., 2005) and Markov Random Fields (MRF) (Li, 2009). VMF is a widely used probability model for clustering large dimensional data and has been previously used in neuroimaging for clustering task-based fMRI data (Lashkari et al., 2010). Lashkari and colleagues developed a clustering scheme for experimental fMRI data wherein the brain regions showing similar responses to experimental stimuli were grouped together (Lashkari et al., 2010). They modeled the responses to experimental stimuli as a VMF distribution and estimated the parameters of the model and underlying cluster labels using an iterative expectation maximization algorithm. Taking advantage of potential strengths of this approach for intrinsic brain connectivity, we use VMF to model rs-fMRI voxel time series and then use an MRF prior (Li, 2009) on the latent cluster labels such that a spatial smoothness is imposed for penalizing neighboring voxels having different cluster labels (Delong, 2011). Finally, a label cost imposed on the number of cluster labels helps in uncovering the optimal number of clusters in the data. The smoothness cost together with the label cost provides the necessary regularization on ill-posed problems such as data clustering or image segmentation. This approach of combining smoothness and label costs was initially proposed in the computer vision literature to segment images into homogeneous regions (Delong, 2011).
In our proposed scheme, the required latent cluster labels for the voxels and model parameters are estimated by an iterative expectation maximization like scheme wherein, (a) given the model parameters, cluster labels are estimated by the state of the art graph cut method based on an α-expansion scheme (Boykov et al., 2001; Delong, 2011) and (b) given the cluster labels for voxels, the model parameters are estimated by maximizing the pseudo log-likelihood (Li, 2009). The MRF prior that is used in this scheme imposes a smoothness constraint on the cluster labels while preserving cluster boundaries. Finding global optimal solutions for the cost functions that arising from such discontinuity preserving constraints is NP-hard (Boykov et al., 2001). In this work, we use the α-expansion scheme (Boykov et al., 2001) which is an efficient state of the art optimization method based on graph cuts to estimate cluster labels from the data. We refer to our method as the VMF-MRF parcellation scheme because the VMF is used to model the rs-fMRI voxel time series and the MRF to impose spatial smoothness on cluster labels and label costs. The performance of VMF-MRF is first evaluated using extensive computer simulations. We demonstrate the robustness of our methods to parameter initialization using stability analysis (Monti et al., 2003). We then use an evidence accumulation approach (Fred and Jain, 2005) for determining the number of clusters and clusters at the group level. We then evaluate the performance on rs-fMRI data by parcellating distal as well as contiguous brain regions. For distal, non-contiguous, brain regions we chose unique spatially distributed regions of interest (ROI) encompassing primary auditory cortex, visual cortex, motor cortex, superior parietal lobule (SPL) and inferior frontal gyrus (Brodmann Area BA 44) based on probabilistic cyto-architectonic maps with known anatomical boundaries. For contiguous brain regions, we first segment the supplementary motor area (SMA) and pre-SMA both at the individual participant and group level, similar to previous studies. We then extend this analysis by segmenting the SMA and pre-SMA along with spatially extended regions encompassing the posterior paracentral lobule. Finally we apply our methods to identify subdivisions of the insula cortex at both the individual-participant and group levels.
Methods
Notation
In the following sections, we represent matrices by upper-case and scalars and vectors by lower-case letters. Random matrices are represented by bold upper-case letters while random vectors and scalars are represented by bold lower-case letters.
Generative model for parcellation
Let be the observed voxel time series, where M is the number of voxels in the ROI. Each voxel time series yi consists of T fMRI observations. We normalize each voxel’s time series yi such that its norm‖yi‖ = 1. Let = be the unknown labels of each voxel where each xi can take a value in the discrete label set ℒ = {1,2,…, L}. Here, we assume that the number of possible labels L is unknown and needs to be determined from the data. Let each T dimensional normalized voxel time series yi given its cluster label xi = l follow T-variate von Mises-Fisher (VMF) distribution (Banerjee et al., 2005) given by
| (1) |
where, c(kl) is the normalizing constant, mean direction μl(‖μl‖ = 1) and concentration kl are the parameters of the distribution. The normalizing constant c(kl) is given by
| (2) |
where, Ir(.) represents the modified Bessel function of order r. The concentration parameter kl controls the concentration of the distribution around the mean μl. The inner product quantifies the correlation between the mean of the distribution and the given observation yi. Therefore, this distribution represents the similarity or correlation between the random variable yi and mean of the distribution μl. We use matlab’s “besseli.m” function to evaluate the Bessel function .
Here, we model the prior distribution of unknown cluster labels X as discrete MRF (Geman, 1984; Li, 2009). Let S = {1,2,…,N} be the set of the indices and N = {Ni, i ∈ S} be the given neighborhood system wherein Ni is the set of sites neighboring the voxel i. A random field X is said to be an MRF on S with respect to a neighborhood system N if and only if (Bishop, 2006; Li, 2009)
| (3) |
| (4) |
where the second property is a Markov property which states that the conditional distribution of i-th voxel is independent of the other voxels given its neighborhood. By the Hammersley-Clifford theorem (Bishop, 2006), an MRF can be represented by a Gibbs distribution
| (5) |
where, Z is the normalizing constant, Us (X)is the energy function which imposes spatial regularization and Ul(X) is the label cost. In this work, we use Potts model (Geman, 1984; Li, 2009) to model neighborhood interactions of the sites. This model penalizes the cost function if neighboring voxels have different cluster labels.
| (6) |
where, the Potts potential function is defined as
| (7) |
where δ(z) = 1, if z = 0; δ(z) = 0, otherwise, βs is a non-negative parameter which controls the amount of spatial regularization. The label cost Ut(X) is given by (Delong, 2011)
| (8) |
where, δp (X) = 1, ∃i : Xi = p; 0 otherwise; βl is a non-negative parameter which controls the amount of label cost. Introducing label costs helps in avoiding over fitting of the data by penalizing complex models (over segmentation) and also in explaining the data with few labels as necessary (Delong, 2011). Introducing both smoothness and label costs provide regularization for ill-posed problems such as image segmentation (Delong, 2011).
The posterior distribution of X given the data Y and model parameters given by
| (9) |
This posterior distribution can be represented by an equivalent Gibbs distribution given by
| (10) |
where, Z′ is the normalizing constant. The posterior potential Ux|Y(Y,X) can be represented in terms of three components: (a) Data cost (UD(Y)), (b) smoothness cost (Us(X)) and (c) label cost Ui(X). The data cost is the negative log-likelihood of the data given by
| (11) |
The smoothness and label costs are as given above.
Estimation of latent labels and parameters
The latent cluster labels and unknown parameters of the above probabilistic model for the parcellation scheme can be estimated by maximizing the posterior probability p(X|Y, Θ) or equivalently minimizing its negative logarithm. Here, we take a two-step iterative approach: (1) Given the current estimate of (Θ(Θ̂(t)), estimate the cluster labels X̂(t+1)(2) Given the current estimate X̂(t+1), estimate the parameters Θ̂(t+1). Steps 1 and 2 are repeated until convergence.
(a) Estimation of latent labels
Given the current estimate of the parameters Θ̂(t), we estimate the cluster labels using the state of the art graph cut method called α-expansion algorithm which is a widely used method in computer vision applications (Boykov et al., 2001). Estimating globally optimal labels is NP-hard problem. The α-expansion algorithm is an efficient local minimum method and can achieve solutions within a known factor of the global minimum if potential functions satisfy certain metric conditions (Boykov et al., 2001; Delong, 2011). Another important advantage of α-expansion algorithm is that it can simultaneously change the labels of arbitrarily large sets of voxels while standard methods such as iterated conditional modes (ICM) and simulated annealing methods (Geman, 1984; Li, 2009) change one voxel label at a time and therefore provide local minimum solutions. Given Θ̂(t), X̂(t+1) is estimated by minimizing the following energy function using α-expansion algorithm:
| (12) |
(b) Estimation of Parameters
Given the current estimate of the labels, the parameters of the VMF distribution are estimated by maximizing the likelihood . Estimating the concentration parameters kl is difficult particularly for high dimensional problems since it involves inverting ratio of two Bessel functions (Banerjee et al., 2005). Here, we use approximate solutions suggested in (Banerjee et al., 2005),
| (13) |
| (14) |
| (15) |
The parameters corresponding to MRF prior βs and βl are difficult to optimize because they are the function of the normalizing factor Z′ which needs to be evaluated over all the possible LN configurations. To overcome this problem, generally the following pseudo likelihood (PL) is maximized with respect to the MRF parameters (Li, 2009).
| (16) |
| (17) |
Where, p(yi|Xi, Ni) is given by equation (1). We define the local MRF p(Xi|Ni) as
| (18) |
Where, δp(XNi) is defined for each neighborhood Ni as δp(XNi) = 1, ∃i in Ni : Xi = p.
The parameters βs, βl are then estimated by minimizing the negative pseudo log-likelihood
| (19) |
We use the Matlab optimization routine “fmincon.m” with the constraint that βs > 0 and βl > 0. Note that in this estimation we plug in the current estimates of VMF parameters μl, kl in equation 17. The estimation of labels and parameters steps is repeated until the fractional change in energy Δ(iter) between two iterations as defined below is less than 0.1%.
| (20) |
where, U(iter) = UD (Y) + Us (X) + Ul(X) evaluated using the labels and parameter values estimated at this iteration.
Stability of VMF-MRF to initialization
To start the proposed VMF-MRF algorithm, initial cluster labels and starting values of the MRF parameters (βs, βt) need to be specified. The cost function that we use for estimating the parameters of MRF is non-convex; therefore the optimization routine results in local optimum solutions and the clusters discovered by the method could be sensitive to (a) initial cluster labels and (b) the starting values of the MRF parameters (βs, βl). To examine the sensitivity of the method to these two factors, we examine the stability of clusters to the initialization of the algorithm using the approach suggested by Monti and colleagues (Monti et al., 2003). Below, we briefly outline the approach, for details please refer to Monti and colleagues (Monti et al., 2003).
Set the initial values for the MRF parameters: .
- For each pair of starting values randomly initialize the cluster labels wherein each pixel is assigned randomly to one of the arbitrarily large number of clusters (20 clusters in this study).
- Run the VMF algorithm until convergence to obtain optimal cluster labels and parameter values.
- Compute the connectivity matrix Mb for each pair of voxels ((m, n),m ≠ n) as
Mb(m,n) = 1, (if m, n) are in the same cluster or 0, otherwise(21)(21) - Repeat the steps (2a) and (2b) B (in this study, we set B = 100) times with different random initializations of cluster labels.
- Compute the consensus matrix ℳ for the pair as
(22) Repeat steps (2) to (3) for very pair of starting values .
The stability of the clusters to the initialization can be assessed by looking at the histogram and the cumulative distribution function (CDF) of the consensus matrices ℳ(Monti et al., 2003). If the clustering is consistent across different random initializations, pair of voxels (m, n) belonging to the same cluster will always cluster together and therefore Mb (m, n) = 1 for all b’s and therefore the (m, n)th entry in the consensus matrix ℳ will be close to 1. However, for (m, n) belonging to different clusters, Mb(m, n) = 0 for all b’s and therefore ℳ(n, n) ≈ 0. Therefore, the histogram of ℳ, for stable clustering, will be bimodal with peaks at 0 and 1 and its CDF will be a step function with steps at 0 and 1 (please refer to Figure 3 in (Monti et al., 2003). Any deviation from the bimodal nature of histogram indicates the instability of the ensuing clusters for the initial values of the parameters. We use this approach to examine the sensitivity of the method to initialization and determine the range of starting parameter values for which the ensuing clusters are stable. We observed that VMF is more sensitive to the starting value of than to . We therefore set the initial values as and . We used this range based on our observations that for , we mostly get singleton clusters and for , the regularization is in adequate and the ensuing clusters are unstable.
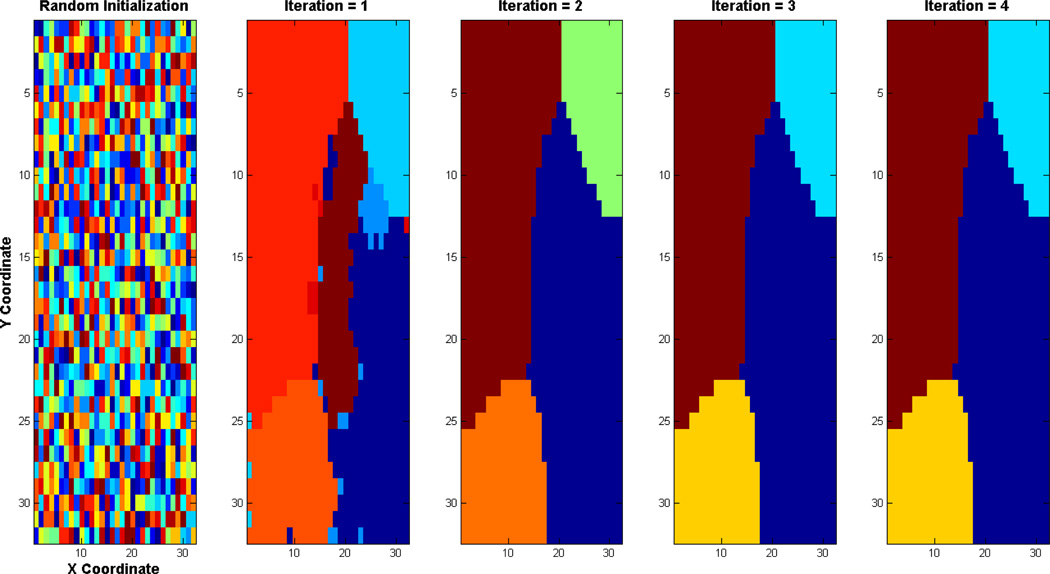
Figure 3. An illustration of VMF-MRF parcellation scheme.
Example based on Case d shown in Figure 1. The method starts with a random initialization wherein cluster label to each pixel is assigned to a number randomly drawn uniformly from 1 to 20 (Left most panel). VMF-MRF starts from this initialization and converges to the final cluster labels shown in the right most last panel. The intermediate iterations are also shown. VMF-MRF recovers the correct number of labels (four) and labeling for this simulation. Note that the color coding of actual and estimated labels is arbitrary.
Parcellation at group level
The boundaries of brain regions obtained by parcellation can vary between participants. To obtain group level segmentation maps we performed group level analyses that capture similarities of clusters across different participants. Our method is based on evidence accumulation approach suggested by Fred and Jain (Fred and Jain, 2005). In this approach, we find an average group level consensus matrix ℳG:
| (23) |
where, M(s) is the consensus matrix for participant s obtained using equation (22) and Ns is the number of participants. To find clusters at group level, we apply hierarchical agglomerative clustering with average-linkage method using ℳG as the similarity matrix. We used the longest cluster lifetime (Fred and Jain, 2005) as a criterion to find the number of clusters at group level. The k-cluster lifetime is defined as the range of thresholds for which the hierarchical clustering results in k number of clusters.
Simulated data using computer based simulations
We first validate the performance of our parcellation methods using simulated datasets for which we know the ground truth of cluster labels. We generated several datasets consisting of arbitrary number of clusters with different shapes and sizes using a two-step procedure. First we synthesize the cluster shapes and sizes using Gibbs sampling procedure wherein we impose spatial regularization using MRF to simulate spatially smooth clusters. Second we generated voxel time series in each cluster such that voxels correlations within a cluster ρin will be different from the voxel correlation across the clusters ρac. We generated datasets with different values of ρin and ρac.
(a) Cluster synthesis
Let there be L = 4 clusters in the data. Let the probability distribution of each voxel in a cluster ‘i’ be normal with mean mi and standard deviation 1. We set , to L equally spaced points between 0 and 8. Let the prior distribution of the voxels be MRF with Potts model as a potential function. We set the parameter of the potential function βs to 2. The conditional distribution of the voxel ‘i’ having a cluster label ‘l’ given the observed data and neighborhood (Ni) cluster labels is given by , where X and Y are the cluster labels and observed data respectively and V(Xi, Xj) is the Potts model. Given this probabilistic model for cluster labels, several realizations from the model are obtained using the following Gibbs sampler (Geman, 1984):
Randomly initialize X to a point in Ls.
-
For i ∈ S do
2.1 Compute pl = P(Xi = l|Y, Ni) for all l ∈ {1,2,‥,L}.
2.2 Set Xi to l with probability pl.
Repeat step 2 for Niter = 50 times.
(b) Time series generation
After the clusters are synthesized, the time series for all voxels are generated using a multivariate normal distribution with mean zero and covariance matrix Σ. The covariance matrix Σ is set such that
| (24) |
Each time sample of voxel time series data (Y(., t), t = 1,2,…, T) is then generated from the multivariate normal distribution N(0, Σ) with Σ as defined above. We test the performance of the parcellation schemes for six different values of ρin and ρac:
Case a: High correlation within the cluster (ρin = 0.7) and low correlation across the clusters (ρac = 0.3).
Case b: High correlation within cluster (ρin = 0.7) and across the clusters (ρac = 0.6).
Case c: Medium correlation within cluster (ρin = 0.5) and low correlation across the clusters (ρac = 0.2).
Case d: Medium correlation within the cluster (ρin = 0.5) and across the clusters (ρac = 0.4).
Case e: Low correlation within the cluster (ρin = 0.3) and very low correlation across the clusters (ρac = 0.1).
Case f: Low correlation within cluster (ρin = 0.3) and across the clusters (ρac = 0.2)
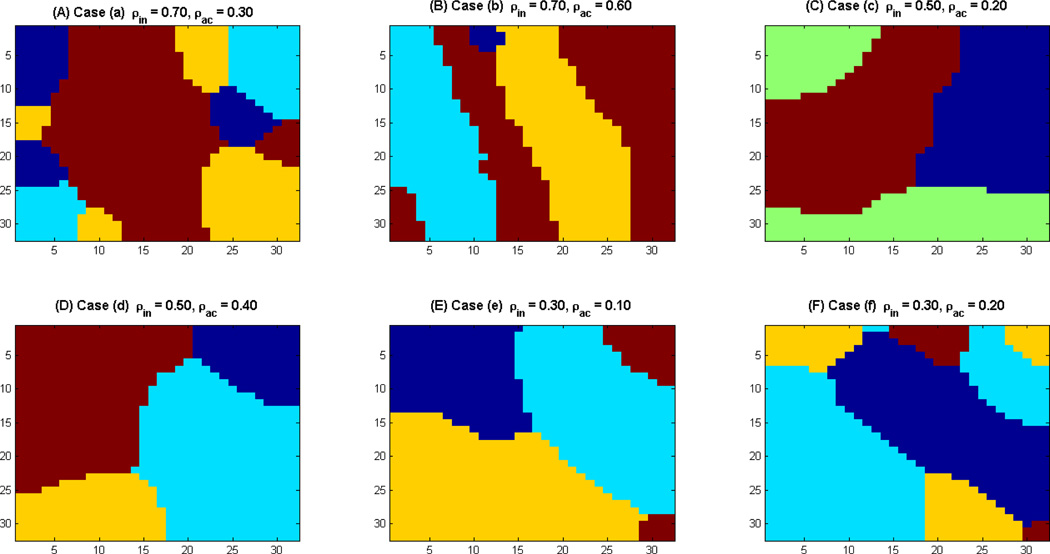
Figure 1 shows the synthetic clusters for the above six cases. Cases a, c and e involve high spatial contrast (ρin > ρac) whereas cases b, d and f involve low spatial contrast (ρin ≈ ρac) and should be more difficult to parcellate. We evaluate the robustness of our method using these test cases.
Figure 1. Synthetic Clusters generated using Gibbs sampling.
(A) Case a: (ρin = 0.7, ρac = 0.3); (B) Case b: (ρin = 0.7, ρac = 0.6); (C) Case c: (ρin = 0.5, ρac = 0.2); (D) Case d: (ρin = 0.5, ρac = 0.4); (E)Case e: (ρin = 0.3, ρac = 0.1); and (F)Case f: (ρin = 0.3, ρac = 0.2).
Resting-state fMRI data
rs-fMRI data were acquired from 21 adult participants. The study protocol was approved by the Stanford University Institutional Review Board. The participants ranged in age from 19 to 22 y (mean age 20.40 y) with an IQ range of 97 to 137 (mean IQ: 112). The participants were recruited locally from Stanford University and neighboring community colleges.
Participants were instructed to keep their eyes closed and try not to move for the duration of the 8-min scan. Functional Images were acquired on a 3T GE Signa scanner (General Electric) using a custom-built head coil. Head movement was minimized during scanning by a comfortable custom-built restraint. A total of 29 axial slices (4.0 mm thickness, 0.5 mm skip) parallel to the AC-PC line and covering the whole brain were imaged with a temporal resolution of 2 s using a T2* weighted gradient echo spiral in-out pulse sequence (Glover and Law, 2001) with the following parameters: TR = 2,000 msec, TE = 30 msec, flip angle = 80 degrees, 1 interleave. The field of view was 20 cm, and the matrix size was 64×64, providing an in-plane spatial resolution of 3.125 mm. To reduce blurring and signal loss arising from field in homogeneities, an automated high-order shimming method based on spiral acquisitions was used before acquiring functional MRI scans. A high resolution T1-weighted spoiled grass gradient recalled (SPGR) inversion recovery 3D MRI sequence was acquired to facilitate anatomical localization of functional data. The following parameters were used: TI = 300 msec, TR = 8.4 msec; TE = 1.8 msec; flip angle = 15 degrees; 22 cm field of view; 132 slices in coronal plane; 256×192 matrix; 2 NEX, acquired resolution = 1.5×0.9×1.1 mm. Structural and functional images were acquired in the same scan session.
Preprocessing of rs-fMRI data
Data were preprocessed using SPM8. The first eight image acquisitions of the task-free functional time series were discarded to allow for stabilization of the MR signal. Each of the remaining 232 volumes underwent the following preprocessing steps: realignment, normalization to the MNI template, and smoothing carried out using a 4-mm full-width half maximum Gaussian kernel to decrease spatial noise. Excessive motion, defined as greater than 3.5 mm of translation or 3.5 degrees of rotation in any plane, was not present in any of the task-free scans. The time series at each voxel were filtered using a band pass filter (0.0083 Hz < f < 0.15 Hz). Linear trend, temporal and global mean were regressed out of each voxel time series.
Multiple spatially segregated cyto-architectonically defined ROIs
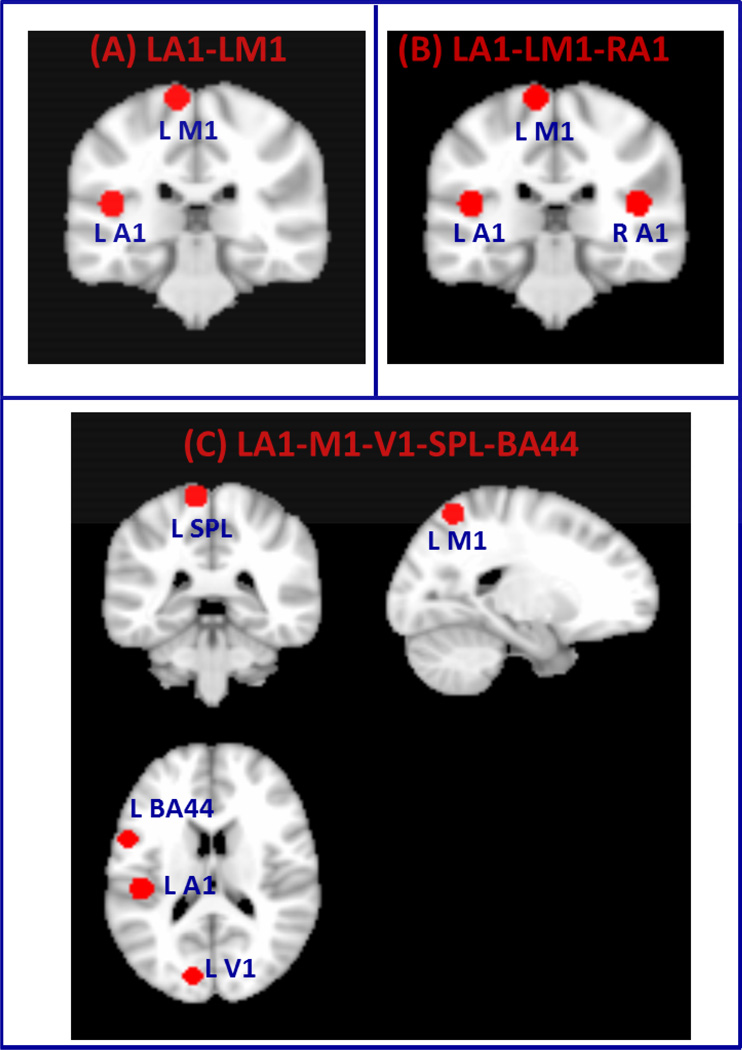
We created three different datasets using rs-fMRI data acquired from five adult participants. In the first dataset, we created two clusters (referred here as “two-cluster data”) from left primary auditory (A1) and motor (M1) cortices (Figure 2A) by drawing 8 mm spheres around the MNI coordinates A1 [−45, −26, 14] and V1 [−10, 35, 72] chosen based on maximum probability maps defined in the Anatomy Toolbox in SPM8 (Eickhoff et al., 2005). In the second dataset (referred here as “three-cluster data”), we create three clusters (Figure 2B) by adding an 8 mm sphere around right A1 [45, −26, 14] (homologous to left A1) in addition to the left A1 and M1 clusters. In the third dataset (referred here as “five-cluster data”), we create three additional clusters by drawing 8mm spheres in the left (i) primary visual cortex (V1) around the MNI coordinate [−12, −85,11], (ii) superior parietal lobule (SPL) around the MNI coordinate [−46 −72, 36] and (iii) inferior frontal gyrus (Brodmann Area BA 44) around the MNI coordinate [−23 −61, 60]. We then create dataset with five clusters by including these three clusters with the two clusters in the first dataset (left auditory and motor). The clusters created for these three datasets are shown in Figure 2C. We then examined the performance of the proposed scheme in segmenting these brain regions using rs-fMRI time series data from these regions. The advantage of this approach is that we know the ground truth of the cluster labels. A similar approach was taken in (Shen et al., 2010) where a synthetic dataset consisting of two clusters was created from a real fMRI data by randomly selecting voxel time series from intra-parietal sulcus and visual cortex (Shen et al., 2010). They applied graph based clustering scheme to parcellate this data into two clusters. Here, we created datasets by selecting voxel time series from primary Left auditory, motor and visual, the left superior parietal lobule (SPL) and left inferior frontal gyrus (Brodmann Area BA 44) regions.
Figure 2. Clusters constructed from fMRI data.
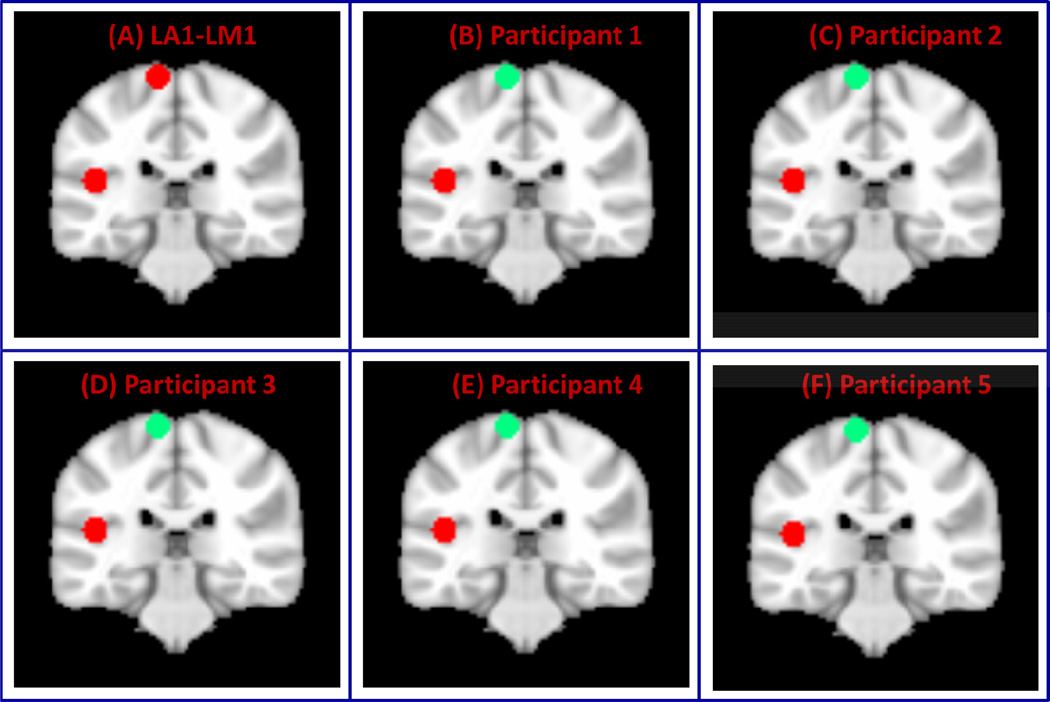
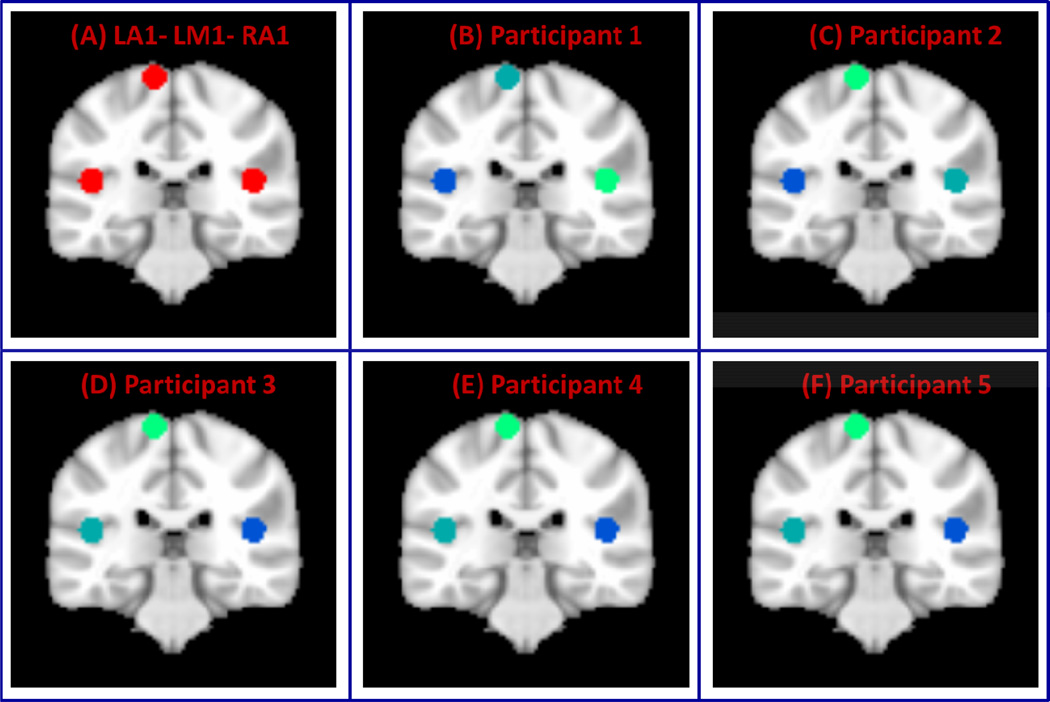
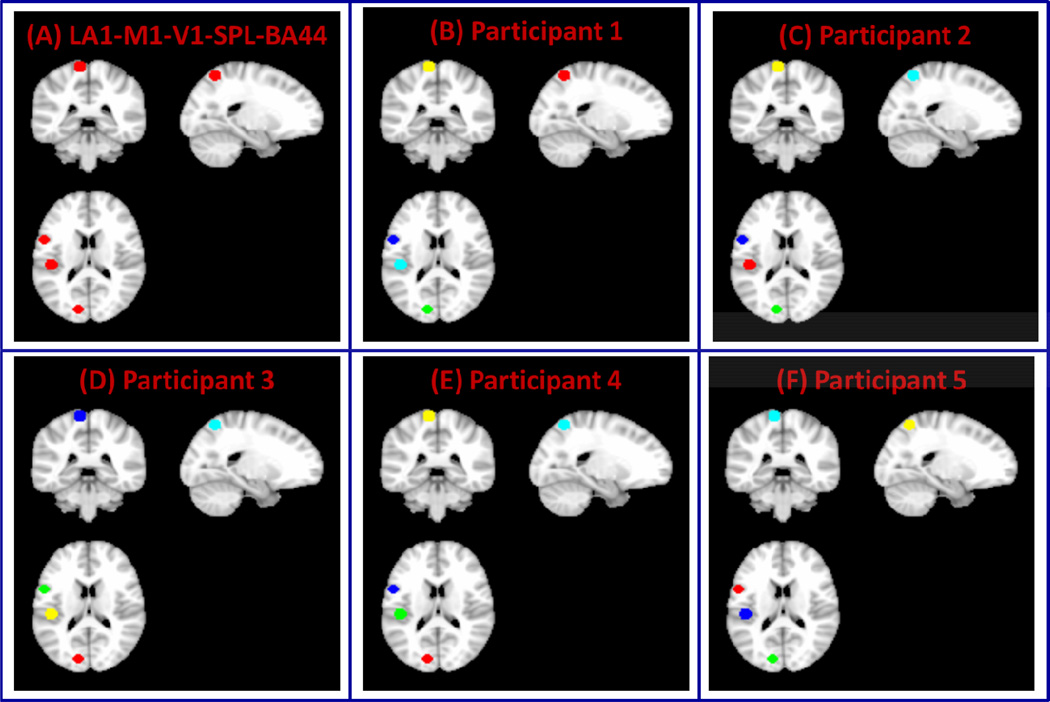
(A) Two clusters formed from left primary auditory (L-A1) and motor cortices (LM1) (B) Three clusters created from L-A1, L-M1 and right primary auditory cortex (RA1) and (C) Five clusters created from L-A1, L-M1, left primary visual cortex (L-V1), left superior parietal lobule (SPL) and left inferior frontal gyrus (Brodmann Area BA 44).
SMA, pre-SMA and posterior paracentral lobule ROIs
To investigate the performance of VMF-MRF, we examined its ability to functionally segment a brain area into spatially contiguous sub regions. We chose the medial frontal cortex because it consists of two spatially contiguous cyto-architectonically distinct regions – the Supplementary Motor Area (SMA) and the pre-SMA (Zilles et al., 1996). Importantly, the SMA and pre-SMA are well demarcated anatomically by a vertical line, passing through the anterior commissure, perpendicular to the anterior-posterior commissure axis (Zilles et al., 1996). SMA and pre-SMA regions were defined using a procedure similar to that used in previous studies (Aron et al., 2007; Johansen-Berg et al., 2004). The automated anatomical labeling (AAL) template (Tzourio-Mazoyer et al., 2002) was used to demarcate the SMA (y < 0) and pre-SMA (y > 0) subdivisions.
To further investigate the performance of VMF-MRF, we examined its ability to functionally segment a large contiguous brain area that includes several spatially contiguous sub regions. We extended our medial frontal cortex ROI (see above) posteriorly to include other areas of the paracentral lobule. Extending the medial frontal cortex ROI posteriorly allowed us not only to examine the performance of our method on a large functionally heterogeneous region but also to investigate whether our method could demarcate the pre-SMA and SMA from the posterior paracentral lobule. To our knowledge, this is the first attempt to delineate the posterior boundary of the SMA.
Insula ROI
Finally, we examined the performance of VMF-MRF in segmenting the insular cortex, a large multimodal integration brain area situated deep within the lateral sulcus between the frontal and temporal lobes. We chose the insula because it is a highly functionally and cytoarchitectonically diverse area of cortex with multiple subdivisions. For this study, we used the insula region as defined by the automated anatomical labeling (AAL) atlas (Tzourio-Mazoyer et al., 2002).
Results
Validation of VMF-MRF using computer simulations
We first illustrate how VMF-MRF works using the synthetic dataset case (ρin = 0.5, ρac = 0.4). VMF-MRF starts with random initialization wherein each pixel is assigned randomly to one of the arbitrarily large number of clusters (20 clusters in this study), as shown in the left most panel of Figure 3. In this case, the method converges in four iterations; the estimated cluster labels in each iteration are shown in Figure 3. The clusters estimated by VMF-MRF (last panel of Figure 3) retrieves the actual clusters which are shown in Figure 1 (Case d).
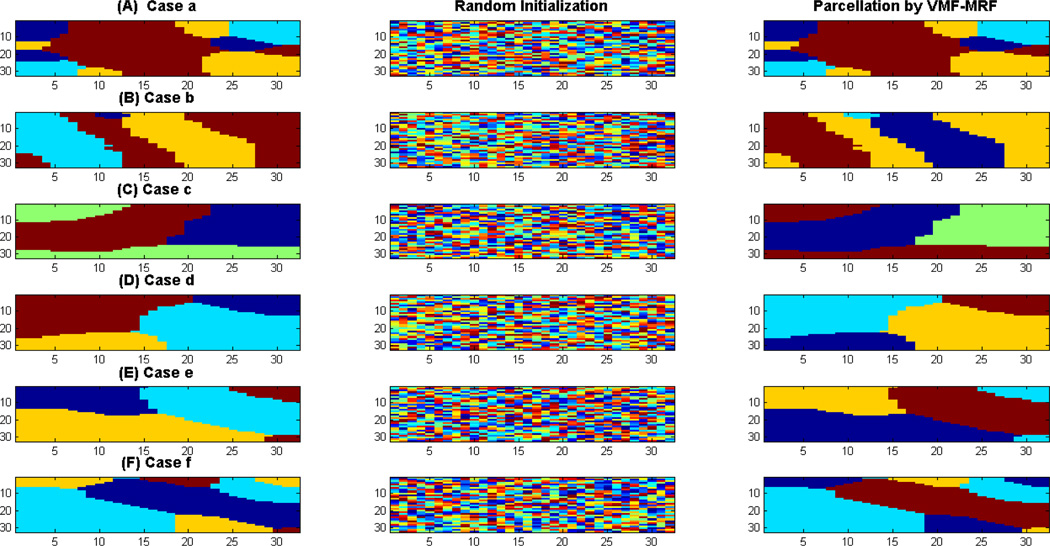
Figure 4 shows the performance of VMF-MRF for six different choices of ρin’s and ρac’s. Cases a, c and e involve high spatial contrast (ρin > ρac) and are relatively easy to cluster whereas cases b, d and f involve low spatial contrast (ρin ≈ ρac) and are more difficult to parcellate. The first column in Figure 4 shows the actual cluster labels for each case, the second column shows the random initialization to the VMF-MRF algorithm and the third shows the cluster labels estimated by VMF-MRF. In each of these cases, VMF-MRF accurately estimates the correct number of clusters and cluster labels. Based on stability analysis (see below), we initialized each pixel or voxel randomly to one of the 20 clusters. Our method is not sensitive to this initial number of cluster labels.
Figure 4. Performance of VMF-MRF on computer simulated synthetic datasets.
Performance shown for six different cases a–f as shown in Figure 1. Left panel in each case shows the correct labeling, middle panel shows the random initialization to VMF-MRF scheme and the right panel shows the clusters recovered by the scheme. Note that the color coding of the actual and estimated clusters is arbitrary. In each case, VMF-MRF is able to recover the correct number of clusters and labels.
Parcellation of multiple spatially segregated cyto-architectonically defined ROIs
To examine the performance of VMF-MRF on real rs-fMRI data, we first used two ROIs in left A1 and M1 (ROIs shown in Figure 2A). As in the case of computer simulated datasets, VMF-MRF was started with random initialization of 20 cluster labels. VMF-MRF accurately estimated two distinct clusters in each of the five participants (Figure 5). To further examine the ability of VMF-MRF to parcellate distal clusters with more than two clusters, we examined a dataset with three clusters: left A1, left M1 and right M1 (ROIs shown in Figure 2B), and a dataset with five clusters: left A1, left M1, left V1, left SPL and left BA 44 (ROIs shown in Figure 2C). As predicted, VMF-MRF accurately estimated three distinct clusters in each of the five participants (Figure 6) for the three-cluster dataset and five clusters in each of the five participants (Figure 7) for the five- cluster dataset.
Figure 5. Performance of VMF-MRF on two-cluster fMRI datasets.
(A) Two clusters created from left auditory and motor cortices (LA1-LM1). (B–F)Clusters obtained after applying VMR-MRF on five participants. For each participant, VMF-MRF started with random initialization wherein each voxel is assigned randomly to one of the 20 clusters. VMF-MRF is able to recover the correct number of clusters (which is two) for each of the five participants.
Figure 6. Performance of VMF-MRF on three-cluster fMRI datasets.
(A) Three clusters created from left auditory, motor and right auditory cortices (LA1-LM1-RA1). (B–F) Clusters obtained after applying VMR-MRF on five participants. For each participant, VMF-MRF started with random initialization wherein each voxel is assigned randomly to one of the 20 clusters. VMF-MRF is able to recover the correct number of clusters (which is three) for each of the five participants. Note that the color coding of actual and estimated labels is arbitrary.
Figure 7. Performance of VMF-MRF on five-cluster fMRI datasets.
(A) Five clusters created from left auditory, motor, and visual cortices, the superior parietal lobule (SPL) and inferior frontal gyrus (Brodmann Area BA 44) (LA1-M1-V1-SPL-BA44). (B–F) Clusters obtained after applying VMF-MRF on five participants. For each participant, VMF-MRF started with random initialization wherein each voxel is assigned randomly to one of the 20 clusters. VMF-MRF is able to recover the correct number of clusters (which is five) for each of the five participants. Note that the color coding of actual and estimated labels is arbitrary.
Parcellation of SMA, pre-SMA and posterior paracentral lobule
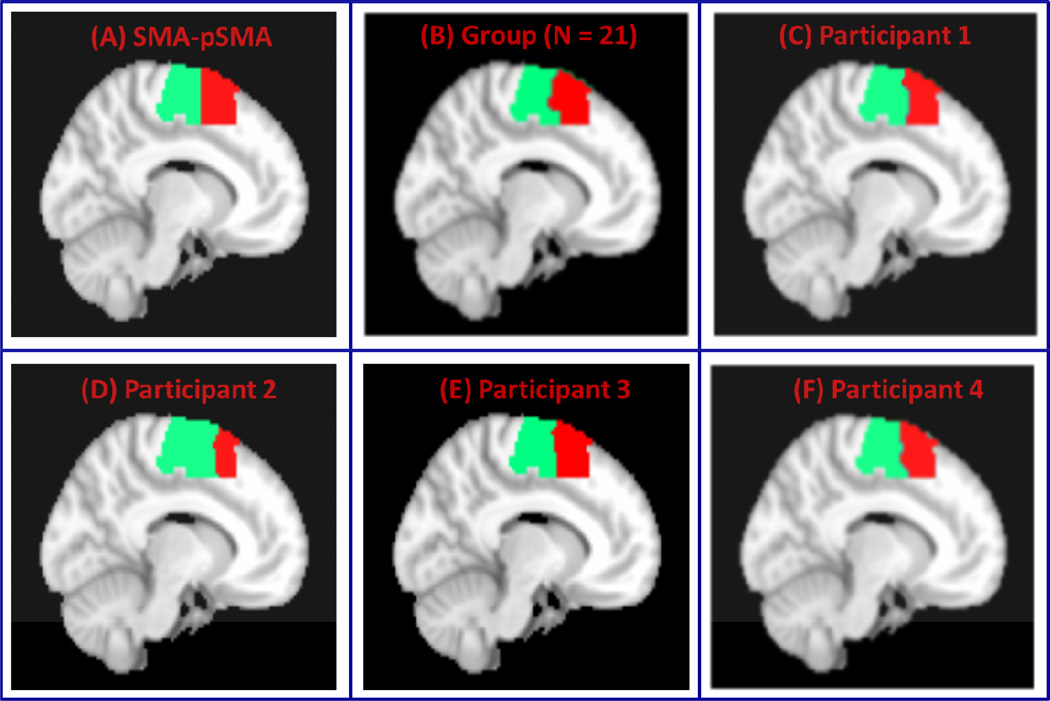
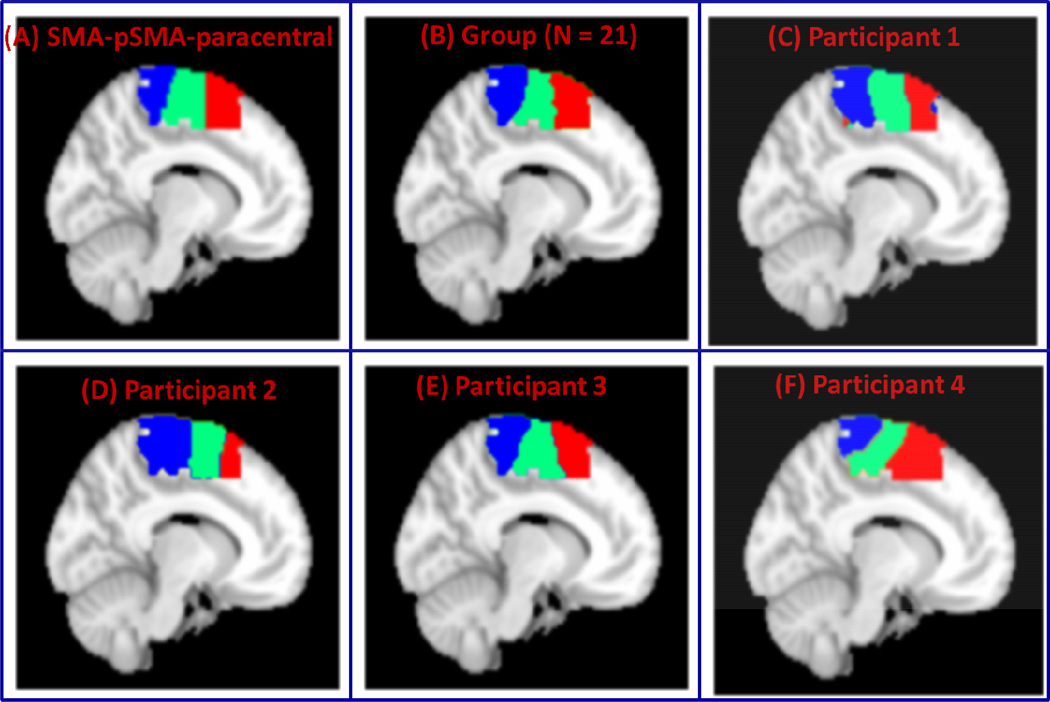
We then applied VMF-MRF to segment a combined medial frontal cortex cluster encompassing the SMA and pre-SMA. Figure 8A shows the SMA and pre-SMA subdivisions we attempted to segment. Figure 8B shows group parcellation using data from 21 participants. Parcellations for four representative participants are shown in Figure 8C–8F. The two clusters are consistent with known borders of the SMA and pre-SMA, which are posterior and anterior to the anterior commissure respectively (Picard and Strick, 2001; Zilles et al., 1996).
Figure 8. VMF-MRF segmentation of SMA-pre-SMA.
(A) SMA-pre-SMA regions with boundary at y=0. (B) Clusters obtained after group (N = 21) analysis. (C–F) Clusters obtained after applying VMR-MRF on four representative participants. For each participant, VMF-MRF started with random initialization wherein each voxel is assigned randomly to one of the 20 clusters. After convergence, VMF-MRF discovered two clusters for each participant as shown in panels C to F.
To examine whether VMF-MRF delineates SMA and pre-SMA regions when adjacent brain regions are included in the parcellation, we applied our method to a ROI that included the SMA, pre-SMA and posterior paracentral lobule. Figure 9A shows boundaries of the three subdivisions we sought to segment. Figure 9B shows the group-level parcellation using data from 21 participants and Figures 9C–9F show parcellation in four representative participants. The three clusters obtained by our method are consistent with the known boundaries of these regions.
Figure 9. VMF-MRF segmentation of SMA-pre-SMA and posterior paracentral lobule.
(A) SMA, pre-SMA and posterior paracentral lobule. (B) Clusters obtained after group (N = 21) analysis. (B–F) Clusters obtained after applying VMR-MRF on four representative participants. For each participant, VMF-MRF started with random initialization wherein each voxel is assigned randomly to one of the 20 clusters. After convergence, VMF-MRF discovered three clusters for each participant as shown in panels C to F.
Parcellation of Insula
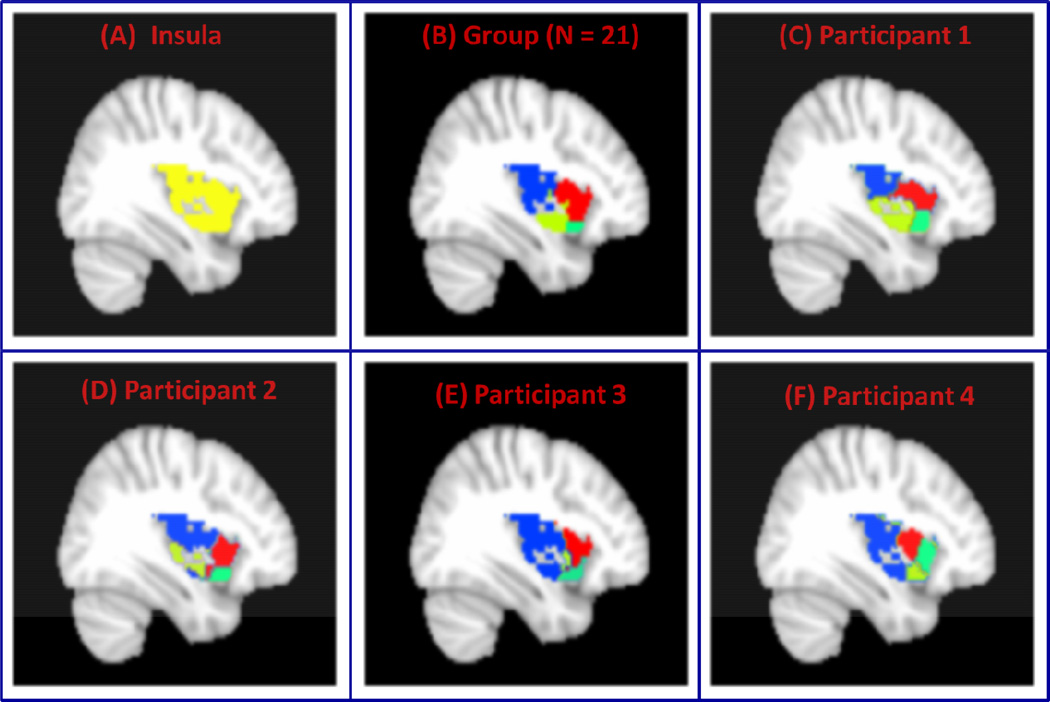
Finally, we applied VMF-MRF to segment Insula shown in Figure 10A. Our VMF-MRF methods were able to segment this region into four subdivisions both at group level using data from 21 participants as shown in Figure 10B and at individual level whose parcellations for four representative participants are shown in Figure 10C– 10F. These four subdivisions include the posterior insula, dorsal anterior insula, ventral mid insula, ventral anterior insula. The partitioning obtained using our method is consistent with and clarifies ambiguities in insula organization reported by previous studies.
Figure 10. VMF-MRF segmentation of Insula.
(A) Insula ROI demarcated using the AAL atlas. (B) Four clusters obtained after group (N = 21) analysis. (B–F) Four clusters obtained after applying VMR-MRF on four representative participants. For each participant, VMF-MRF started with random initialization wherein each voxel is assigned randomly to one of the 20 clusters. After convergence, VMF-MRF discovered four clusters for each participant as shown in panels C to F.
Stability of VMF-MRF to initialization
We examined the sensitivity of the algorithm to the initialization of parameters and cluster labels as defined in the Methods section. We examined the stability of our method on each of the datasets presented above and set the initial parameter values based on this analysis. Here we report the stability analyses for three-cluster, five-cluster and SMA-pSMA datasets for participant 1. The results are similar for other datasets and participants (data not shown).
Supplementary Figure S1 shows the histograms of consensus matrices ℳ for starting values of and for the 3-cluster dataset of participant 1. The histograms for initial parameter values are bimodal and the corresponding CDFs are very close to step functions indicating that the ensuing clusters are stable. Table 1 shows the number of times the algorithm converges to a particular number of clusters. This table suggests that the algorithm consistently converges to three clusters most of the time for the initial parameter values . For the initial values , the method is inconsistent and converges to clusters greater than 3. This inconsistency is reflected in their respective histograms and CDFs. The results are similar for the other four participants (data is not shown).
Table 1. Stability analysis for three-cluster data of participant 1.
The table shows the number times (out of 100 times) a particular number of clusters obtained for each initial value of . The clusters are more stable for (see Supplementary Figure S1) and for , 99 out of 100 times the method resulted in 3 clusters.
| No of Clusters | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 2 | 0 | 1 | 12 | 42 | 30 | 11 | 4 | |
| 4 | 2 | 36 | 38 | 24 | 0 | 0 | 0 | |
| 6 | 0 | 86 | 11 | 3 | 0 | 0 | 0 | |
| 8 | 0 | 96 | 4 | 0 | 0 | 0 | 0 | |
| 10 | 1 | 99 | 0 | 0 | 0 | 0 | 0 | |
Supplementary Figure S2 shows the stability analyses for the five-cluster data of participant 1. The histograms are bimodal and CDFs are closer to step function for the parameter range . In this range, the method consistently converges to the correct number of clusters, which is five, for most of the time (Table 2). The method consistently discovers, for this and other participants (data not shown), the correct number of stable clusters for a range of initial parameter values and different random initialization of cluster labels.
Table 2. Stability analysis for five-cluster data of participant 1.
The table shows the number times (out of 100 times) a particular number of clusters obtained for each initial value of . The clusters are more stable for (see Supplementary Figure S2) and for 89 out of 100 times the method resulted in 5 clusters.
| No of Clusters | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 2 | 2 | 5 | 25 | 61 | 7 | 0 | 0 | |
| 4 | 0 | 4 | 6 | 88 | 2 | 0 | 0 | |
| 6 | 0 | 0 | 18 | 82 | 0 | 0 | 0 | |
| 8 | 0 | 2 | 16 | 82 | 0 | 0 | 0 | |
| 10 | 0 | 2 | 9 | 89 | 0 | 0 | 0 | |
Supplementary Figure S3 shows the stability analyses for SMA-pSMA parcellation for participant 1. The histograms for the initial parameter values are bimodal and their respective CDFs are close to step functions indicating stable clusters in this range. Table 3 shows that in this parameter range, the method discovers the correct number of clusters, which is two. For the initial parameter values , the clustering is stable as reflected by their respective histograms and CDFs.
Table 3. Stability analysis for SMA-pSMA data of participant 1.
The table shows the number times (out of 100 times) a particular number of clusters obtained for each initial value of . The clusters are more stable for (see Supplementary Figure S3) and for , 100 out 100 times the method resulted in 2 clusters.
| No of Clusters | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 2 | 3 | 14 | 25 | 34 | 18 | 5 | 1 | |
| 4 | 52 | 41 | 7 | 0 | 0 | 0 | 0 | |
| 6 | 93 | 7 | 0 | 0 | 0 | 0 | 0 | |
| 8 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 10 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |
Discussion
We have developed a novel probabilistic algorithm to parcellate both distal and contiguous voxels based on rs-fMRI time series. Our algorithm uses a spatio-temporal probabilistic scheme wherein voxel time series are modeled as a VMF distribution, which is widely used to represent high dimensional data. The latent cluster labels are modeled as an MRF to impose spatial smoothness on the labels. A label cost is imposed to penalize complex models thereby allowing us to segment brain regions by automatically estimating the number of clusters in the data while at the same time exploiting spatial correlations inherent in rs-fMRI data. We demonstrate the usefulness and performance of our method using both simulated and real rs-fMRI data. We also performed extensive stability analysis to examine the sensitivity of our methods to initialization parameters.
Regularization
Problems in image segmentation are generally ill-posed and require regularization to provide meaningful solutions (Delong, 2011). MRFs are used extensively in image segmentation problems to provide spatial regularization on cluster labels by penalizing cluster configurations with neighborhood voxels having different cluster labels (Li, 2009). Imposing label costs in addition to smoothness cost overcomes the problem of over segmentation (Delong, 2011). Imposing both these costs provides regularization for ill-posed computer vision problems involving segmentation. The energy function (Equation 12) that we used for segmentation has a similar interpretation as the information theoretic criteria used for selecting models (Delong, 2011). Information theoretic model selection procedures such as Akaike (AIC) and Bayesian (BIC) information criteria helps avoid over fitting by penalizing complex models. Smoothness and label costs play a similar role in the proposed parcellation method by penalizing complex models and over-segmentation.
Relationship of VMF-MRF with EM and K-means algorithms
The traditional K-means and EM algorithms for mixture models that are generally used in data clustering problems can be easily seen to be special cases of our proposed methods (Delong, 2011). In the case of uniform priors (βs = 0, βl = 0) where there is no regularization, the proposed method is equivalent to EM algorithm for estimating the parameters of mixture of VMF distributions with hard assignment of cluster labels (Banerjee et al., 2005). In the limiting case of infinite concentration parameters (kl → ∞), the proposed method is the same as the K-means algorithm with similarity metric which is a “cosine” function (Banerjee et al., 2005). Our regularization approach has several advantages over the K-means and EM algorithms which are very sensitive to initialization even if the right number of models (or clusters) K is known and to over fitting if the initial number of clusters K is large (Delong, 2011). Regularization, on the other hand, helps in avoiding major problems associated with methods having to fix K. As shown by Delong and colleagues (Delong, 2011), using label costs as regularization and starting with a large number of models converges to a small number of good models; furthermore, as long as the initial set of randomly sampled models is large, regularization is more robust to local minima and more stable to initialization when compared to K-means algorithm with no regularization (see Figures 10 and 11 in Delong, 2011). Our results on both simulated and rs-fMRI data are consistent with these observations and show that our method can recover the correct number of clusters starting from a large number of random initial labels.
Stability of VMF-MRF to initialization
As noted above on theoretical grounds, our proposed methods have several advantages over methods such as K-means which do not use any regularization. However, an issue that needs to be addressed here concerns sensitivity of VMF-MRF to initialization of regularization parameters and initial cluster labels. We conducted additional analyses to examine the sensitivity of our methods to initialization of regularization parameters (βs and βl) and initial cluster labels. Optimizing model parameters (βs, βl) in discrete MRFs is, in general, a difficult problem since one needs to evaluate the partition function at all the possible configurations of the cluster labels. Therefore, these parameters are, in general, manually tuned based on the problem at hand. However, to avoid manual tuning of parameters for each problem, we used pseudo likelihood (Equations 16–18) as the cost function for choosing these parameters. The cost function (negative pseudo log-likelihood) is non-convex and therefore suboptimal solutions can be obtained because of local minima. Therefore, the parcellation obtained can be sensitive to the initial starting values of regularization parameters and cluster labels. To examine the sensitivity of our VMF-MRF methods to initialization, we carried out an extensive stability analysis using the approach suggested by Monti and colleagues (Monti et al., 2003). Stability analysis on various datasets used in our study suggests that our method is robust to a range of initial parameter values and random initialization of cluster labels (see Tables 1–3 and Supplementary Figures 1–3). In the present study, we therefore selected the initial parameter values for which the clusters are most stable. A different approach is to combine all the stable clustering results (100 clustering results for each initial parameter values) to obtain the final clusters using hierarchical clustering with the consensus matrix as a similarity measure to obtain final clusters (Fred and Jain, 2005; Monti et al., 2003). We investigated this approach and found similar results to those reported here.
Parcellation at the group level
The boundaries of functionally segmented brain regions vary between participants and group level maps are necessary for demonstrating and clarifying consistency of segmentation maps across individuals. To obtain group level segmentation maps we use the evidence accumulation method suggested by (Fred and Jain, 2005) for capturing similarities of clusters across different participants. Our approach is based on hierarchical clustering using the average consensus matrix computed from each individual participant’s consensus matrix. A cluster lifetime model is used to determine the number of clusters at the group level (Fred and Jain, 2005). Further research is needed to integrate our approach with bootstrapping procedures (Bellec et al., 2010) to uncover stable clusters at both the individual and group levels. More research is also needed to quantify similarities and variations of clustering within and between groups.
Performance of VMF-MRF on computer simulated datasets
We evaluated the performance of VMF-MRF on several computer simulated datasets consisting of clusters with arbitrary shapes and sizes synthesized using Gibbs sampling (Figure 1). It is important to note here that the probabilistic model used for synthesizing datasets is different from the probabilistic model used for estimating the cluster labels in order to avoid the bias in evaluating the performance of the method. For synthesizing the clusters, we used a Gibbs sampling scheme wherein the distribution of each pixel in a cluster is Gaussian with cluster specific mean and standard deviation of 1 and the spatial regularization was imposed using a MRF. After the clusters are synthesized, the time series of each pixel is generated using a multivariate Gaussian distribution with mean zero and covariance defined such that inter cluster correlations are different from intra cluster correlations. On the other hand in VMF-MRF model, pixel time series is modeled as a VMF distribution, spatial regularization is imposed using MRF and a label cost is imposed to identify the optimal number of clusters from the data. The model parameters and cluster labels are then estimated using an iterative EM like algorithm. Our simulations show that VMF-MRF performs well on a wide range of datasets having different correlations for pixel time series within and across the clusters. For example, cases a, c and e shown in Figure 1 are relatively easy to cluster because the correlations of pixel time series within the cluster are larger compared to the pixel correlations across the clusters whereas cases b, d and f are difficult to cluster because the differences in correlations within and across the clusters is small. In each of these six cases, VMF-MRF accurately estimates the correct number of clusters and class labels (Figure 4) starting from random initializations. Our concurrent use of smoothness and label costs resulted in accurate identification of the true number of cluster labels while at the same time maintaining accurate cluster boundaries.
Parcellation of multiple spatially segregated cyto-architectonically defined ROIs
Next we evaluated the performance of VMF-MRF on rs-fMRI data constructed from multiple distal brain areas. We created clusters that are geometrically distal and non-contiguous, allowing us to test our methods on arbitrary number of and functionally distinct brain regions. First we chose two ROIs from left primary auditory and motor cortices. When VMF-MRF was applied to this data, it accurately recovered the correct number of clusters and voxel labels (Figure 5) for all participants we examined. VMF-MRF started with random initialization assuming a large number of clusters (20 in this case) and converged to the correct number of clusters and voxel labels. To extend this analysis further and examine how the method scales to datasets having more than two clusters, we performed two additional analyses. First, we added a third ROI in right primary motor cortex (Figure 2B) and second, we created a dataset with five ROIs consisting of left auditory, visual and motor cortices as well as association cortical regions, SPL and BA 44 (Figure 2C). VMF-MRF accurately uncovered three and five clusters respectively in these data sets (Figures 6 and 7) for all participants.
These datasets are more realistic since they are constructed from rs-fMRI compared to computer simulated datasets, the ground truth about cluster labels is known because the regions are functionally distinct and their boundaries are defined by cyto-architectonic maps. Furthermore, the time-series in these datasets need not follow a Gaussian distribution as in the case of simulated data and also incorporate autocorrelations whereas time samples in simulated datasets are independently distributed. The analyses on these rs-fMRI datasets together with stability analyses suggest that VMF-MRF accurately recovers the correct number of clusters and boundaries in real fMRI data and further validates the robustness of our methods.
Parcellation of SMA, pre-SMA and posterior paracentral lobule
The SMA and pre-SMA together have been widely used to construct test datasets and validate different types of segmentation procedures (Behrens et al., 2006; Johansen-Berg et al., 2004;Kim et al., 2010; Nanetti et al., 2009). We therefore applied VMF-MRF to parcellate spatially contiguous SMA and pre-SMA regions based on rs-fMRI data. As expected, VMF-MRF discovered two clusters starting from an arbitrarily large number of clusters in each of the participants we examined (Figure 8C–8F). Group level clustering on 21 participants also resulted in two clusters (Figure 8B). Although the precise boundary between the SMA and pre-SMA is not known, consistent with previous studies, we found that their boundary was found to be close to y =0. This region has previously been segmented using rs-fMRI using the K-means algorithm (Kim et al., 2010) which requires the number of clusters to be specified a priori. In contrast, VMF-MRF estimates the actual number of clusters directly from the data. Another important difference between our method and that of Kim and colleagues (Kim et al., 2010) is in the features used in parcellating this region. Kim and colleagues used differences in global connectivity of each voxel in SMA-pre-SMA regions with respect to the entire brain as features for parcellations, whereas in our work we used only local time series from the SMA and pre-SMA regions. Our methods also uncovered the SMA and pre-SMA when more posterior regions of the paracentral lobule were added to the ROI we sought to segment. VMF-MRF identified three distinct subdivisions at both the individual participant and group levels and provides new data on the posterior boundaries of the SMA (Figure 9B–9F). To our knowledge, this is the first attempt to delineate the posterior boundaries of the SMA as previous studies have not included extended regions of the posterior paracentral lobule.
Parcellation of Insular cortex
Finally, we applied VMF-MRF to segment the right insula (Figure 10A), a brain region important for cognitive and affective control (Chang et al., 2012; Lindquist et al., 2012; Menon and Uddin, 2010; Supekar and Menon, 2012; Uddin et al., 2011). VMF-MRF identified four clusters both at group (Figure 10B) and individual participant levels (Figure 10C–10F). These four subdivisions consist of the posterior insula, dorsal anterior insula, ventral mid insula and ventral anterior insula. The partitioning obtained using our method is consistent with and clarifies ambiguities in insula organization reported by previous studies. Using a K-means based clustering approach, Deen et.al reported three distinct subdivisions of the insula which closely matched our partitions (Deen et al., 2011). Recently, Kelly and colleagues used a similar K-means/spectral clustering method and reported a multi-resolution organization of the insula comprising subdivisions that ranged between 2 and 15 (Kelly et al., 2012). Their results with four clusters are highly consistent with our VMF-MRF partitioning solution which automatically detected the most appropriate number of clusters. In a meta-analytic clustering study, Cauda and colleagues reported two subdivisions of the insula along the anterior-posterior axis (Cauda et al., 2011). Using repeated K-means and diffusion tensor imaging data, Nanetti and and colleagues also reported two subdivisions (Nanetti et al., 2009). Chang and colleagues report three insular subdivisions using K-means clustering of rs-fMRI (Chang et al, 2012). The inconsistencies across these previous studies can be attributed to methodological differences in how the number of clusters is determined. It is important to note here that previous neuroimaging studies examining organization of the human insular cortex have used clustering methods that require manual setting of the number of clusters. This may explain the wide range of insula subdivisions reported in the literature. Critically, unlike previous studies our procedures allow for automated detection of clusters using novel stability selection and group similarity analysis procedures. Our data-driven approach of automatically selecting the number of clusters provides a significant advantage over previously reported methods, particularly when parcellating brain regions such as the insula where the precise number of subdivisions are not yet well understood.
Conclusions
We have developed a novel method for segmenting brain regions into functionally homogeneous subdivisions based on rs-fMRI data. Our method uses a probabilistic model wherein voxel time series is modeled as a von Mises Fisher distribution, a Markov Random Field Prior on latent cluster labels is used to impose spatial regularization on the cluster labels and a prior on the label costs is imposed to automatically select the number of cluster labels from the data. Our methods are a significant advance over existing methods because they do not require a priori specification of the number of clusters. We have validated the proposed method on several simulated datasets and resting-state fMRI data. Furthermore, we develop a novel stability approach to examine the sensitivity of the method to parameter initialization. Thus, a particular advantage of our approach is that functional segmentation can be performed at both the individual participant and group levels, facilitating more precise investigations of the heterogeneous connectivity and functions of different brain areas in normal as well as developmental and clinical populations (Uddin et al., 2010).
Supplementary Material
Acknowledgements
This research was supported by grants from the National Institutes of Health (HD047520, HD059205, HD057610, and NS071221). We thank Dr. Lucina Uddin for useful feedback.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, smallworld human brain functional network with highly connected association cortical hubs. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AR, Behrens TE, Smith S, Frank MJ, Poldrack RA. Triangulating a cognitive control network using diffusion-weighted magnetic resonance imaging (MRI) and functional MRI. J Neurosci. 2007;27:3743–3752. doi: 10.1523/JNEUROSCI.0519-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Dhillon IS, Ghosh J, Sra S. Clustering on the unit hypersphere using von Mises-Fisher distributions. Journal of Machine Learning Research. 2005;6:1345–1382. [Google Scholar]
- Barnes KA, Cohen AL, Power JD, Nelson SM, Dosenbach YB, Miezin FM, Petersen SE, Schlaggar BL. Identifying Basal Ganglia divisions in individuals using resting-state functional connectivity MRI. Front Syst Neurosci. 2010;4:18. doi: 10.3389/fnsys.2010.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes KA, Nelson SM, Cohen AL, Power JD, Coalson RS, Miezin FM, Vogel AC, Dubis JW, Church JA, Petersen SE, Schlaggar BL. Parcellation in left lateral parietal cortex is similar in adults and children. Cereb Cortex. 2012;22:1148–1158. doi: 10.1093/cercor/bhr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Jenkinson M, Robson MD, Smith SM, Johansen-Berg H. A consistent relationship between local white matter architecture and functional specialisation in medial frontal cortex. Neuroimage. 2006;30:220–227. doi: 10.1016/j.neuroimage.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Bellec P, Rosa-Neto P, Lyttelton OC, Benali H, Evans AC. Multi-level bootstrap analysis of stable clusters in resting-state fMRI. Neuroimage. 2010;51:1126–1139. doi: 10.1016/j.neuroimage.2010.02.082. [DOI] [PubMed] [Google Scholar]
- Bishop C. Pattern Recognition and Machine Learning. Springer; 2006. [Google Scholar]
- Boykov Y, Veksler O, Zabih R. Fast approximate energy minimization via graph cuts. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2001;23:1222–1239. [Google Scholar]
- Brodmann K. Vergleichende Lokalisationslehre der Groszhirnrinde in ihren Prinzipien dargestellt auf Grund des ZellenbauesBrodmann's Localization in the Cerebral Cortex. Nature Publishing Group. 1994 [Google Scholar]
- Cauda F, D'Agata F, Sacco K, Duca S, Geminiani G, Vercelli A. Functional connectivity of the insula in the resting brain. Neuroimage. 2011;55:8–23. doi: 10.1016/j.neuroimage.2010.11.049. [DOI] [PubMed] [Google Scholar]
- Chang LJ, Yarkoni T, Khaw MW, Sanfey AG. Decoding the Role of the Insula in Human Cognition: Functional Parcellation and Large-Scale Reverse Inference. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NU, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2011 doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen B, Pitskel NB, Pelphrey KA. Three systems of insular functional connectivity identified with cluster analysis. Cereb Cortex. 2011;21:1498–1506. doi: 10.1093/cercor/bhq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong AO, Isac A, Hossam N, Boykov Y. Fast approximate energy minimization with label costs. International Journal of Computer Vision. 2011 [Google Scholar]
- Eickhoff SB, Stephan KE, Mohlberg H, Grefkes C, Fink GR, Amunts K, Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Fred ALN, Jain AK. Combining multiple clusterings using evidence accumulation. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2005;27:835–850. doi: 10.1109/TPAMI.2005.113. [DOI] [PubMed] [Google Scholar]
- Geman SGD. Stochastic relaxation, Gibbs distribution and the Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell. 1984;6:721–741. doi: 10.1109/tpami.1984.4767596. [DOI] [PubMed] [Google Scholar]
- Glover GH, Law CS. Spiral-in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magn Reson Med. 2001;46:515–522. doi: 10.1002/mrm.1222. [DOI] [PubMed] [Google Scholar]
- Golland Y, Golland P, Bentin S, Malach R. Data-driven clustering reveals a fundamental subdivision of the human cortex into two global systems. Neuropsychologia. 2008;46:540–553. doi: 10.1016/j.neuropsychologia.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Robson MD, Drobnjak I, Rushworth MF, Brady JM, Smith SM, Higham DJ, Matthews PM. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc Natl Acad Sci U S A. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C, Toro R, Di Martino A, Cox CL, Bellec P, Castellanos FX, Milham MP. A convergent functional architecture of the insula emerges across imaging modalities. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C, Uddin LQ, Shehzad Z, Margulies DS, Castellanos FX, Milham MP, Petrides M. Broca's region: linking human brain functional connectivity data and non-human primate tracing anatomy studies. Eur J Neurosci. 2010;32:383–398. doi: 10.1111/j.1460-9568.2010.07279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee JM, Jo HJ, Kim SH, Lee JH, Kim ST, Seo SW, Cox RW, Na DL, Kim SI, Saad ZS. Defining functional SMA and pre-SMA subregions in human MFC using resting state fMRI: functional connectivity-based parcellation method. Neuroimage. 2010;49:2375–2386. doi: 10.1016/j.neuroimage.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashkari D, Vul E, Kanwisher N, Golland P. Discovering structure in the space of fMRI selectivity profiles. Neuroimage. 2010;50:1085–1098. doi: 10.1016/j.neuroimage.2009.12.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SZ. Markov random field modeling in image analysis. 3rd ed. London: Springer; 2009. [Google Scholar]
- Lindquist KA, Wager TD, Kober H, Bliss-Moreau E, Barrett LF. The brain basis of emotion: a meta-analytic review. Behav Brain Sci. 2012;35:121–143. doi: 10.1017/S0140525X11000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V, Uddin LQ. Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct. 2010;214:655–667. doi: 10.1007/s00429-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. [Google Scholar]
- Nanetti L, Cerliani L, Gazzola V, Renken R, Keysers C. Group analyses of connectivitybased cortical parcellation using repeated k-means clustering. Neuroimage. 2009;47:1666–1677. doi: 10.1016/j.neuroimage.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, Velanova K, Donaldson DI, Phillips JS, Schlaggar BL, Petersen SE. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67:156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard N, Strick PL. Imaging the premotor areas. Curr Opin Neurobiol. 2001;11:663–672. doi: 10.1016/s0959-4388(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Shen X, Papademetris X, Constable RT. Graph-theory based parcellation of functional subunits in the brain from resting-state fMRI data. Neuroimage. 2010;50:1027–1035. doi: 10.1016/j.neuroimage.2009.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Menon V. Developmental maturation of dynamic causal control signals in higherorder cognition: a neurocognitive network model. PLoS Comput Biol. 2012;8:e1002374. doi: 10.1371/journal.pcbi.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Amin H, Rykhlevskaia E, Nguyen DA, Greicius MD, Menon V. Dissociable connectivity within human angular gyrus and intraparietal sulcus: evidence from functional and structural connectivity. Cereb Cortex. 2010;20:2636–2646. doi: 10.1093/cercor/bhq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar KS, Ryali S, Menon V. Dynamic reconfiguration of structural and functional connectivity across core neurocognitive brain networks with development. J Neurosci. 2011;31:18578–18589. doi: 10.1523/JNEUROSCI.4465-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Snyder AZ, Shimony JS, Fox MD, Raichle ME. Noninvasive functional and structural connectivity mapping of the human thalamocortical system. Cereb Cortex. 2010;20:1187–1194. doi: 10.1093/cercor/bhp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Centenary of Brodmann's map--conception and fate. Nat Rev Neurosci. 2010;11:139–145. doi: 10.1038/nrn2776. [DOI] [PubMed] [Google Scholar]
- Zilles K, Schlaug G, Geyer S, Luppino G, Matelli M, Qu M, Schleicher A, Schormann T. Anatomy and transmitter receptors of the supplementary motor areas in the human and nonhuman primate brain. Adv Neurol. 1996;70:29–43. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.