Summary
Human CST (CTC1-STN1-TEN1) is an RPA-like complex that is needed for efficient replication through the telomere duplex and genome-wide replication restart after fork stalling. Here we show that STN1/CST has a second function in telomere replication during G-overhang maturation. Analysis of overhang structure after STN1 depletion revealed normal kinetics for telomerase-mediated extension in S-phase but a delay in subsequent overhang shortening. This delay resulted from a defect in C-strand fill-in. Short telomeres exhibited the fill-in defect but normal telomere duplex replication, indicating that STN1/CST functions independently in these processes. Our work also indicates that the requirement for STN1/CST in telomere duplex replication correlates with increasing telomere length and replication stress. Our results provide the first direct evidence that STN1/CST participates in C-strand fill-in. They also demonstrate that STN1/CST participates in two mechanistically separate steps during telomere replication and identify CST as a novel replication factor that solves diverse replication-associated problems.
Keywords: CTC1, STN1, TEN1, polymerase alpha, telomere, 3′ overhang
Introduction
Mammalian telomeres consist of kbp of T2AG3/C3TA2 repeats bound by a six protein complex called shelterin (Palm and de Lange, 2008). The DNA ends in a 3′ G-strand overhang of 30-110 nt that serves as the substrate for telomerase (Chai et al., 2006; Zhao et al., 2008; Zhao et al., 2009). Telomeres pose a unique challenge to the replication machinery due to their repetitive nature and unusual terminal structure (Gilson and Geli, 2007; Stewart et al., 2012). The duplex region is replicated by the conventional replication machinery, however a number of additional proteins are needed for efficient passage of the replication fork (Sfeir et al., 2009; Vannier et al., 2012). In humans, telomere replication occurs throughout S-phase and telomerase extension of the G-strand is tightly linked to duplex replication (Zhao et al., 2009). The daughter telomere generated by leading strand synthesis is processed almost immediately to generate the overhang needed for telomerase action (Chow et al., 2012). An overhang is naturally present on the telomere replicated by lagging strand synthesis. After G-strand extension by telomerase, the complementary C-strand is filled in by DNA pol α primase (pol α). Although telomerase extends the overhang soon after duplex replication, C-strand fill-in occurs some hours later. The process involves several rounds of primer synthesis resulting in gradual overhang shortening over an additional 3-4 hours (Zhao et al., 2009). It is currently unclear how pol α is recruited or regulated during the fill-in reaction given the likely absence of a replisome. Here we identify mammalian CST (CTC1-STN1-TEN1) as a key player in C-strand fill-in.
CTC1 and STN1 were originally identified as a pol α stimulatory factor (AAF) that increases pol α processivity and affinity for ssDNA templates (Casteel et al., 2009; Goulian et al., 1990). Recently, CST was found to be important for telomere maintenance with depletion leading to longer G-overhangs and telomere loss or disruption (Chen et al., 2012; Miyake et al., 2009; Stewart, 2012; Surovtseva et al., 2009; Wu et al., 2012). Mammalian CST resembles the Cdc13-Stn1-Ten1complex that is responsible for telomere protection in budding yeast (ScCST) in that the STN1 and TEN1 subunits are conserved, both complexes resemble RPA and both bind ssDNA (Chen et al., 2012; Miyake et al., 2009; Price et al., 2010). However in mammalian cells, shelterin rather than CST is primarily responsible for telomere protection. CST instead plays a role in replication both at the telomere and elsewhere in the genome. CST is not a conventional replication factor as it does not co-localize with replication foci (Miyake et al., 2009) and it appears to function in duplex DNA replication only during replication stress (Stewart, 2012). At the telomere, it facilitates replication of the telomere duplex, most likely by rescuing replication after fork stalling. Elsewhere in the genome, CST is involved in the restart of DNA synthesis via new origin firing.
In budding yeast, ScCST controls G-strand extension through positive and negative regulation of telomerase (Giraud-Panis et al., 2010). Because Cdc13 and Stn1 both interact with pol α (Chandra et al., 2001; Puglisi et al., 2008; Qi and Zakian, 2000; Sun et al., 2011), ScCST is also proposed to recruit pol α for complementary C-strand synthesis. However, this role remains to be demonstrated directly. Given that mammalian CTC1-STN1 (AAF) and Xenopus CST both stimulate pol α activity (Goulian et al., 1990; Nakaoka et al., 2012), mammalian CST seemed a likely candidate to direct telomeric C-strand fill-in. To address this possibility we examined the cell-cycle regulation of G-overhang structure. We now present the first direct evidence that CST participates in C-strand synthesis. We first demonstrate that depletion of STN1 causes a defect in C-strand fill-in during late S/G2 phase. We then show that this defect is separable from the effect of STN1 depletion on telomere duplex replication. Our results indicate that CST functions in two distinct aspects of telomere replication: passage of the replication fork through the telomeric duplex and C-strand fill-in synthesis after telomerase action.
Results
Effect of STN1 depletion on G-overhang and telomere length
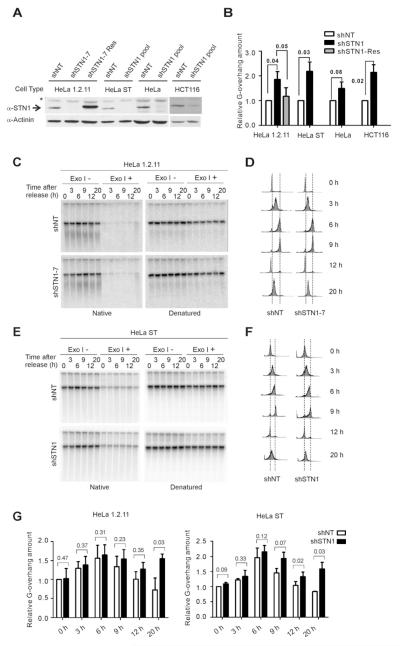
We and others previously found that depletion of CTC1 or STN1 in HeLa cells results in a modest but consistent increase in G-overhang size but has little effect on telomere length (Miyake et al., 2009; Price et al., 2010; Stewart, 2012; Surovtseva et al., 2009). To further investigate the role of STN1 in G-overhang and telomere length regulation, we depleted STN1 in cell lines with different telomere lengths and/or telomerase levels. These included HCT116 (3-6 kb telomeres), HeLa 1.2.11 (10-20 kb telomeres), HeLa (3-5 kb telomeres) and HeLa ST that overexpress telomerase (25-45 kb telomeres (Cristofari and Lingner, 2006)) (Figures 1A and S1A). For experiments with HeLa, HeLa ST and HCT116, we used pools of cells expressing shRNA to STN1 (shSTN1) or a non-target control (shNT). Experiments with HeLa 1.2.11 were performed with previously characterized single cell clones (shSTN1-7, shSTN1-6 or shNT) and a cell line where STN1 expression was rescued with a FLAG-tagged sh-resistant STN1 allele (shSTN1-7 Res) (Stewart, 2012). STN1 mRNA depletion was 75-82% for HeLa, HeLa 1.2.11 and HeLa ST and ~65% for the HCT116 pool.
Figure 1. STN1 depletion delays G-overhang shortening.
(A) Western blots showing STN1 knockdown and expression of sh-resistant FLAG-STN1; *, cross-reacting band. (B-G) Effect of STN1 depletion on overhang signal analyzed by in-gel hybridization. (B) Quantification of overhang signal from asynchronous cultures (C-G) Overhang signal from synchronous cultures after release into S. (C & E). Representative gels showing overhang signal in HeLa 1.2.11 clones (C) or HeLa ST pools (E) DNA was hybridized with (TA2C3)4 probe before and after denaturation. (D & F) FACS data showing DNA content of cells from (C & E). (G) Quantification of overhang signal from HeLa 1.2.11 or HeLa ST cells (mean ± SEM, n = 3 exps., p-values are shown).
G-overhang status was examined by in-gel hybridization of probe to the overhang under non-denaturing conditions. Quantification revealed that STN1 knockdown caused a 1.5-2 fold increase in overhang signal in each cell type (Figures 1B and S1B-C). This increase was largely rescued by expression of sh-resistant STN1. To determine whether the increase in overhang signal reflected a change in telomerase activity, we performed TRAP assays on extracts from HeLa ST and HeLa 1.2.11 cells. These revealed no significant difference in activity (Figure S1D-E). STN1 depletion also had little effect on telomere length (Figure S1F-I). The telomeres from shSTN1 HeLa 1.2.11, HeLa and HCT116 cells remained essentially the same length after 40-60 PD. As expected, the HeLa ST cells underwent gradual telomere elongation but the rate of telomere growth was unaffected by STN1 depletion. Thus, our results confirmed previous observations (Miyake et al., 2009) but see also (Chen et al., 2012) and indicate that STN1 is unlikely to be a significant determinant of telomere length in HeLa or HCT116 cells. Overall, our findings indicate that the increase in G-overhang after STN1 depletion is unlikely to be caused by an elevation in telomerase activity and it occurs without net telomere elongation.
STN1 promotes G-overhang shortening in late S/G2
During S-phase, telomerase-expressing cells exhibit a transient increase in overhang length due to the time lag between G-strand extension by telomerase and C-strand fill-in by pol α (Zhao et al., 2009). To address the role of CST in overhang length regulation, we asked if this transient change in overhang length was affected by STN1 depletion. HeLa 1.2.11 or HeLa ST cultures were blocked at G1/S, released into S and harvested at 3 hr intervals. Relative overhang signal was then analyzed by in gel hybridization (Figure 1C-G). The control shNT cells exhibited the expected transient increase in overhang length during early to mid S phase as the signal peaked at ~6 hrs after release and then gradually declined during late S/G2 phase (Zhao et al., 2009). Interestingly, in STN1 depleted cells, the increase in overhang length followed similar dynamics. However, the decline in length during late S/G2 showed a consistent delay. Although this delay was modest in the HeLa 1.2.11 cells, it was statistically significant in the HeLa ST cultures. In these cultures the overhang signal from the shNT cells decreased ~26% between 6 and 9 hrs post release while in the shSTN1 cells the signal decreased by only 5-6%. In both HeLa strains, the overhangs of shNT cells were restored to their original average length by G1 of the next cell cycle (12 hrs after release). However, the overhangs of shSTN1 cells remained longer, and then became further elongated during the following S phase. The delay in overhang shortening after STN1 depletion did not appear to result from a decrease in overall growth rate because FACS analysis indicated that passage through the cell cycle was unaffected (Figure 1D & F).
The delay in overhang shortening in STN1 depleted HeLa 1.2.11 was confirmed by direct analysis of overhang size after the overhangs were released from the telomere duplex by degradation of the duplex DNA with DSN (duplex specific nuclease, supplemental results and Figure S2A-B). We also showed that the overhangs return to their original length if the G1 of the next cell cycle is prolonged (supplemental results and Figure S2D-G). The latter finding demonstrates that overhang processing can occur in G1. It may also explain why in HeLa cells, STN1 depletion does not cause progressive overhang elongation with increased population doubling. Overall, our results show that STN1 depletion delays overhang shortening in late S/G2 but has no effect on the timing or extent of overhang elongation in early to mid S. These findings indicate that STN1 is unlikely to limit G-strand extension by telomerase or C-strand resection by nuclease as these are early events in telomere replication (Chow et al., 2012; Zhao et al., 2009). Instead, our results point to a role for STN1 in the overhang shortening that occurs as cells exit S phase.
Leading and lagging daughters are both affected by STN1 depletion
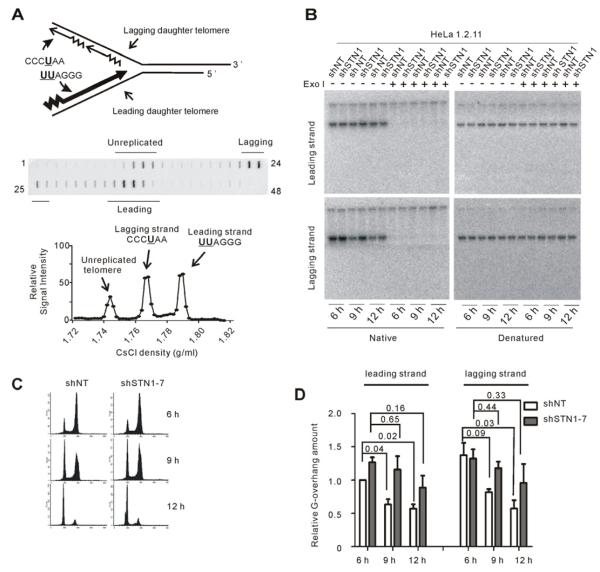
Replication of telomeric DNA by leading and lagging strand synthesis generates dissimilar termini on the two daughters that are later subjected to different DNA processing events (Chow et al., 2012). To examine whether the delay in overhang shortening occurs at both daughters, we used BrdU labeling and CsCl density gradient centrifugation to isolate newly replicated leading and lagging daughter telomeres prior to overhang analysis (Chai et al., 2006). The telomeric G-strand is always synthesized as the leading strand while the C-strand is synthesized as the lagging strand (Figure 2A). Thus, leading daughters incorporate twice as much BrdU (UUAGGG) and band at a higher density in CsCl as compared to lagging daughters (CCCUAA).
Figure 2. The delay in G-overhang shorting occurs at both leading and lagging daughters.
(A) Top, cartoon showing BrdU incorporation into daughter telomeres generated by leading versus lagging strand replication. Bottom, experiment showing separation of newly replicated daughters. Telomeric DNA in fractions was detected by slot blot. (B) Overhang signal from leading and lagging daughters isolated after release into S. In-gel hybridization with (TA2C3)4 probe. (C) FACS data showing DNA content of cells from (B). (D) Quantification of data mean ± SEM, n = 3 exps., p-values are shown). Normalization was to shNT leading strand telomeres at 6 hr.
HeLa 1.2.11 shNT and shSTN1 cells were blocked at G1/S, released into BrdU and collected 6, 9 and 12 hours later. DNA was restriction digested and subjected to density gradient centrifugation. Fractions containing the leading and lagging daughters were identified by slot blot (Figure 2A, Figure S2C) and the residual DNA in those fractions was subject to overhang analysis (Figures 2B). In agreement with previous results, the overhangs on the leading daughters of the control shNT cells were generally shorter than those of the lagging daughters at the 6 hr time point (Figure 2D) (Zhao et al., 2009), which is after telomerase extension but prior to C-strand fill-in (see below). The overhangs on both daughters then became shorter as the cells progressed into late S/G2 phase and by the following G1 they were of comparable size. While the overall decrease in overhang length for the purified shNT daughter telomeres was similar to that observed when the entire telomere population was analyzed (Fig. 1G), the difference between the 6 and 9 hr time points was more striking. This is probably because signal from contaminating unreplicated and partially replicated telomeres had been removed (Figure S2C). In contrast to the control samples, the overhangs from shSTN1 cells showed little decrease in length until the 12 hr time points. This delay in overhang shortening was clearly visible on both daughters. We therefore conclude that the event(s) involving STN1 that are responsible for overhang shortening must be common to both leading and lagging daughters.
STN1 depletion causes a delay in C-strand fill-in
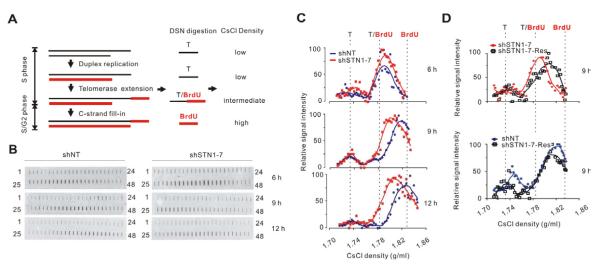
The delay in overhang shortening could be explained if STN1 is needed for C-strand fill-in following G-strand extension by telomerase. To test for such a role, we monitored overhang maturation on lagging daughters by examining overhang density following growth in BrdU (Zhao et al., 2009). The change in density can be detected by CsCl density gradient centrifugation of overhangs that have been released from the telomeric duplex by DSN digestion (Zhao et al., 2008). During telomere duplex replication, the parental G-strand does not incorporate BrdU so the overhangs remain unlabeled and of low density until they are extended by telomerase (Figures 2A and 3A). After telomerase incorporates BrdU, the lagging overhangs consist of ~50% unlabeled DNA and ~50% BrdU-labeled, telomerase synthesized DNA, and are of intermediate density. After C-strand fill-in, the remaining overhangs are fully BrdU labeled and are of high density (Figure 3A).
Figure 3. STN1 promotes C-strand fill-in.
(A) Strategy to monitor C-strand fill-in at lagging daughter telomeres. (B-D) Analysis of overhang density. HeLa 1.2.11 clones were labeled with BrdU for the indicated times after release into S. Overhangs were released from lagging daughters with DSN and analyzed in CsCl gradients. (B) Detection of overhangs in gradient fractions by slot blot with probe to the overhang. (C) Quantification of overhang signals from shNT or shSTN1-7 clones plotted versus density (representative experiment). Data are from blots shown in (B). (D) As for (C) but with shSTN1 cells rescued with sh-resistant STN1.
HeLa 1.2.11 cells were blocked at G1/S, released into BrdU for 6, 9 or 12 hrs, DNA isolated and the newly replicated leading and lagging daughters separated in CsCl gradients. Fractions containing the lagging daughters were pooled and the duplex DNA digested with DSN. The released overhangs were then subject to a second round of density gradient centrifugation. Overhang-containing fractions were identified by slot blot and fraction density determined from the refractive index (Figures 3B-3C). When we compared the overhangs isolated from shNT and shSTN1 cells 6 hrs after release into S, we found them to be of the same intermediate density (Figure 3C). This result again indicated that STN1 depletion had no effect on G-strand extension by telomerase (see supplemental materials> for further discussion). By 9 hrs after release, the G-overhangs in shNT cells were converted to a higher density due to C-strand fill-in (Zhao et al., 2009), and they became fully BrdU substituted by 12 hrs post release. In contrast, the overhangs of shSTN1 cells remained at an intermediate density at 9 hrs and showed only a slight density increase by 12 hrs. This lack of density shift at 9 hrs was largely rescued by expression of sh-resistant STN1 (Figure 3D). Taken together, these data indicate that shSTN1 cells experience a delay in C-strand fill-in during late S/G2 phase and suggest that STN1 may participate in the fill-in process.
The role of STN1 in C-strand fill-in is independent of its role in telomere duplex replication
We previously showed that STN1 depletion in HeLa 1.2.11 cells slows replication through the duplex region of the telomere by ~1.5 hrs (Stewart, 2012) most likely due to the role of CST in replication restart after fork stalling. While this delay in duplex replication could partially explain the delay in C-strand fill-in observed 9 hrs after release of shSTN1 cells into S, it is unlikely to cause the delayed fill-in at 12 hrs. Nonetheless, we sought to separate the effect of STN1 depletion on duplex replication from the effect on C-strand fill-in by examining fill-in in cells where the rate of duplex replication was unaffected by STN1 knockdown.
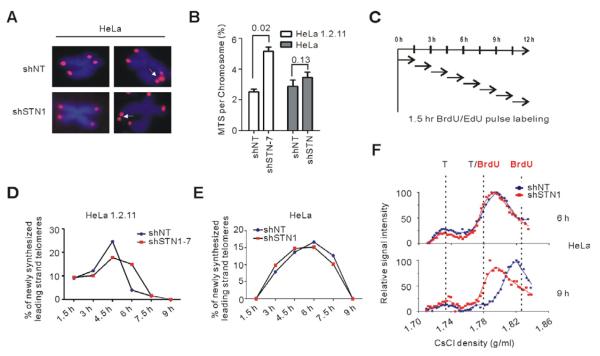
FISH analysis with HeLa 1.2.11 previously revealed an increase in multiple telomere signals (MTS) on individual chromatids after STN1 or CTC1 depletion (Price et al., 2010; Stewart, 2012). These MTS appeared to reflect difficulty in replicating the long (10-20 kb) telomere duplex. When we performed telomere FISH using HeLa cells with short 3-5 kb telomeres, we observed fewer MTS (Figure 4A-B) suggesting that STN1 depletion might have less effect on telomere duplex replication in cells with shorter telomeric tracts. To test this possibility, we compared the rates of telomere duplex replication in the two HeLa strains.
Figure 4. STN1 participates in telomere duplex replication and C-strand fill-in.
(A) Telomere FISH on HeLa with short telomeres. Arrows indicate occasional MTS. (B) Quantification of MTS (mean ± SEM, n = 3 exp., p-values are shown). (C) Experimental time-line. HeLa or HeLa 1.2.11 cells were released into S then incubated with BrdU or EdU for consecutive 1.5 hr intervals. (D-E) Amounts of telomere replication throughout S. Graphs from representative experiments show percent of leading daughters that completed replication relative to total telomere signal for each time period in HeLa 1.2.11 clones (D) or HeLa pools (E). (F) Detection of delayed C-strand fill-in in HeLa with short telomeres. Overhangs were detected by slot blot, signals quantified and plotted versus density (representative experiment).
Cells were blocked in G1/S, released into S and samples pulse labeled with either BrdU or EdU for consecutive 1.5 hr intervals (Figure 4C). To quantify the amount of telomeric DNA replicated during each 1.5 interval, DNA from the BrdU-labeled cells was isolated and newly replicated leading and lagging daughters were separated from unreplicated daughters in CsCl gradients. The leading strand peak was used to quantify the fraction of telomeres completing replication during any time interval (Figure S3D) because this peak contains fully replicated telomeres and minimal contamination with replication intermediates (Figure 2A) (Chow et al., 2012). To examine the rate of bulk genomic DNA replication during each time period, we quantified the amount of EdU uptake by FACS (Figure S3A-S3C).
When we compared the amount of newly replicated telomere in HeLa 1.2.11 versus HeLa with short telomeres, we found the two HeLa strains responded differently to STN1 depletion. As expected, in HeLa 1.2.11 replication through the telomere was slower in the shSTN1 cells despite the rate of bulk genomic DNA replication remaining essentially unchanged (Figures 4D and S3C, (Stewart, 2012)). The shNT and shSTN1 cells appeared to initiate telomere replication at a similar rate, but replication then proceeded faster in the shNT cells. This resulted in a ~1.5 hr difference in the time taken for shNT and shSTN1 cells to complete replication of all of their telomeres. The HeLa with short telomeres entered S-phase somewhat less synchronously after the G1/S block but, like HeLa 1.2.11, the shNT and shSTN1 cells underwent bulk genomic DNA replication at the same rate and initiated telomere replication at the same time (Figures 4E and S4C). However, in contrast to HeLa 1.2.11, telomere replication in the HeLa shNT and shSTN1 cells peaked and declined at similar time points (Figure 4E and Figure S4D). These results imply that STN1 depletion affects telomere duplex replication in a manner dependent on telomere length.
Since STN1 depletion did not significantly affect the rate of telomere duplex replication in HeLa with short telomeres, we examined whether these cells still exhibited a delay in C-strand fill-in. As before, we monitored fill-in by using CsCl density gradients to determine overhang density on newly replicated lagging strand telomeres. Comparison of overhang density revealed that, as observed for the HeLa 1.2.11 cells, most of the overhangs from the shNT and shSTN1 HeLa with short telomeres were of intermediate density 6 hrs after release (Figure 4F) indicating that telomerase extension was complete (Zhao et al., 2009). Moreover, while the density of the overhangs from the shNT cells had shifted to higher density by 9 hrs post release, the overhangs from the shSTN1 cells remained at the intermediate density, again indicating a delay in C-strand fill-in. These results demonstrate that STN1 depletion causes a defect in C-strand fill-in even when telomere duplex replication has proceeded at a normal rate. We therefore conclude that human STN1/CST is needed for C-strand synthesis after extension of the lagging strand telomere by telomerase. While it is not possible to use overhang density to monitor C-strand fill-in on telomeres replicated by leading strand synthesis, the delay in overhang shortening observed by in-gel hybridization implies that CST is also needed for C-strand synthesis at the leading strand telomere.
Discussion
It has long been recognized that telomeres must utilize a unique mechanism to recruit and regulate pol α because the replisome will not be present to direct C-strand fill-in following telomerase action. In some ciliates, the problem is solved through formation of a “telomere synthesis” complex that contains both telomerase and pol α (Ray et al., 2002). However, such a complex has not been observed in mammals and the mechanism of pol α regulation at telomeres has remained obscure. Our results provide new insight into this longstanding problem by demonstrating that STN1/CST is needed for the C-strand fill-in reaction. Our finding is particularly interesting given that that STN1/CST was recently shown to participate in telomere duplex replication (Gu et al., 2012; Stewart, 2012). We show here that the effect of STN1 depletion on C-strand fill-in is separable from the effect on telomere duplex replication as the fill-in defect occurs in cells with short telomeres where duplex replication is unaffected. Thus, STN1/CST participates in two independent steps in telomere replication that each requires a specialized approach to resolve challenges to the replication machinery.
Our results also provide new information about G-overhang maturation. First, we demonstrate that overhang processing is not restricted to S/G2 but can continue during the subsequent G1. Second, we provide direct evidence that STN1/CST is needed for C-strand fill-in at telomeres replicated by leading as well as lagging strand synthesis despite the two daughter telomeres being subject to quite different processing reactions during initial overhang generation (Chai et al., 2006; Chow et al., 2012). While this manuscript was in revision, another study showed that STN1 depletion causes overhang elongation at both leading and lagging daughters and that overhang shortening in late S/G2 is delayed (Huang et al., 2012). However, this study did not directly address the underlying cause of overhang elongation or the delay in shortening. Additional work will be needed to determine the precise role of CST in directing C-strand fill-in but given that CST/AAF can modulate pol α processivity and affinity for ssDNA templates in vitro (Goulian et al., 1990; Nakaoka et al., 2012). CST is likely to also function in this context in vivo. CST interacts with the shelterin subunits most closely associated with the 3′overhang (TPP1 in humans and Pot1b in mice (Chen et al., 2012; Wu et al., 2012)). These interactions are likely to deliver CST to the G-strand after telomerase action leaving CST ideally positioned to recruit and/or regulate pol α. To date, we have not detected a stable interaction between CST and pol α (unpublished observations, see also (Nakaoka et al., 2012)) so interactions with pol α may be transient or cell cycle regulated
During telomere duplex replication, CST probably helps restart replication after fork stalling (Stewart, 2012). This role is also likely to involve the ability of CST to modulate pol α activity. CST might recruit pol α to help restart stalled forks where the replisome has become damaged and lost the polymerase. Alternatively, CST might facilitate firing of dormant replication origins that lie within the telomere downstream of the stall site (Drosopoulos et al., 2012). The latter scenario fits with our finding that CST promotes genome-wide origin firing during recovery from HU-induced fork stalling (Stewart, 2012). Either scenario fits with our finding that STN1/CST is more important for efficient telomere duplex replication when telomeres are very long since one would expect the frequency of replication fork stalling to increase with telomere length.
Overall our findings indicate that CST is a novel replication factor that is used to solve a variety of replication problems where the replisome is absent or may be damaged. Interestingly, mutations in CTC1 cause Coats plus, a severe disorder with pleiotropic clinical symptoms (Armanios, 2012). The wider range of symptoms observed in Coats plus as compared the short telomere syndromes caused by mutations in telomerase or shelterin subunits may reflect the fundamental role of CST in resolving diverse replication-related problems.
Experimental Procedures
Quantification of G-overhang amount by non-denaturing in-gel hybridization
Purified DNAs were restriction digested then briefly run on 1% agarose gels so the telomeres remained in a tight band. Gels were dried, hybridized with 32P-labeled (TA2C3)4 probe to the G-overhang. Gels were then denatured, re-hybridized with the same probe and the signal used to normalize for gel loading.
Separation of leading and lagging daughters, analysis of overhang density and replication rates
HeLa cells were released into S after a double thymidine block and pulse labeled with BrdU (100 mM) or EdU (50 mM) as indicated. Genomic DNA was isolated by high salt precipitation (see supplemental methods). Leading and lagging strand daughter telomeres were separated as described (Chai et al., 2006.). For overhang density analysis, fractions containing lagging daughters were pooled, digested with DSN (Duplex Specific Nuclease, Evrogen, Russia) at 37°C for 2 hrs, mixed with CsCl and centrifuged for 20 hr at 60,000 rpm in a VTI80 rotor (Beckman). Following fractionation, the telomeric overhang was detected by slot blot with high specific activity T3C3(TA2C3)3 probe (Zhao et al., 2008). The rates of bulk genomic and telomeric DNA replication were determined by quantifying EdU uptake or the fraction of leading daughters completing replication within a specific time period (see supplemental methods>) (Stewart, 2012).
Supplementary Material
Highlights.
STN1 depletion delays telomere overhang shortening after telomerase-extension.
STN1 is needed for C-strand fill-in synthesis by DNA polymerase α.
STN1 prevents defects in telomere duplex replication at long telomeres.
CST has independent roles in telomere duplex replication and C-strand fill-in.
Acknowledgements
We thank Mary Chaiken for reagents, help with experiments and manuscript preparation, Alex Barnhill for help with MTS scoring and Birgit Ehmer for assistance with cell sorting. This work was supported by NIH grants GM041803 to C.M.P. and AGO1228 to W.E.W. J.A.S. was supported by F32GM097833 and T32CA117846 and C.K. by T32CA117846.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Armanios M. An emerging role for the conserved telomere component 1 (CTC1) in human genetic disease. Pediatr Blood Cancer. 2012;59:209–210. doi: 10.1002/pbc.24200. [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE. Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 2012;26:1167–1178. doi: 10.1101/gad.187211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J Cell Biol. 2012;197:253–266. doi: 10.1083/jcb.201112083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V. How telomeres are replicated. Nature Reviews Molecular Cell Biology. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- Goulian M, Heard CJ, Grimm SL. Purification and properties of an accessory protein for DNA polymerase alpha/primase. J Biol Chem. 1990;265:13221–13230. [PubMed] [Google Scholar]
- Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012;31:2309–2321. doi: 10.1038/emboj.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Dai X, Chai W. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell research. 2012 doi: 10.1038/cr.2012.132. E. pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem. 2012;287:619–627. doi: 10.1074/jbc.M111.263723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Ray S, Karamysheva Z, Wang L, Shippen DE, Price CM. Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol Cell Biol. 2002;22:5859–5868. doi: 10.1128/MCB.22.16.5859-5868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Chaiken MF, Wang F, Price CM. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain P, Wright W, Price CM. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012;31:3537–3549. doi: 10.1038/emboj.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yang Y, Wan K, Mao N, Yu TY, Lin YC, DeZwaan DC, Freeman BC, Lin JJ, Lue NF, et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell research. 2011;21:258–274. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T. Telomeric 3′ Overhangs Derive from Resection by Exo1 and Apollo and Fill-In by POT1b-Associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008;36:e14. doi: 10.1093/nar/gkm1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.