Abstract
In the past decade, a series of technological advances have revolutionized our ability to interrogate cancer genomes, culminating in whole-genome sequencing, which provides genome-wide coverage at a single base-pair resolution. To date, the tumor genome has been sequenced in nearly 40 cases of acute myeloid leukemia (AML). On average, each AML genome contains approximately 400 mutations, including 6-26 coding mutations. The majority of these mutations are ‘background’ mutations that were acquired during normal aging of hematopoietic stem cells. Though comprehensively identifying ‘driver’ mutations remains a challenge, a number of novel driver mutations in AML have been identified through whole-genome sequencing. The digital nature of next-generation sequencing has revealed clonal heterogeneity in the majority of AML at diagnosis. Importantly, in some cases, a minor subclone contributed to relapse, suggesting the strategies to assess clonal heterogeneity are needed to optimize therapy. As sequencing technologies improve and costs decrease, it is likely that whole-genome sequencing of cancer cells will become commonplace in the diagnostic work-up of patients with AML and other cancers.
Keywords: acute myeloid leukemia, AML, clone, coding mutation, driver mutation, genetics, genome, sequencing, subclone
Introduction
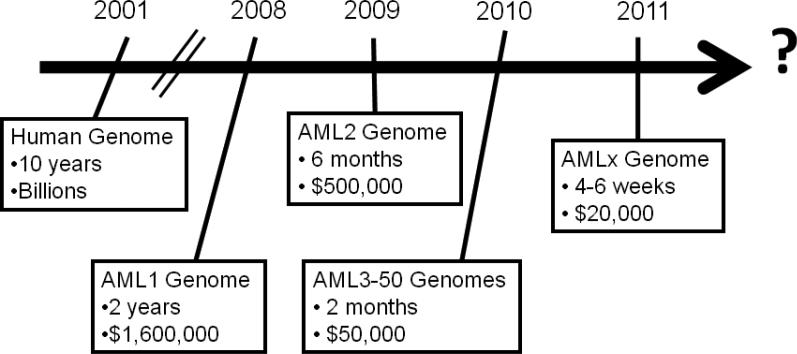
In the past, the size and complexity of the human genome (3 billion base pairs) made sequencing of human cancer genomes impractical. Two major advances helped overcome these obstacles. First, the generation of the draft sequence of the human genome by the Human Genome Project in 2001 provided a map of the human genome [1]. Second, technological advances in DNA sequencing dramatically reduced the cost and time required to sequence genomes (Figure 1). Whereas the Human Genome Project took over 10 years and several billion dollars to sequence the first human genome, current estimates are 4-6 weeks and $10,000 per human genome ($20,000 for paired tumor/normal genomes). Thus, we are rapidly approaching the time when sequencing the genomes of patients with cancer will be practical in the clinical setting. This chapter will focus on the use of next-generation sequencing technologies to characterize AML genomes.
Figure 1. Timeline for sequencing of AML genomes.
Cost and time estimates are for paired leukemia/normal genomes and include data production, bioinformatic analysis, validation, and interpretation. Current cost and time estimates to sequence a human genome are based on data from The Genome Institute at Washington University.
Next-generation sequencing of cancer
Massively parallel sequencing results in the generation of millions of short (50 to 100 nucleotide) DNA sequences simultaneously. These sequences are then mapped back to the human reference genome to generate a picture of the cancer genome. For studies of cancer, it is key to sequence both the tumor and non-malignant tissue from the individual. There are 3-4 million inherited sequence variants per human genome (and hundreds of copy number variants). Consequently, the great majority of sequence variants identified in a cancer genome are inherited polymorphisms and not acquired mutations. Thus, a comparison of a tumor genome to its paired normal genome is required to efficiently identify acquired (somatic) sequence variants.
Next-generation sequencing has been used in several different ways to interrogate cancer genomes. The goals, advantages, and limitations of each approach are summarized below. Ultimately, combinations of approaches (eg, whole-genome and transcriptome sequencing) may be required to comprehensively study cancer cells.
Whole-genome sequencing
The goal of this approach is to sequence the entire genome. The advantages of whole-genome sequencing include: (1) the entire genome is surveyed, not just coding genes; and (2) structural variants, including deletions, amplifications, chromosomal translocations, and uniparenteral disomy are readily identified. The major limitations are cost and the complexity of the data analysis. As cost and bioinformatic approaches to analyze sequence data advance, whole-genome sequencing is likely to become the dominant platform for mutation discovery.
Exome sequencing
The goal of this approach is to selectively sequence the 1% - 2% of the genome containing coding genes, microRNAs, and other non-coding RNAs. The major advantages of exome sequencing are reduced cost and relatively deep sequence coverage, since only 1% - 2% of the genome is analyzed. However, exome sequencing will not detect mutations in regions outside of the exome (>98% of the genome) and will not detect most structural variants, such as chromosomal translocations with intronic breakpoints.
Transcriptome sequencing
The goal of this approach is to sequence all transcribed genes (reviewed in [2]). Advantages of transcriptome sequencing include: (1) quantitative information about gene expression levels are obtained; (2) post-transcriptional changes in gene expression, such as alternative splicing, are detected; and (3) fusion transcripts produced by chromosomal rearrangements can be detected. Transcriptome sequencing does not detect mutations in non-coding regions of the genome, and it cannot detect mutations that cause the loss of one or both copies of a gene, or mutations that accelerate RNA turnover (eg, frameshift or nonsense mutations that cause “nonsense mediated decay”). Moreover, transcriptome sequencing is biased towards abundantly expressed transcripts; thus, sequencing coverage of genes expressed at lower levels can be low or absent.
Other sequencing applications
There are emerging next-generation sequencing applications to characterize genome-wide epigenetic modifications in cancer cells. This is particularly relevant to the study of AML, since genes regulating DNA methylation are frequently mutated in AML, including DNMT3A. Next-generation sequencing techniques have been developed to map genome-wide DNA methylation at a single-base-resolution [3] and to assess chromatin structure by mapping the location of post-translationally modified (eg, methylated and acetylated) histones on the genome [4].
Defining the AML genome
In a landmark study published in 2008, Ley and colleagues reported the sequence of the first primary cancer genome, a case of FAB M1-AML [5]. Since then, the sequence of 38 additional AML genomes has been reported, including 8 cases of secondary or therapy-related AML [6-11]. In addition, the exome was sequenced in 14 cases of M5-AML [12].
These studies have begun to define several important global features of AML. First, there are only a relatively small number of somatically acquired single nucleotide variants (SNVs or “point mutations”) in AML cells. Within coding regions, the number of somatic SNVs ranged from 6-26 with an average of 14 [9]. Total somatic SNVs ranged from 116 - 706 per AML genome, with an average of 420. This suggests that AML does not result from genomic instability; rather, it may be associated with a small number of oncogenic “driver” mutations. Second, most acquired AML-associated SNVs are private, suggesting that many of these mutations are likely to be random personal “passenger” mutations.
A current challenge in the field is to identify those mutations present in an AML genome that contribute to leukemogenesis. One approach is to identify those genes mutated in multiple AML samples, since it is unlikely that this would occur by chance (without selective pressure). In an analysis of 108 AML genomes, Welch and colleagues identified 24 genes that were mutated in at least 3 independent samples [9]. This list contains several novel genes, including three members of the cohesin complex, STAG2, SMC3, and SMC1A, which are involved in sister chromatid separation during anaphase and CTCF-mediated transcriptional regulation [13]. This approach likely underestimates driver mutations, and studies are underway to extend this analysis to look for recurring mutations in gene families or signaling pathways.
Somatic mutations accumulate in hematopoietic stem cells with aging and account for the majority of mutations present in AML
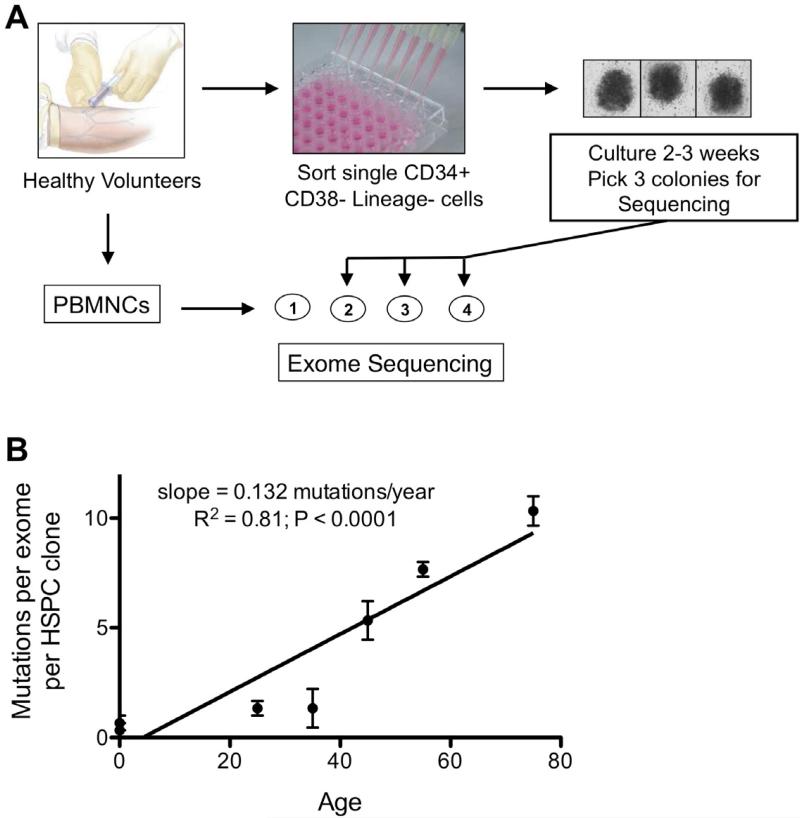
With each cell division, there is a finite probability of somatic mutation secondary to errors in DNA replication [14]. In the case of hematopoietic stem cells (HSCs), since they have self-renewal capacity, mutations acquired during cell division would be propagated to daughter HSCs. Thus, it is predicted that HSCs will accumulate mutations as a function of age. To test this prediction, we isolated and expanded individual hematopoietic stem/progenitor cells (HSPCs) from 7 healthy individuals of different ages (Figure 2A) [9]. We then randomly selected three different HSPCs from each donor and performed exome sequencing. In addition, we sequenced unselected peripheral blood mononuclear cells from each donor to serve as source of ‘germline’ DNA (it is estimated that ~ 1,000 HSCs contribute to hematopoiesis at any one time) [15-17]. Somatic mutations were detected in nearly all HSPC clones. Of note, no two HPSC clones shared the same mutations, indicating that each clone was derived from a unique HSC. As predicted, the number of mutations per HSPC clone increased with age (Figure 2B). By age 50 years, HSCs have accumulated, on average 6 somatic mutations in coding genes. By comparison, on average there are 10 somatic mutations in coding genes in the leukemic genome of a 50-year-old patient with AML [9]. Thus, the majority of mutations in AML represent “background’ mutations that were acquired during normal aging of HSCs, and the majority of these mutations likely do not contribute to leukemogenesis.
Figure 2. Sequencing of HSPC clones from healthy donors.
A. Peripheral blood or umbilical cord mononuclear cells (MNCs) from 7 healthy donors were isolated, and an aliquot was reserved for exome sequencing. CD34+ CD38- lineage- hematopoietic stem/progenitors cells (HSPCs) were sorted into a 96-well plate and expanded for 3 weeks on stromal cells with a cocktail of cytokines. Three individual clones were selected for exome sequencing for each donor. B. Shown is the number of single nucleotide mutations per exome for the 7 healthy donors. The 2 umbilical cord donors are designated with an age of zero.
Clonal heterogeneity in AML
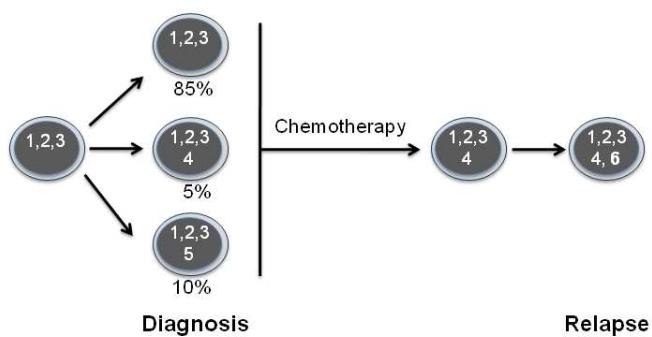
An unexpected finding of next-generation sequencing of AML is the magnitude of clonal heterogeneity. The digital nature of next generation sequencing allows for the accurate quantitation of mutant allele frequencies and the estimation of tumor clonal populations. Ding and colleagues sequenced the genomes of 8 cases of relapsed AML both at diagnosis and after relapse [11]. In most cases, clonal heterogeneity was detected in the diagnostic AML sample, with 1-5 clones detected. Of note, whole genome sequencing is currently able to detect clones that are present at 5% or greater. Hence, the magnitude of clonal diversity in AML is likely underestimated. Importantly, a minor clone often became the dominant clone at relapse (Figure 3). These observations have several important clinical implications, including: (1) current molecular prognostic markers (eg, DNTM3A mutations) need to be of sufficient sensitivity to interrogate minor clones; and (2) targeted therapies may need to be directed at mutations present in both the founder and minor clones.
Figure 3. Clonal heterogeneity in AML.
In this example, three AML clones were identified at diagnosis. The founder clone contains mutations 1, 2, and 3 and is the major clone (representing 85% of the total leukemic cell population). Two additional minor subclones were identified that contain at least one additional mutation (represented as mutation 4 and 5). During chemotherapy, there is selective expansion of one of the minor subclones and acquisition of further mutations (represented as mutation 6), leading to relapse.
Current limitations of whole-genome sequencing
Though clearly a valuable and promising approach to analyze AML (and other cancers), there are some technical limitations of whole-genome sequencing. The ability of next-generation sequencing to detect sequence variants is dependent on the “read-depth,” the number of unique times a single nucleotide is sampled. Whole-genome sequencing typically aims to cover each nucleotide with an average of 30 - 40 separate reads. However, at this coverage, a substantial number of sequence variants identified are false positives due to mapping errors, polymerase errors, and low frequency of variants within the sample. Thus, secondary validation of variants remains a necessary part of all sequencing projects. In addition, mapping sequence reads to the approximately 50% of the human genome that contains repetitive sequences is imperfect, raising the possibilities that mutations are being missed. These problems will be improved as bioinformatics can draw on additional sequenced human genomes and as decreased costs permit deeper sequence, but efficiently mapping sequences to repetitive regions of the genome will likely require technical improvements that permit longer sequence read-lengths.
There are inherent limitations of whole-genome sequencing that need to be considered when analyzing cancers. Most importantly, whole-genome sequencing does not provide information about epigenetic modifications and alterations in gene expression. Thus, to fully characterize acquired genomic changes in an AML patient, assays to measure RNA expression (eg, transcriptome sequencing) and epigenetic changes in combination with whole-genome sequencing are required.
Genome sequencing remains expensive and the requisite infrastructure, expertise, and time to complete sequence analysis are significant barriers to the routine use of whole-genome sequencing of AML in the clinical setting. However, as the number of sequenced AML genomes rises, the technical and bioinformatic infrastructure required for clinical analysis will become more accessible. Current cost estimates at the Washington University Genome Institute for whole-genome sequencing of a paired normal/leukemia sample is $20,000. This is a fully loaded cost that includes sequencing, bioinformatic analysis, and validation. The current time to sequence and analyze a paired tumor and normal genome is 4 - 6 weeks. The cost of whole-genome sequencing needs to be considered in the context of the increasingly complex diagnostic evaluation for AML. As the cost of sequencing continues to fall and the number of molecular assays continues to rise, we will rapidly reach the inflection point where whole-genome sequencing will be the most cost-effective diagnostic tool for AML patients.
Finally, there are important practical and ethical considerations concerning the clinical application of whole genome sequencing. Based on the cancer genomes analyzed to date, it is clear that a tremendous number of genetic variants will be identified in each AML genome, of which the vast majority will be inherited variants. Some of these will be in genes known to be involved in cancer susceptibility, and some will be associated with unsuspected genetic diseases (eg, hereditary hemochromatosis). Best practices to communicate this information to patients and their families are being considered and developed by several groups.
Conclusions
The Human Genome Project and advances in sequencing technologies have revolutionized our ability to characterize cancers at a molecular level. Using whole-genome sequencing, it is now possible to interrogate cancers genome-wide at a single base-pair resolution. Application of whole-genome sequencing to AML already has yielded important discoveries, including the identification of recurring gene mutations in AML (eg, IDH1 and DNTM3A) and the clonal heterogeneity of the majority of AML at presentation. Ongoing whole genome sequencing of additional AML samples likely will identify the great majority of relevant mutations in AML in the near future. With continued improvements in sequencing, it is likely that whole-genome sequencing will become a routine part of the diagnostic work-up of patients with AML and other cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement:
No relevant financial relationships with any commercial interest.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 3.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neff T, Armstrong SA. Chromatin maps, histone modifications and leukemia. Leukemia. 2009;23:1243–1251. doi: 10.1038/leu.2009.40. [DOI] [PubMed] [Google Scholar]
- 5.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprech TL, Liu F, Xia J. The origin and evolution of mutations in acute myeloid leukemia. Cell. doi: 10.1016/j.cell.2012.06.023. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 13.Millau JF, Gaudreau L. CTCF, cohesin, and histone variants: connecting the genome. Biochem Cell Biol. 2011;89:505–513. doi: 10.1139/o11-052. [DOI] [PubMed] [Google Scholar]
- 14.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catlin SN, Busque L, Gale RE, Guttorp P, Abkowitz JL. The replication rate of human hematopoietic stem cells in vivo. Blood. 2011;117:4460–4466. doi: 10.1182/blood-2010-08-303537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abkowitz JL, Catlin SN, Guttorp P. Evidence that hematopoiesis may be a stochastic process in vivo. Nat Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- 17.Abkowitz JL, Golinelli D, Harrison DE, Guttorp P. In vivo kinetics of murine hemopoietic stem cells. Blood. 2000;96:3399–3405. [PubMed] [Google Scholar]